Abstract
Cyclooxygenase-2 (COX-2) is linked to worse prognosis in patients with malignant gliomas and other tumor types. Amplification/overexpression of epidermal growth factor receptor (EGFR) is commonly seen in these tumors. We have previously shown that EGFR signaling, through activation of p38-mitogen activated protein kinase (MAPK), protein kinase C-δ (PKC-δ), and Sp1, plays an important role in the regulation of COX-2 expression in glioma cells. Here, we report that the Src kinase also has a role in this signaling cascade upstream of p38-MAPK/PKC-δ. In addition, more detailed analysis revealed the involvement of FOXM1, a member of the forkhead box family of transcriptional activators, in EGF-dependent COX-2 induction. FOXM1 protein levels increase after stimulation with EGF although its role in modulating COX-2 expression does not depend on this increase. While a conventional FOXM1 responsive element resides in a distal region (−2872/−2539 relative to the transcriptional start site) of the COX-2 promoter, this is not required for EGF-dependent induction of COX-2. Instead, FOXM1 is capable of interacting with Sp1 at the Sp1 binding site (−245/−240 relative to the start site) of the COX-2 promoter and appears to act in cooperation with Sp1 to mediate EGF-induced COX-2 expression. Definition of this novel interaction provides us with a clearer understanding of the mechanistic basis for the induction of COX-2 with EGF and guides our evaluation of potential newer therapeutic targets that have relevance in this disease.
Keywords: cyclooxygenase-2, FOXM1, Sp1, epidermal growth factor receptor, glioma
INTRODUCTION
Cyclooxygenases (COXs) are critical enzymes required for prostaglandin synthesis. While COX-1 is ubiquitously expressed, COX-2 is highly regulated allowing local modulation of prostaglandin levels in response to various stimuli. In fact, COX-2 is upregulated in many cancers and has been implicated in the development and progression of different malignancies (for review see (1)). COX-2 also appears to enhance resistance to cytotoxic therapies including chemotherapy and radiation increasing the difficulty of treating cancers that overexpress it (2-5). In glial neoplasms, COX-2 levels correlate with tumor grade and its overexpression is a negative prognostic factor in glioblastoma multiformes (GBMs) (6, 7). Thus, improving our understanding of COX-2 regulation in gliomas may aid in the development of potential novel therapies for these aggressive brain tumors.
FOXM1 is a member of evolutionally conserved Forkhead box family of transcriptional factors. This transcription factor is involved in the promotion of proliferation and functions as a cell cycle regulator (8, 9). Its expression is finely regulated and highly linked to a cell’s proliferative potential (10). FOXM1 is ubiquitously expressed in the developing embryo and in some adult tissues that show high proliferative capacity (11). In line with these observations, its expression appears to be suppressed in differentiated cells. FOXM1 expression is often upregulated in human malignancies including tumors arising from the lung (12, 13), prostate (14), breast (15), liver (10) and brain (GBMs) (16, 17). Such alterations may be associated with cancer genesis and progression as well as therapeutic resistance.
Human FOXM1 includes three alternatively spliced isoforms (FOXM1a, FOXM1b and FOXM1c) arising from differential splicing of two alternative exons (VA1 and VA2). VA1, present in FOXM1a and FOXM1c, does not affect DNA-binding specificity or transcriptional activation ability, while VA2, present only in FOXM1a, abolishes this factor’s transcriptional regulatory activity (18). FOXM1b, which contain neither alternative exons, is most highly expressed in malignant gliomas. This isoform is overexpressed in most GBM clinical specimens as well as glioma cell lines (19). Reports suggest that aberrant expression of FOXM1b may play a role in promoting astrocyte transformation and development of GBMs (20). Like with COX-2, FOXM1 levels correlate directly with glioma grade and inversely with patient survival (21).
We previously showed that COX-2 expression is strongly regulated by epidermal growth factor receptor (EGFR) in human glioma cells (22). A signaling cascade involving p38-MAP kinase (p38-MAPK)/protein kinase C-δ (PKC-δ) and terminating with Sp1-dependent transcriptional activation of the COX-2 promoter was defined as the major pathway responsible for EGFR-dependent COX-2 expression (22, 23). Wang et al. had also demonstrated that FOXM1 expression resulted in transcriptional activation of the COX-2 promoter correlating with induction of lung cancers in an established mouse model (13). Given this finding and the potential correlation between FOXM1 and COX-2 expression in gliomas, we sought to explore whether FOXM1 may play a regulatory role in COX-2 expression in glioma cells. In this current study, we find that activation of EGFR increases FOXM1 expression in a process requiring both Src and p38-MAPK. Interestingly, while FOXM1 expression is required for EGFR-dependent COX-2 expression, it appears to mainly be activating transcription through cooperation with the Sp1 transcription factor.
MATERIALS AND METHODS
Plasmids
The wild-type COX-2 promoter DNA from bases −1122 to +27 (COX2/P1.1) relative to the transcriptional start site was inserted upstream of the luciferase open reading frame in an MuLV-based, self-inactivating retroviral vector, called pTJ105 (kindly provided by T.J. Murphy, Emory University). The wild-type COX-2 promoter DNA from position −2872 to −818 was obtained by PCR of human genomic DNA, verified by sequencing and subcloned into pCR2.1 TA cloning vector (Invitrogen, Carlsbad, CA). The 2087 base COX-2 promoter sequence was released by Sac1 digestion, fused with the COX2/P1.1 promoter DNA to produce a COX-2 promoter from bases −2872 to +27 (COX2/P2.9) and inserted upstream of luciferase in pTJ105. The COX-2 promoter DNA from base −2539 to +27 (COX2/P2.6) was obtained by digesting the COX2/P2.9 retroviral vector with Hinc11/Hind111 and inserting this fragment upstream of luciferase in pTJ105. Similarly, the P2.9 and P2.6 promoter fragments were also inserted upstream of luciferase in pGL3 (Promega, Madison, WI, USA). The 6× Foxm1b TATA (6×FOX)-luciferase reporter vector was kindly provided by Pradip Raychaudhuri (University of Illinois, Chicago) (24). MKK3be cDNA was excised from pcDNA3 with EcoRI (22, 25), constitutively-active Src cDNA was excised from pCSrcA-HA14 (kindly provided by Yue Feng, Emory University) with BamH1/Xho1 (26) and FOXM1b cDNA was excised from pCMV-T7FOXM1 (kindly provided by Pradip Raychaudhuri) with Hind111/EcoR1 (27). Each fragment was then inserted into pRetroX-Tight-Pur (Clontech, Mountain View, CA, USA) to permit tetracycline-inducible expression of each cDNA.
Cell lines, culture conditions and transfections
All cell lines (SF767, LN229, SF767/LN229 derivatives, Phoenix) were cultured as previously described (23). The SF767 and LN229 glioma cell lines were originally obtained from Mark Israel (Dartmouth Medical School) in 1999. All derivatives were subsequently generated in our laboratory from the original parental lines. Phoenix cells were obtained from T.J. Murphy in 2006. LN229/ER was engineered to overexpress wild-type EGFR as previously described (28). The U87MG control and EGFRvIII cells lysates were provided by C. Hadjipanayis from cells originally obtained from W. Cavenee (University of California, San Diego) in 2008 (29). The MuLV-based COX-2 promoter/luciferase constructs (pCOX2/P1.1, pCOX2/2.6 and pCOX2/P2.9) were stably introduced into SF767 derivatives for functional COX-2 promoter analysis via generation of retrovirus as previously described (23). The pGL3-based COX-2 promoter/luciferase constructs were transiently transfected into SF767 derivatives via lipofection using Lipofectamine 2000 (Invitrogen), according to manufacturer’s recommendation. To generate cell lines with inducible expression of MKK3, constitutively-active Src and FOXM1b, pRetroX-Tet-On Advanced (Clontech) and the pRetroX-Tight-Pur vector-containing MKK3, constitutive-active Src and FOXM1b were sequentially introduced/infected into SF767, similar to the procedure described above, and selected in the presence of 1 mg/ml G418 and 2 μg/ml puromycin to obtain the SF767tetMKK3, SF767tetSrc and SF767tetFOXM1 cell lines. Doxycycline (1 μg/ml) was used to induce expression of the gene of interest in these cell lines. Standard aseptic culture techniques were used to propagate the various cells used in this report. All cell lines are periodically checked for mycoplasma infection by PCR (last tested in 2012). Cell line authentication by STR analysis was performed on 2/2013 for all parental glioma cell lines studied. FOXM1, Src, p38-MAPK, COX-2 and Sp1 knockdowns were accomplished with commercially available siRNA (Integrated DNA Technologies, Coralville, IA) using Lipofectamine 2000 (Invitrogen) according to manufacturer’s recommendations. The Stealth RNAi medium GC duplex-negative control (Invitrogen) was used to control for sequence-independent effects of introducing short RNA duplexes into cells.
