Abstract
In cells, microtubules (MTs) are nucleated at MT-organizing centers (MTOCs). The centrosome-based MTOCs organize radial MT arrays which are often not optimal for polarized trafficking. A recently discovered subset of non-centrosomal MTs nucleated at the Golgi has proven to be indispensable for the Golgi organization, post-Golgi trafficking and cell polarity. Here, we summarize the history of this discovery, known molecular prerequisites of MT nucleation at the Golgi and unique functions of Golgi-derived MTs.
Keywords: Golgi, microtubules, CLASP, AKAP450, microtubule-organizing center, cell polarity
Introduction
A large part of vesicular trafficking is accomplished by molecular motors moving along microtubules (MTs), 25nm-thick dynamic cytoskeletal polymers. Intrinsic polarity of MTs allows for directional movement of molecular motors that deliver vesicular cargos to their specific destinations, which is critical for the complex organization of the cytoplasm. Because molecular motors move specifically toward either the plus or minus end of a MT, the origin and destination of MT-dependent transport is defined by the location of MT ends. In cells, MTs are nucleated at MT-organizing centers (MTOCs). It was traditionally thought that, in an interphase cell, the majority of MTs are nucleated at the centrosome-based MTOC, so that MT minus-ends are embedded at the centrosome while the dynamic plus-ends extend radially toward the cell periphery. However, such intrinsic organization of centrosomal MTs is often not optimal for the directional cargo delivery between two cellular locations, such as polarized trafficking. In most cases, interphase MTs are not strictly radial; a few mechanisms responsible for specific MT architecture have been described. A number of such mechanisms account for the remodeling of centrosomal MTs via selective MT stabilization/destabilization or centrosome positioning. Another easy solution for building non-radial MT tracks is the organization of specialized MT arrays that are not associated with the centrosome.
Indeed, complex MT networks integrating non-centrosomal MTs have been described in many types of differentiated cells, especially those characterized by extensive trafficking. These acentrosomal MTs can arise from release/severing and subsequent repositioning of centrosomal MTs (e.g. axonal and dendritic MTs in neurons (reviewed in (Baas 1998), or basoapical vertical MTs in epithelial cells (Mogensen et al. 2000). Alternatively, nucleation of MTs at alternative, non-centrosomal MTOCs has been described (e.g. MTs nucleated at the melanosome membrane in melanocytes (Malikov et al. 2004), nuclear envelope-derived MTs in myotubes (Tassin et al. 1985), and apical membrane-associated MT nucleation in polarized epithelia (Feldman and Priess 2012)). Moreover, recent studies have proven that a significant non-centrosomal MT array can be formed in a wide range of non-differentiated as well as differentiated cell types via MT nucleation at Golgi membranes. Here, we will summarize the existing evidence on the regulation and functions of this MT sub-population.
Discovery of Golgi-derived MTs as a distinct MT sub-population
The first evidence of MT fragments appearing in association with the Golgi apparatus after full depolymerization of MTs and subsequent short recovery in cultured hepatocytes was found in 2001 by the Christian Poüs group (Chabin-Brion et al. 2001). This study also showed that placing isolated Golgi membranes in a tubulin solution resulted in the formation of MTs bound to Golgi vesicles.
This pioneering study opened a new perspective on the organization and regulation of the interphase MT network; however, it was not fully appreciated until recently. One of the reasons was the fact that the evidence was acquired in experimental conditions of full MT depolymerization when the concentration of tubulin dimers in the cytoplasm significantly exceeds that of physiological levels, which might cause non-specific MT nucleation. Registering Golgi-derived MT in untreated cells is a significant challenge because the Golgi is often located in the vicinity of the centrosome, and the MT density in this region precludes clear distinction of which MTOC a single MT comes from. To overcome this difficulty, we have adapted a MT plus end-tracking approach which has been successfully used previously to detect centrosomal MTs in mitotic or interphase cells (Piehl et al. 2004). Sites of MT nucleation can be detected by tracking polymerizing MTs back to their sites of origin in retinal pigment epithelial (RPE) cells co-expressing the fluorescently tagged MT +TIP EB3 and a Golgi marker (see (Zhu and Kaverina 2011) for protocol). This approach readily detects Golgi-associated MT nucleation in addition to the radial centrosomal MT array in untreated interphase cells exhibiting steady state MT dynamics (Efimov et al. 2007).