Growth factor and inhibitor treatments
Cells were serum starved for 24 hours prior to stimulation with EGF (90 ng/ml, Roche Diagnostics, Indianapolis, IN), except as otherwise noted. Dimethylsulfoxide stock solutions of inhibitors against the following kinases, p38-MAPK (SB202190), Src (PP2 and SU6656), MEK (U0126), all PKC isoforms (bisindolylmaleimide I) and PKC-δ (rottlerin) (Calbiochem, San Diego, CA). Mithramycin A (Sigma), as inhibitor of the Sp family of transcription factors, was also made as a stock solution in dimethylsulfoxide. Working concentrations for each inhibitor are noted in the figure legend with vehicle (dimethylsulfoxide) used as a negative control.
RNase protection assay
RNase protection assay (RPA) was done by standard techniques as previously described (30). RPAs were performed to detect of COX-2 mRNA expression with cyclophilin as a normalization control as described elsewhere (22). Protected RNAs were resolved on a 5% urea/polyacrylamide gel and detected by phosphor imaging on a Typhoon 9210 using ImageQuant software from Molecular Dynamics (GE Healthcare).
Immunoblotting and antibodies used
All blots were done according to standard procedures as previously described (23). Blots were probed with antibodies against COX-2 (Cayman Chemical, Ann Arbor, MI), p38-MAPK/phosphorylated p38-MAPK, PKC-δ/phosphorylated PKC-δ (Thr505) and Src (Cell Signaling Technology, Beverly, MA), and HA, Sp1 and FOXM1 (Santa Cruz Biotechnology, Santa Cruz, CA). As loading controls, blots were also probed with antibodies against EIF5α (Santa Cruz), where appropriate. Blots were detected using anti-rabbit or mouse IgG secondary antibody conjugated with horseradish peroxidase and chemiluminescent substrate. Quantitation of scanned blots was performed on ImageJ (public domain software, National Institute of Health).
Matrigel invasion assay
Invasion assay was performed on SF767tetFOXM1 cells after transient introduction of control or COX-2 siRNA. 24 hours after transfection, cells were cultured in serum-free DMEM with or without doxycycline (1 μg/ml) for 24 hours, washed out with PBS and recultured in serum-free DMEM without doxycycline for 6 hours before seeding into the 24-well Matrigel invasion chambers (BD Biosciences, San Jose, CA). 1×104 cells in 500 μL serum-free DMEM containing 1% BSA were loaded into the top inserts while DMEM with 5% fetal bovine serum was used in the lower chamber as a chemoattractant. After incubation for 24 hours at 37°C, non-invasive cells were removed from the top of the Matrigel with a cotton tipped swab. Invasive cells that migrated through 8-μm pores to the underside of the membrane were fixed and stained using Diff-Quick stain kit (IMEB Inc., San Marcos, CA). Pictures were taken in 5 different areas of each membrane at 40× objective magnification. Invasive cells were counted on the pictures.
Luciferase reporter assay
The human COX-2 promoter/luciferase reporter constructs were introduced into the SF767 glioma cell line either stably by retroviral transduction or transiently by transfection as described above. After overnight serum starvation, glioma cells were treated under the described conditions as specifically indicated and harvested. Luciferase activity was measured as described previously (22). Reading of luciferase activity was normalized to total protein level as measured by Bradford assay.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was done as previously described (22). Immunoprecipitation was done with antibodies against Sp1 or FOXM1 (Santa Cruz). The amount of COX-2 promoter DNA in the DNA:protein complex was determined by semiquantitative PCR. The primer pair for the PCR product that encompasses the Sp1 site (−245/−240) was previously described (22). For the region containing the FOXM1 binding site, 5′-aactcacatgaatggcttatcacttc-3′ (forward) and 5′-agcgtctgaaatttgttgggaa-3′ (reverse) were used to generate a 287 base PCR product from −2707 to −2421 relative to the COX-2 start site that encompasses the FOXM1 binding sites at −2609/−2584. The PCR products were resolved on a 1.5% agarose gel, stained with ethidium bromide and detected on a UV light box. A portion of the DNA recovered from an aliquot of sheared chromatin was used as the “input” sample. Normal rabbit serum was used as a negative control.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays (EMSA) were performed by standard technique as previously described (22). A 22-mer containing the Sp1 consensus site (conSp1, Santa Cruz) was used. Complexes were resolved on a 5% nondenaturing polyacrylamide gel and detected by phosphor imaging as described above. Antibodies against Sp1, Sp3 or FOXM1 (Santa Cruz) were premixed with nuclear extracts for 30 min at room temperature before mixing with the labeled probe for the supershift assay.
Coimmunoprecipitation assay
SF767 cells ± EGF stimulation for 2 hours were fixed with 1% formaldehyde at 37°C for 10 mins and then harvested for immunoprecipitation following the ChIP protocol. Immunoprecipitation with antibody against Sp1 was resolved by SDS-PAGE and immunoblotted with antibodies against Sp1 or FOXM1. Detection was performed with secondary antibody linked to horseradish peroxidase and chemiluminescence.
RESULTS
FOXM1 is induced by EGF and required for EGFR-dependent COX-2 expression
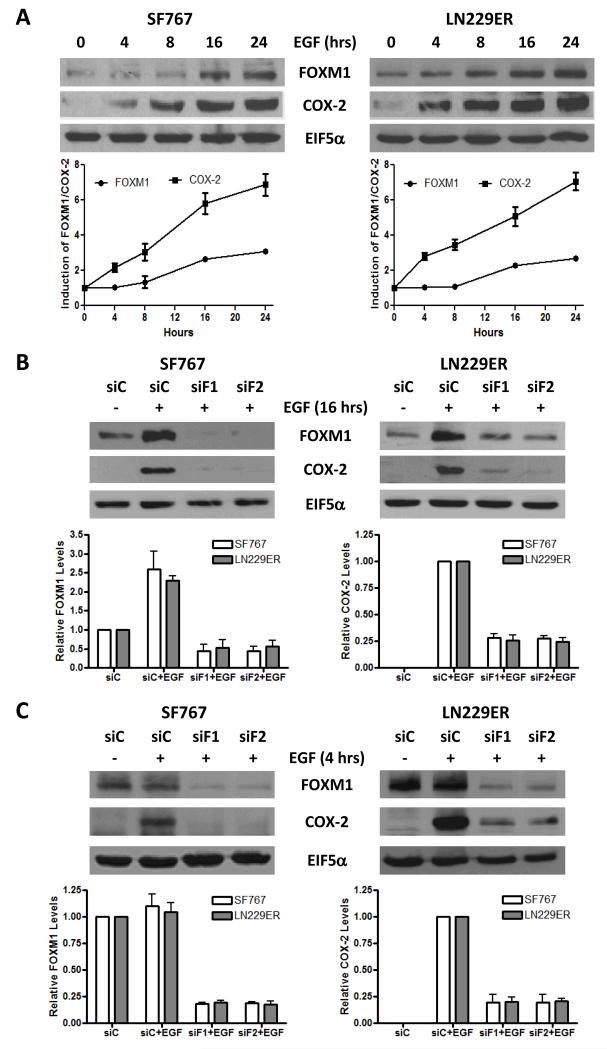
We previously demonstrated that EGF regulates COX-2 expression through the p38-MAPK/PKC-δ/Sp1 signaling axis in human glioma cells (22, 23). Similarly, expression of the constitutively-active EGFRvIII results in COX-2 upregulation (Supp. Fig. 1). Since FOXM1 can transcriptionally regulate the COX-2 promoter in a lung cancer model (13), we examined the role of FOXM1 in EGFR-dependent induction of COX-2 in glioma cells. SF767, which show high basal expression of EGFR, and LN229ER, cells engineered to overexpress EGFR, were treated with EGF for varying times after culturing overnight in serum-free media. In each case, EGF significantly induced FOXM1 by 8-16 hours post-stimulation although this induction appeared to trail COX-2 induction (Fig. 1A). Next, after suppression of FOXM1 with siRNA in these same cells, EGF-dependent COX-2 induction at 16 hours was blocked in the FOXM1-silenced cells (Fig. 1B). Finally, even as early as 4 hours after EGF stimulation, FOXM1 knockdown resulted in loss of EGF-induced COX-2 expression (Fig. 1C). These results indicate that FOXM1 is required for EGF-dependent induction of COX-2 and suggest that while increased expression of FOXM1 is not crucial for EGF-dependent COX-2 induction, a basal level of FOXM1 expression is needed.
Figure 1. FOXM1 is involved in EGF-induced COX-2 expression.
Immunoblots analysis of the SF767 and LN229ER glioma cells that were treated in the following manner: (A) Stimulated with EGF for the indicated times and stimulated with or without EGF for (B) 16 hours or (C) 4 hours in cells transfected with either control (siC) or two different FOXM1-targeting (siF1 and siF2) siRNA 40 hours prior to ligand stimulation. Blots were probed with antibodies against FOXM1, COX-2 and EIF5α (normalization control), as indicated. The displayed blots are representative of 3 independent experiments. Displayed graphs show the quantitation from pooled results of the independent experiments. Error bars are one standard error of the mean (SEM).
Src and p38-MAPK mediate EGF-dependent FOXM1 and COX-2 induction and FOXM1 is required for Src- or p38-MAPK-dependent induction of COX-2
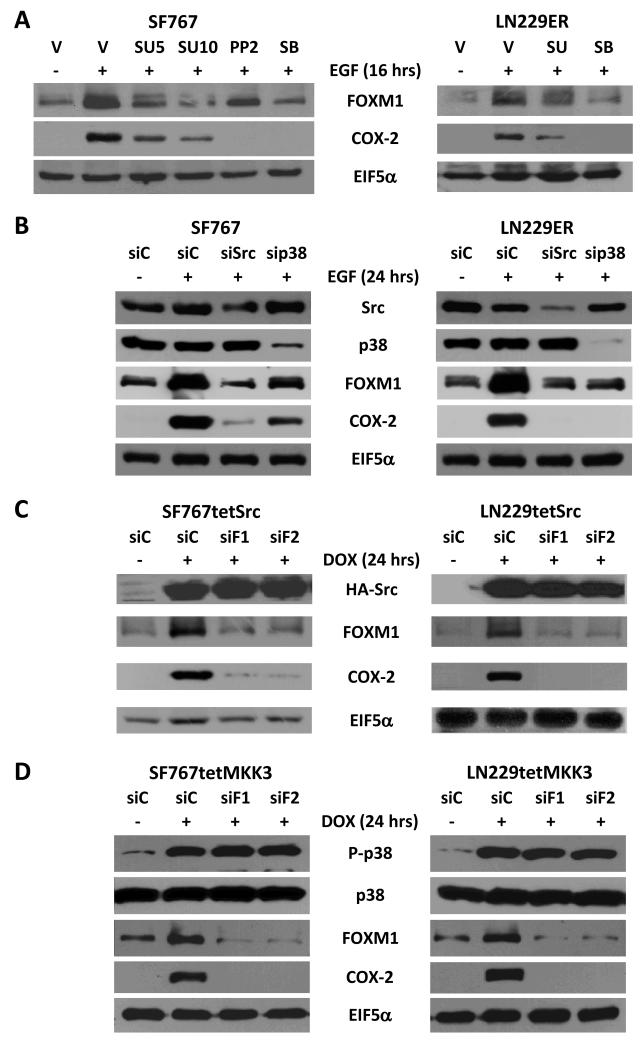
We have previously shown a significant role for p38-MAPK in COX-2 induction by EGF (22). Since, Src kinase is another downstream mediator of EGF signaling, we also examined whether Src has a role in the induction of COX-2 expression. We had previously established that 5 and 10 μM of SU6656 was sufficient to inhibit downstream signaling from the Src kinase (31). In addition, 20 μM of SB202190 is sufficient to inhibit signaling from p38-MAPK based on loss of ATF2 phosphosporylation (a downstream marker of p38-MAPK function) after EGF stimulation with inhibitor treatment (data not shown). We found that chemical inhibition of Src with SU6656 (at 5 and 10 μM) or PP2 (another Src inhibitor with well-characterized activity profile) (at 2 μM) and p38-MAPK with SB202190 (at 20 μM) after EGF treatment in SF767 and LN229ER cells resulted in significant attenuation of COX-2 induction (Fig. 2A). This question was further assessed using genetic inhibition of Src and p38-MAPK with a specific siRNAs against these kinases. Knockdown of Src and p38-MAPK was similarly able to attenuate FOXM1 and COX-2 induction (Fig. 2B). Src’s role in the regulation of COX-2 expression was further confirmed using SF767 and LN229 derivatives engineered to stably express an activated Src gene under the control of a tetracycline-inducible promoter (SF767tetSrc and LN229tetSrc). In these cells, induction of Src with doxycycline resulted in a significant increase in COX-2 expression (Fig. 2C). Similarly, inhibition of Src kinase and p38-MAPK chemically with SU6656/PP2 for Src and SB202190 for p38-MAPK or genetically with siRNA targeting Src or p38-MAPK attenuated EGF-dependent FOXM1 induction (Fig. 2A-B). Furthermore, induction of either Src (using SF767tetSrc and LN229tetSrc) or MKK3 (using SF767tetMKK3 or LN229tetMKK3 that was previously described (23)) resulted in significant induction of FOXM1 expression (Fig. 2C-D). In each case, siRNA knockdown of FOXM1 expression led to a significant decrease in COX-2 induction (Fig. 2C-D). These data indicate that the Src kinase is another signaling molecule downstream of EGFR required for COX-2 induction, both Src and p38-MAPK are important factors in EGF-induced FOXM1 expression and FOXM1 is a critical factor for both Src- and p38-MAPK-induced COX-2 expression.
Figure 2. Src and p38-MAPK induces FOXM1 and COX-2 expression and COX-2 regulation requires FOXM1.
Immunoblots analysis of indicated cells that were treated in the following manner: (A) SF767 and LN229ER glioma cells treated with vehicle (V, DMSO), the Src kinase inhibitors, SU6656 (SU) at the indicated concentrations (5 or 10 μM) and PP2 (2 μM) or the p38-MAPK inhibitor SB202190 (SB, 20 μM) for 2 hours prior to a 16 hour stimulation with or without EGF (90 ng/ml). (B) SF767 and LN229ER cells transfected with either control (siC) or siRNA targeting Src (siSrc) or p38-MAPK (sip38) 40 hours prior to a 24 hour stimulation with or without EGF (90 ng/ml). (C) SF767tetSrc and LN229tetSrc cells engineered with a tet-inducible activated Src tagged with hemagglutinin A (HA) are transfected with either siC or two different FOXM1 (siF1 and siF2) siRNA 40 hours prior to a 24 hour treatment with or without doxycycline (DOX, 1 μg/ml). (D) SF767tetMKK3 and LN229tetMKK3 cells engineered with a tet-inducible MKK3 are transfected with siC, siF1 or siF2 40 hours prior to a 24 hour treatment with or without DOX. Blots were probed with antibodies against FOXM1, COX-2, Src, p38-MAPK (p38), phosphorylated p38-MAPK (P-p38) and EIF5α (normalization control), as indicated. The displayed blots are representative of 2-3 independent experiments. Quantitation of pooled results from independent experiments is shown on Supp. Fig. 2.
Src kinase is upstream of the p38-MAPK/PKC-δ/Sp1 signaling cascade
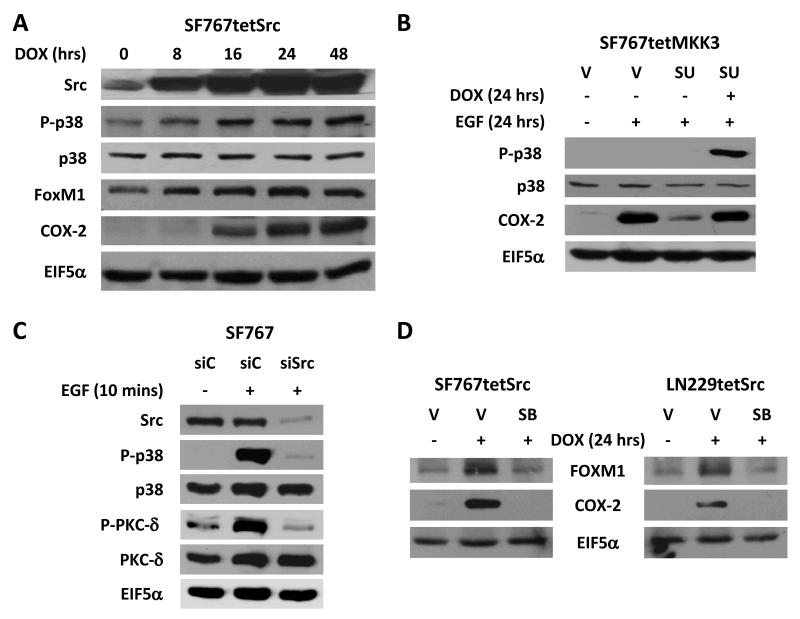
Based on our data above and previous work (22, 23), both Src kinase and p38-MAPK are required for EGF-dependent COX-2 expression. However, Src kinase’s role relative to the p38-MAPK/PKC-δ/Sp1 signaling cascade in the induction of COX-2 remained unclear. After induction of activated Src in SF767tetSrc cells with doxycycline, p38-MAPK becomes phosphorylated and COX-2 is induced with kinetics that just lag the induction of Src (Fig. 3A). This data suggest that p38-MAPK is a downsteam effector of Src kinase. This was further confirmed when selective activation of p38-MAPK could reverse the inhibitory effect of the Src inhibitor SU6656 on EGF-dependent induction of COX-2 in SF767tetMKK3 cells (Fig. 3B). While p38 does not appear to be activated by EGF (Fig. 3B), this is, in fact, not the case with p38 activation peaking at 10 minutes and almost completely attenuated by 1 hour after EGF stimulation (Supp. Fig. 4A). In addition, this early p38 activation at 10 minutes post-EGF stimulation is very nicely inhibited by chemical inhibition of Src kinase activity (Supp. Fig. 4B). Genetic suppression of Src by siRNA-mediated knockdown was also able to attenuate signaling to p38-MAPK after EGF stimulation (Fig. 3C). When the converse experiment was performed, the p38 inhibitor SB202190 was able to efficiently block COX-2 induction by Src in SF767tetSrc and LN229tetSrc cells (Fig. 3D). Because our previous work placed PKC-δ and Sp1 downstream of p38-MAPK in the COX-2 signaling cascade (23), we predicted that they would lie downstream of Src as well. Again, genetic suppression of Src by siRNA-mediated knockdown was able to attenuate signaling to PKC-δ after EGF stimulation (Fig. 3C) and, in each case, Src-dependent induction of COX-2 could be blocked by chemical inhibition of PKC-δ (with bisindolylmaleimide I or rottlerin) or Sp1 (with mithramycin) (Supp Fig. 5). These data suggest that Src is upstream of p38-MAPK/PKC-δ/Sp1 in the COX-2 signaling cascade.
Figure 3. Src kinase is upstream of the p38-MAPK/PKC-δ signaling cascade.
Immunoblots analysis of the indicated cells that were treated in the following manner: (A) SF767tetSrc cells were treated with doxycycline (DOX, 1 μg/ml) for the indicated times to induce Src expression. (B) SF767tetMKK3 cells were pretreated with vehicle (V, DMSO) or SU6656 (SU, 5 μM) for 1 hour prior to stimulation with or without EGF or EGF+DOX for 24 hours. (C) SF767 cells transfected with either siC or siSrc 40 hours prior to a 10 minute stimulation with or without EGF (90 ng/ml). (D) SF767tetSrc and LN229tetSrc cells were pretreated with V or SB202190 (SB, 20 μM) for 1 hour prior to treatment with or without DOX for 24 hours. Blots were probed with antibodies against Src, FOXM1, P-p38, p38, phosphorylated PKC-δ (P-PKC-δ, at Thr505), PKC-δ COX-2 and EIF5α (normalization control), as indicated. The displayed blots were representative of 2 independent experiments. Quantitation of pooled results from independent experiment is shown on Supp. Fig. 3.
Overexpression of FOXM1 increases COX-2 expression in a biologically relevant fashion but does not further enhance EGF-induced COX-2 expression
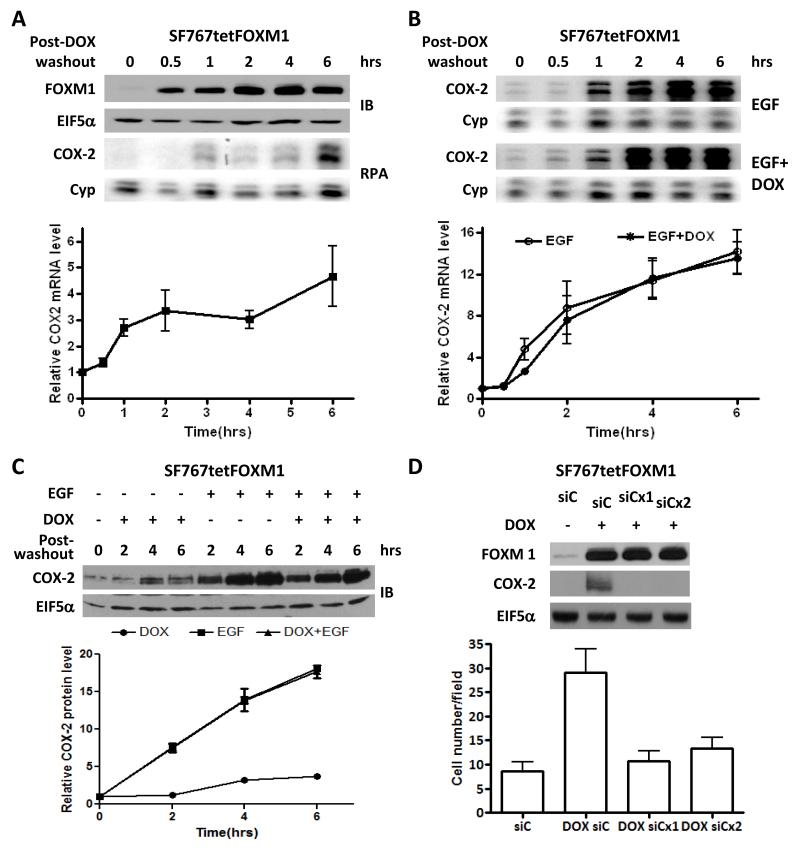
Increased expression of FOXM1 can raise COX-2 levels in some lung cancer cells (13). While FOXM1 is required for EGF-induced COX-2 expression in glioma cells (see previous data above), we were uncertain as to whether overexpression of FOXM1 alone can similarly induce COX-2 in these cells. To test this, SF767tetFOXM1 cells (with FOXM1 under the control of a tet-inducible promoter) were treated with doxycycline for 2 hours and then washed out. Here, FOXM1 was induced for the duration of this experiment (6 hours) and correlated with increased levels of COX-2 mRNA (Fig. 4A). However, increased FOXM1 expression was not able to further enhance EGF-induced COX-2 expression (Fig. 4B). FOXM1 was also able to increase not only COX-2 mRNA levels but also COX-2 protein expression (Fig. 4C). Finally, forced FOXM1 expression have been previously shown to increase glioma cell invasion (19, 20). Similarly, FOXM1 overexpression in our system increased invasive potential of SF767 cells as measured by a Matrigel invasion assay and this increase was attenuated by COX-2 knockdown suggesting that COX-2 is a biologically relevant target of FOXM1 (Fig. 4D).
Figure 4. Overexpression of FOXM1 induces COX-2 in a biologically relevant fashion but does not further enhance EGF-induced COX-2 expression.
(A) Immunoblot (IB) and RNase protection assay (RPA) analysis of SF767tetFOXM1 cells were treated with DOX for 2 hours, washed with PBS × 3 and then incubated in serum-free media without DOX (post-washout) for the indicated times. (B) RPA analysis of SF767tetFOXM1 after EGF treatment for the indicated times post-DOX washout after 2 hours of DOX pretreatment. (C) Immunoblot analysis of SF767tetFOXM1 cells after pretreatment with or without DOX followed by washout and treatment with or without EGF for the indicated times. (D) Immunoblot analysis of SF767tetFOXM1 cells after transient introduction of control (siC) or COX-2 (siCx1 or siCx2) siRNA and treatment with or without DOX (1 μg/ml) as detailed in the Methods section. Cells were then assessed in the Matrigel invasion assay with the graph representing average number of invasive cells counted per 40× objective field. From 5 fields were counted for each duplicate sample per experiment. The invasion assay results are the average of 3 independent experiments. Blots were probed with antibodies against FOXM1, COX-2 and EIF5α (normalization control), as indicated. RPA detected COX-2 RNA levels with cyclophilin (Cyp) as a normalization control. The displayed blots and RPA phosphor images were representative of 3 independent experiments. Graphs (A and B) display the average COX-2 expression on RPA from 3 independent experiments each normalized to Cyp and the 0 hour time arbitrarily set at 1. Error bars are one SEM.
The COX-2 promoter contains a FOXM1 responsive element
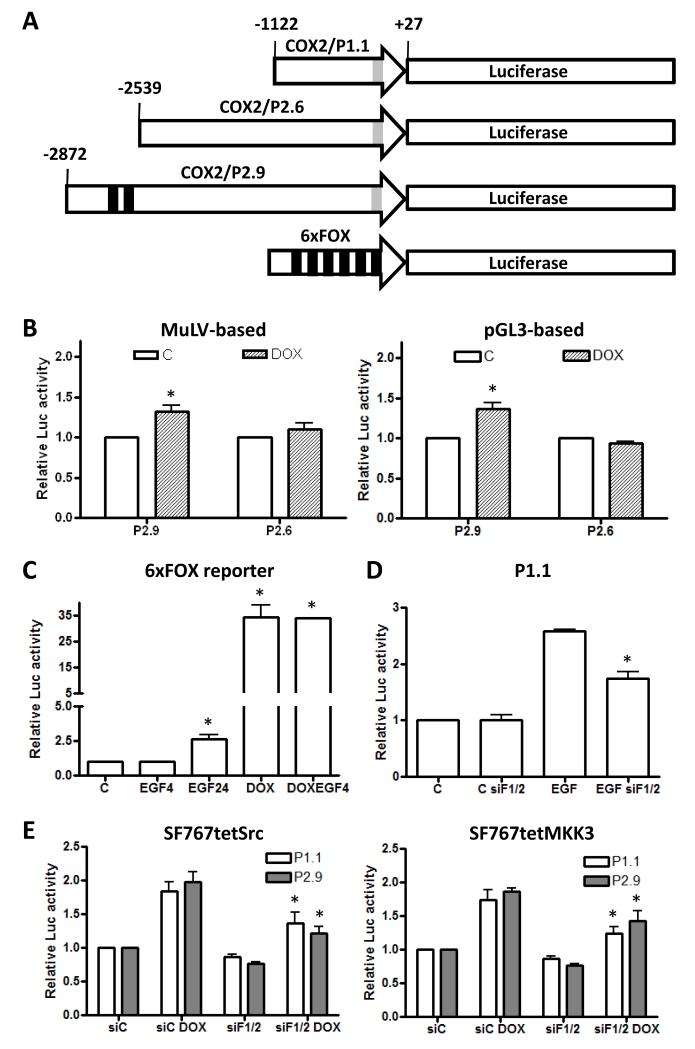
Because overexpression of FOXM1 induces COX-2 expression in glioma cells, we sought to identify the FOXM1 responsive element in the COX-2 promoter. Examination of the human COX-2 promoter region identified potential FOXM1 responsive elements at the −2609/−2584 base region relative to the COX-2 start site. To test if this site is active, luciferase reporter constructs containing the human COX-2 promoter to the −2872 (COX2/P2.9) and −2539 (COX2/P2.6) positions (Fig. 5A) were either stably introduced into the SF767tetFOXM1 cells using MuLV-based luciferase constructs or transiently transfected into the LN229tetFOXM1 cells using pGL3-based luciferase constructs. After FOXM1 induction with doxycycline, luciferase was consistently induced, albeit at a low level, in the cells containing the COX2/P2.9 reporter but not the COX2/P2.6 reporter (Fig. 5B). This result suggests that FOXM1 responsive element(s) resides between −2872 and −2539 positions relative to the transcriptional start in the human COX-2 promoter consistent with the potential FOXM1 sites the −2609/−2584 base region.
Figure 5. Identification of a FOXM1 responsive element in the COX-2 promoter.
(A) Luciferase reporter constructs with different lengths of the COX-2 promoter [−1122/+27 (P1.1), −2539/+27 (P2.6) and −2872/+27 (P2.9)] or 6 tandem FOXM1 binding sites (6×FOX) are depicted. The locations of the FOXM1 (in black) and Sp1 (in gray) binding sites are marked. (B) The P2.9 and P2.6 reporter constructs (MuLV-based or pGL3-based, as indicated) were introduced into SF767tetFOXM1 (MuLV-based) and LN229tetFOXM1 (pGL3-based) cells engineered with a tet-inducible FOXM1. Cells were treated ± DOX for 2 hours, washed with PBS × 3, cultured in serum-free media without DOX for 6 hours and then harvested. As a control to show that P2.6 behaves as expected, this construct was also assessed in cells treated with EGF (Supp. Fig. 6) (C) SF767tetFOXM1 cells were transiently transfected with the 6×FOX reporter, treated with vehicle (C), EGF (90 ng/ml) for 4 or 24 hours, DOX for 24 hours and DOX+EGF for 24 hours and then harvested. (D) The P1.1 luciferase reporter was stably introduced into SF767 cells, transiently transfected with control (siC) or a mix of two FOXM1-targeting (siF1/F2) siRNA, treated 40 hour after transfection with or without EGF for 6 hours and then harvested. (E) The P1.1 and P2.9 luciferase reporters were stably introduced into SF767tetSrc and SF767tetMKK3 cells, transiently transfected with siC or siF1/F2 siRNA, treated 40 hour after transfection with or without DOX for 24 hours and then harvested. Graphs show the average of 3 independent experiments with each sample performed in triplicate. Luciferase levels are normalized to total protein in lysate and expressed as fold induction over basal. (*) indicate statistically significant difference compared with control by student’s t-test with p-value < 0.05. Error bars are one SEM.
While FOXM1 can be phosphorylated by ERK1/2 leading to increased transcriptional activity (32), we have not found any obvious change in phosphorylation of FOXM1 in our system after EGFR activation when FOXM1 is immunoprecipitated after in vivo 32P-orthophosphate labeling (data not shown). Since EGFR activation clearly signals to ERK1/2 in glioma cells, we sought to determine whether this mechanism of activating FOXM1 is important in these cells. To test this possibility, the luciferase reporter containing six tandem FOXM1 binding sites (6×FOX reporter) was transiently introduced into SF767tetFOXM1 cells. Luciferase activity did not appreciably change after addition of EGF for 4 hours despite normal activation of ERK1/2 at this time point (Fig. 5C and data not shown). However, luciferase activity does increase significantly (by approximately 2.5-fold) after EGF stimulation for 24 hours (Fig. 5C). As a positive control, FOXM1 induction with doxycycline led to a 35-fold increase in luciferase activity (Fig. 5C). These data suggest that EGF is unlikely to be modulating FOXM1 activity through ERK1/2 signaling in glioma cells. Since FOXM1 is not significantly induced after four hours of EGF stimulation but does show significant induction after 24 hours of stimulation, our data is more consistent with increased FOXM1 levels and not post-translational modification with phosphorylation as the predominant mechanism for FOXM1-dependent transcriptional activation.
FOXM1 does not act through its responsive element in EGF-dependent activation of the COX-2 promoter
Although FOXM1 levels are not significantly increased at four hours following EGF stimulation, our data clearly show that knockdown of FOXM1 reduced EGF-induced COX-2 expression at that time point (Fig. 1). This finding suggests that increase in FOXM1 expression may not be a major mechanism by which FOXM1 contributes to EGF-induced COX-2 expression, especially at early time points. To explore potential mechanisms, the luciferase reporter construct COX2/P1.1 (Fig. 5A) (22), which excludes the conventional FOXM1 binding sites residing between −2872 and −2539, was stably introduced into SF767 cells. EGF stimulation clearly increased luciferase activity and this could be significantly reduced by FOXM1 knockdown (Fig. 5D). To test if FOXM1 is similarly required for p38-MAPK- and Src-dependent activation of the COX-2 promoter, the COX-2/P1.1 and COX-2/P2.9 luciferase reporters were stably introduced into SF767tetSrc and SF767tetMKK3 cells. In each case, induction of luciferase activity by MKK3 or Src was similar and this could again be significantly reduced by FOXM1 knockdown (Fig. 5E). Thus, FOXM1 has a role in EGF-dependent activation of the COX-2 promoter independent of the conventional FOXM1 binding sites residing between −2872 and −2539.
FOXM1 can interact with Sp1 and bind at the Sp1 responsive element
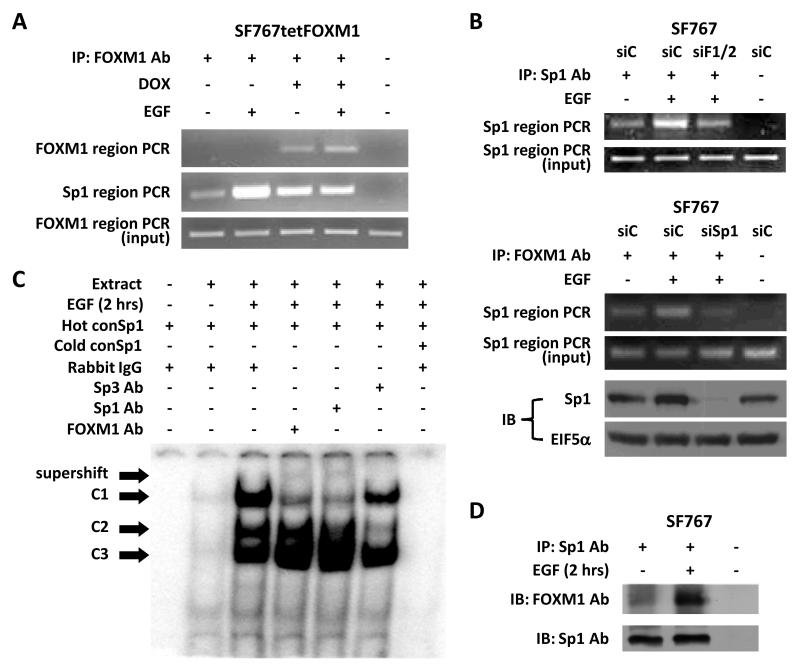
Next, we sought to explore potential mechanisms for how FOXM1 was involved by COX-2 promoter activation independent of the FOXM1 binding sites. Because FOXM1 has been shown to cooperate with Sp1 to regulate c-Myc expression (33), we decided to test if FOXM1 can also cooperate with Sp1 to regulate COX-2 expression in glioma cells. ChIP assay revealed that FOXM1 could associate with the distal COX-2 promoter region at −2707/−2421 (which encompass the putative FOXM1 sites at −2609/−2584) after DOX induction of FOXM1 and the proximal COX-2 promoter region at −322/−39 (which encompass the Sp1 site at −245/−240) especially after EGF stimulation (Fig. 6A). Overall, these data suggest that FOXM1 binds to the Forkhead consensus sites only when overexpressed but will show increased association with the Sp1 site after either EGF stimulation or FOXM1 overexpression. Additional ChIP assay revealed that EGF-induced association of Sp1 to its binding site was significantly reduced in the setting of FOXM1 knockdown (Fig. 6B). Correspondingly, EGF-induced association of FoxM1 to the Sp1 binding site was also reduced in the setting of Sp1 knockdown (Fig. 6B). These results support a role for FOXM1 in the EGF-induced association of Sp1 to its binding site on the COX-2 promoter.
Figure 6. FOXM1 can interact with Sp1 and bind at the Sp1 responsive element.
(A) SF767tetFOXM1 cells were treated with or without DOX for 16 hours, washed with PBS × 3, cultured in serum-free medium with or without EGF for 5 hours and then harvested for chromatin immunoprecipitation (ChIP) assay. Immunoprecipitation was done with an antibody against FOXM1. PCR was performed as described in the Methods section spanning −2707/−2421 (FOXM1 site, upper panel) and −322/−39 (Sp1 site, middle panel) of the COX-2 promoter. A portion of DNA recovered from an aliquot of sheared chromatin was used for the FOXM1 PCR as the “input” sample (lower panel). (B) SF767 cells were transfected with control (siC), a mix of two FOXM1-targeting (siF1/2) siRNAs or an Sp1-tageting siRNA (siSp1), as indicated, treated 40 hours after transfection with or without EGF for 5 hours and harvested for ChIP assay. Immunoprecipitation was done with an antibody against Sp1 or FOXM1, as indicated. Sp1 region PCR was performed on the immunoprecipitate (upper panel of each set) or input DNA (lower panel of each set). For the set utilizing siSp1, immunoblot (IB) probing for Sp1 and EIF5 are shown. (C) Electrophoretic mobility shift assay (EMSA) using nuclear extracts from SF767 cells treated with or without EGF for 2 hours. The consensus Sp1 oligonucleotide (conSp1) was labeled with 32P. Cold conSp1 was used at a 40-fold molar excess. Three complexes (C1, C2 and C3) were seen (marked by arrows). Nuclear extracts were preincubated with antibodies against FOXM1, Sp1 and Sp3 to assess for complex supershift (marked by arrow). (D) SF767 cells were treated with or without EGF for 2 hours, fixed with 1% formaldehyde at 37°C for 10 mins and then harvested for immunoprecipitation. Immunoprecipitation was performed using an antibody against Sp1 and then evaluated by immunoblot using antibodies against FOXM1 and Sp1. The last lane represents the IgG control. The displayed gels, phosphor images and blots were representative of 2-3 independent experiments.
To further confirm this idea, EMSA was performed using nuclear extracts from SF767 and an Sp1 consensus oligonucleotide (conSp1). Extract from cells treated with EGF increases the formation of three major complexes (C1, C2 and C3) as previously reported (Fig. 6C) (22). Consistent with our previous results, preincubation of nuclear extract with Sp1 antibody reduced formation of the C1 complex and resulted in a slower migrating band representing supershifted C1 while preincubation with Sp3 antibody significantly reduced formation of the C2 complex (Fig. 6C). With FOXM1 antibody preincubation, the C1 complex was also reduced, again, with corresponding formation of a supershifted C1 (Fig. 6C). In a coimmunoprecipitation assay, FOXM1 was detectable in complex with Sp1 and this association was significantly increased after stimulation with EGF (Fig. 6D). These results suggest that the C1 complex contains Sp1/FOXM1 and EGF treatment will to increase interactions between Sp1/FOXM1 and the Sp1 binding site. Finally, we found that increased association of FOXM1 with the Sp1 site in the COX-2 promoter after EGF stimulation could be decreased by the p38-MAPK inhibitor SB202190 but not the MEK inhibitor U0126 suggesting that p38-MAPK but not ERK1/2 is involved in the regulation of the Sp1-FOXM1 complex (Supp. Fig. 7).
DISCUSSION
In our previous studies, we showed that the Sp1 transcription factor has a crucial role at mediating EGF-dependent COX-2 induction in glioma cells (22, 23). Stimulation of human glioma cells with EGF leads to activation of p38-MAPK and PKC-δ, which directly phosphorylates and activates Sp1 leading to increased expression of COX-2. In the present study, we found that the Src kinase is also involved in this signaling cascade and functions upstream of p38-MAPK. In addition, we found that EGF induces expression of both FOXM1 and COX-2 in human glioma cells in a process requiring both Src kinase and p38-MAPK. However, careful examination of the kinetics of induction revealed that increased COX-2 expression occurred at an earlier time point than increased FOXM1 expression. While this might suggest that COX-2 induction is not dependent on FOXM1, silencing of FOXM1 clearly reduces EGF-dependent COX-2 induction, indicating that FOXM1 is, in fact, required for this induction.
When FOXM1 was overexpressed, this alone could induce COX-2 expression but did not further enhance EGF-dependent COX-2 induction. COX-2 promoter analyses show that overexpression of FOXM1 increased luciferase activity with the luciferase reporter containing the P2.9 but not the P2.6 fragment, suggesting that a FOXM1 response element is located between −2872 and −2539 relative to the COX-2 transcriptional start site. Evaluation of the COX-2 promoter sequence revealed two core FOXM1 consensus sites (TAAACA or TGTTTA) 14 base pairs apart found between position −2609 and −2584. Our ChIP assay data show that FOXM1 binds to this region only when overexpressed with little to no association with basal levels of FOXM1. This suggests that FOXM1 binds to and transactivates the COX-2 promoter when overexpressed. The FoxM1 DNA binding domain has an unusually low affinity for its binding site with a dissociation constant (KD) of 7000 ± 300 nM for single copies of the Forkhead consensus sequence (34). This value is one and four orders of magnitude lower than the affinity reported for the FoxO3a-DBD and FoxD3-DBD, respectively. The low affinity of FOXM1 for single copies of the consensus sequence may explain the difficulty in detecting its binding to the Forkhead site in the COX-2 promoter under basal conditions. Our data show that FOXM1 expression is significantly increased by 16 hours after stimulation with EGF. Such an increase in FOXM1 protein may contribute to late activation (16-24 hours post-EGF stimulation) of the COX-2 promoter by a typical mechanism in which FOXM1 binds to its consensus sequence leading to transactivation.
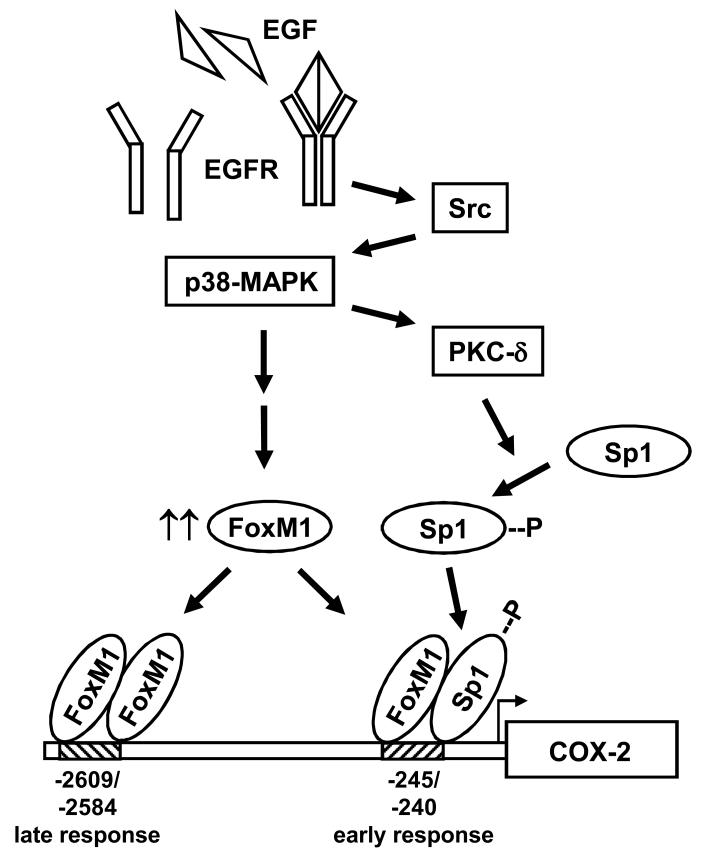
Although overexpression of FOXM1 leads to transcriptional activation of the COX-2 promoter through binding to its consensus, we also found that siRNA knockdown of FOXM1 impairs early EGF-dependent COX-2 induction (4 hours post-EGF stimulation), at a point when FOXM1 protein levels remain unchanged. Therefore, another mechanism must be responsible for this early EGF-dependent COX-2 induction. This conclusion was further supported when we found that EGF-, Src- and MKK3-dependent activation of the COX-2/P1.1 promoter fragment, which excludes the consensus FOXM1 binding sites in the distal promoter region, was still inhibited after FOXM1 knockdown. These results now suggest that FOXM1 also contributes to COX-2 promoter activation by an atypical mechanism that does not require a Forkhead consensus sequence. Wierstra and Alves showed that FOXM1c can directly bind Sp1 and transactivate the human c-myc P1 and P2 promoters synergistically with Sp1 (33). Based on this finding and our previous work that demonstrated a significant role for Sp1 in activating the COX-2 promoter, we hypothesized that FOXM1 may also be cooperating with Sp1 to regulate COX-2 expression in glioma cells. This turned out to be the case when we found by ChIP assays that while FOXM1 was not associating with the Forkhead consensus sites (at −2609/−2584) at 4 hours after EGF stimulation, it was associating with the COX-2 promoter fragment that contained the Sp1 binding site (at −245/−240) (Fig. 6A). This result suggests that FOXM1 is part of the Sp1 complex that associates with the Sp1 binding site and transactivates the COX-2 promoter. The findings that an anti-FOXM1 antibody could supershift the Sp1 complex on EMSA (Fig. 6C) and that FOXM1 and Sp1 could be coimmunoprecipated (Fig. 6D) provided additional support for this model. An illustration of our current working model for EGF-dependent COX-2 induction is shown (Fig. 7). In this model, early EGF-dependent activation of the COX-2 promoter occurs predominantly through the association of FOXM1 and Sp1 and binding to the Sp1 site while later, or sustained EGF-dependent activation of the COX-2 promoter occurs through increased expression of the FOXM1 transcription factor and association with its own consensus sequence. We have further confirmed this model by demonstrating that overexpression of FOXM1 in the absence of EGFR activation increases COX-2 expression, presumably through stimulation of its own upstream responsive element, and this increase is not influenced by Sp1 knockdown (Supp. Fig. 8).
Figure 7. Schematic of the signaling cascade involved in EGF-induced COX-2 expression.
Engagement of EGF by its receptor results in sequential activation of Src, p38-MAPK and PKC-δ. Activation of these kinases ultimately results in Sp1 phosphorylation and increased expression of FOXM1. These transcription factors will then associate their respective binding sites to activate the COX-2 promoter with either an early (4-6 hours) or a late (16-24 hours) response following EGF stimulation.
Our data show that EGF can induce FOXM1 expression in gliomas, which agrees with the finding that gefitinib can reduce expression of FOXM1 in breast carcinoma cells (35). Induction of activated Src and MKK3 results in increased FOXM1 expression indicating that the EGFR signaling cascade required for FOXM1 induction includes both Src and p38-MAPK. One regulatory pathway with particular relevance to gliomas is sonic hedgehog signaling and its downstream target Gli-1, which can transcriptionally regulate FOXM1 (36). In addition, proteasome inhibition can reduce FOXM1 expression and transcriptional activity suggesting that a component of the regulation of this transcription factor may involve a negative regulator that is stabilized by proteasome inhibition and inhibits FOXM1 activity and expression (37). The precise mechanism of FOXM1 induction by EGFR signaling remains to be elucidated and is beyond the scope of this current study.
In summary, we find that EGF induces FOXM1 expression in a process requiring Src kinase and p38-MAPK and such induction may, in part, contribute to the late phase of EGF-dependent COX-2 induction through a conventional mechanism whereby FOXM1 binds to its consensus site. In addition, FOXM1 can interact and cooperate with Sp1, independent of the FOXM1 binding sites, to transactivate the COX-2 promoter through the Sp1 binding site. This association between FOXM1 and Sp1 is responsible for the early phase of COX-2 induction after EGF stimulation and may, in fact, be the major mechanism by which FOXM1 is involved in COX-2 regulation. Since FOXM1 is not expressed in many normal tissues (18), it may be a good therapeutic target to inhibit COX-2 expression and potentially angiogenesis in glioma.
Supplementary Material
ACKNOWLEDGEMENT
We thank P. Raychaudhuri for providing pCMV-T7FOXM1 and 6×FOX-luciferase reporter, Y. Feng for providing pCSrcA-HA14, T.J. Murphy for providing pTJ105 (MuLV luciferase vector), and C. Hadjipanayis for providing lysates from U87MG control and EGFRvIII-expressing cells. This work was supported in part by a Cancer Center grant from the National Cancer Institute (P30-CA138292).
Footnotes
Conflict of Interest: None
REFERENCES
- 1.Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–86. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 2.Dandekar DS, Lokeshwar BL. Inhibition of cyclooxygenase (COX)-2 expression by Tet-inducible COX-2 antisense cDNA in hormone-refractory prostate cancer significantly slows tumor growth and improves efficacy of chemotherapeutic drugs. Clin Cancer Res. 2004;10:8037–47. doi: 10.1158/1078-0432.CCR-04-1208. [DOI] [PubMed] [Google Scholar]
- 3.Kuipers GK, Slotman BJ, Wedekind LE, Stoter TR, Berg J, Sminia P, et al. Radiosensitization of human glioma cells by cyclooxygenase-2 (COX-2) inhibition: independent on COX-2 expression and dependent on the COX-2 inhibitor and sequence of administration. Int J Radiat Biol. 2007;83:677–85. doi: 10.1080/09553000701558985. [DOI] [PubMed] [Google Scholar]
- 4.Nakata E, Mason KA, Hunter N, Husain A, Raju U, Liao Z, et al. Potentiation of tumor response to radiation or chemoradiation by selective cyclooxygenase-2 enzyme inhibitors. Int J Radiat Oncol Biol Phys. 2004;58:369–75. doi: 10.1016/j.ijrobp.2003.09.061. [DOI] [PubMed] [Google Scholar]
- 5.Kang KB, Wang TT, Woon CT, Cheah ES, Moore XL, Zhu C, et al. Enhancement of glioblastoma radioresponse by a selective COX-2 inhibitor celecoxib: inhibition of tumor angiogenesis with extensive tumor necrosis. Int J Radiat Oncol Biol Phys. 2007;67:888–96. doi: 10.1016/j.ijrobp.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 6.Joki T, Heese O, Nikas DC, Bello L, Zhang J, Kraeft SK, et al. Expression of cyclooxygenase 2 (COX-2) in human glioma and in vitro inhibition by a specific COX-2 inhibitor, NS-398. Cancer Res. 2000;60:4926–31. [PubMed] [Google Scholar]
- 7.Shono T, Tofilon PJ, Bruner JM, Owolabi O, Lang FF. Cyclooxygenase-2 expression in human gliomas: prognostic significance and molecular correlations. Cancer Res. 2001;61:4375–81. [PubMed] [Google Scholar]
- 8.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–6. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–9. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–61. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 13.Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, Kalin TV. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008;27:4137–49. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Ligr M, McCarron JP, Daniels G, Zhang D, Zhao X, et al. Natura-alpha targets forkhead box m1 and inhibits androgen-dependent and -independent prostate cancer growth and invasion. Clin Cancer Res. 2011;17:4414–24. doi: 10.1158/1078-0432.CCR-11-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29:2983–95. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11:477–87. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer cell. 2011;20:427–42. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao KM, Sha M, Lu Z, Wong GG. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J Biol Chem. 1997;272:19827–36. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 20.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–9. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–42. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K, Shu HK. EGFR activation results in enhanced cyclooxygenase-2 expression through p38 mitogen-activated protein kinase-dependent activation of the Sp1/Sp3 transcription factors in human gliomas. Cancer Res. 2007;67:6121–9. doi: 10.1158/0008-5472.CAN-07-0141. [DOI] [PubMed] [Google Scholar]
- 23.Xu K, Chang CM, Gao H, Shu HK. Epidermal growth factor-dependent cyclooxygenase-2 induction in gliomas requires protein kinase C-delta. Oncogene. 2009;28:1410–20. doi: 10.1038/onc.2008.500. [DOI] [PubMed] [Google Scholar]
- 24.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes & development. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S, Jiang Y, Li Z, Nishida E, Mathias P, Lin S, et al. Apoptosis signaling pathway in T cells is composed of ICE/Ced-3 family proteases and MAP kinase kinase 6b. Immunity. 1997;6:739–49. doi: 10.1016/s1074-7613(00)80449-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Lu Z, Ku L, Chen Y, Wang H, Feng Y. Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. The EMBO journal. 2003;22:1801–10. doi: 10.1093/emboj/cdg171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–61. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Chang CM, Yuan M, McKenna WG, Shu HK. Resistance to small molecule inhibitors of epidermal growth factor receptor in malignant gliomas. Cancer Res. 2003;63:7443–50. [PubMed] [Google Scholar]
- 29.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu K, Robida AM, Murphy TJ. Immediate-early MEK-1-dependent stabilization of rat smooth muscle cell cyclooxygenase-2 mRNA by Galpha(q)-coupled receptor signaling. J Biol Chem. 2000;275:23012–9. doi: 10.1074/jbc.M001611200. [DOI] [PubMed] [Google Scholar]
- 31.Xu K, Kitchen CM, Shu HK, Murphy TJ. Platelet-derived growth factor-induced stabilization of cyclooxygenase 2 mRNA in rat smooth muscle cells requires the c-Src family of protein-tyrosine kinases. J Biol Chem. 2007;282:32699–709. doi: 10.1074/jbc.M705272200. [DOI] [PubMed] [Google Scholar]
- 32.Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118:795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- 33.Wierstra I, Alves J. FOXM1c and Sp1 transactivate the P1 and P2 promoters of human c-myc synergistically. Biochem Biophys Res Commun. 2007;352:61–8. doi: 10.1016/j.bbrc.2006.10.151. [DOI] [PubMed] [Google Scholar]
- 34.Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, et al. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 2010;38:4527–38. doi: 10.1093/nar/gkq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS, et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther. 2009;8:582–91. doi: 10.1158/1535-7163.MCT-08-0805. [DOI] [PubMed] [Google Scholar]
- 36.Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62:4773–80. [PubMed] [Google Scholar]
- 37.Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PLoS One. 2009;4:e6593. doi: 10.1371/journal.pone.0006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.