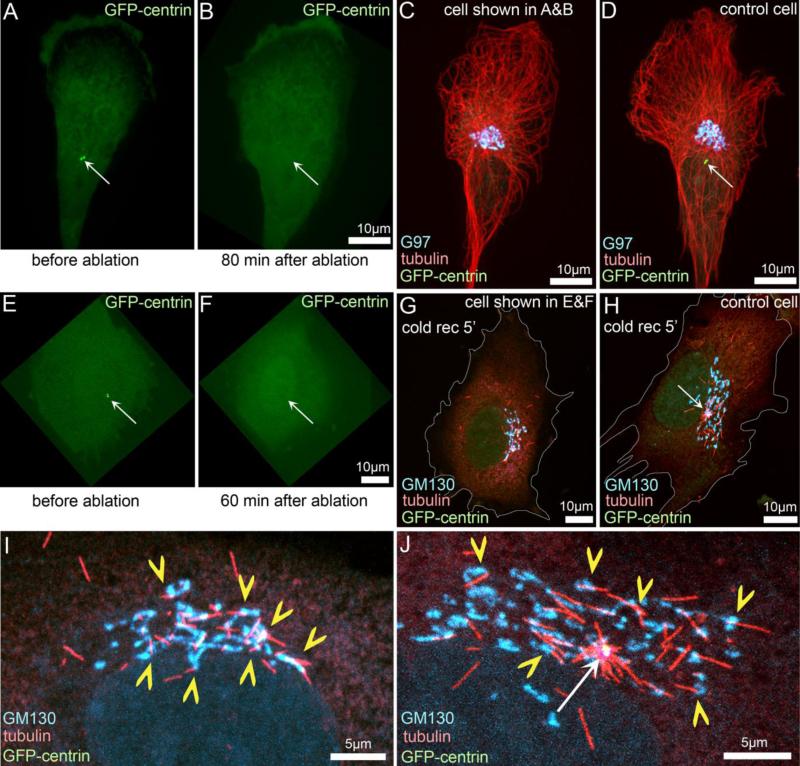
Another concern at the time came from a hypothesis that MTs can be released from the centrosome and their minus end could subsequently be captured by the Golgi membrane (Rios et al. 2004). Functionally, such MTs would still fulfill the requirements of non-centrosomal MTs; however, these MTs would not be true Golgi-derived MTs - and would be regulated by centrosomal release machinery rather than through a Golgi-associated MT nucleation mechanism. This concern arose because the evidence of MT nucleation at the Golgi was first obtained in fixed cells at distinct time points after nocodazole washout. Since then, our live cell imaging assays following MT regrowth after nocodazole washout directly visualized formation of MTs at Golgi stacks distant from the centrosome (Efimov et al. 2007). Moreover, the number of MTs formed at the Golgi in cells where the centrosome has been ablated by laser microsurgery does not change, both in steady state (Efimov et al. 2007) and in MT re-growth assays (Fig. 1). Furthermore, CENP-F−/− mouse embryonic fibroblasts cells nucleate MTs only from the Golgi but not from the centrosome (Moynihan et al. 2009), confirming that the centrosome is indeed dispensable for formation of Golgi-derived MTs.
Figure 1. Golgi-derived MTs are formed independently of the centrosome.
The centrosome was laser-ablated in immortalized human pigment epithelial cells (hTert-RPE1) that stably express centrin1-GFP (Uetake et al. 2007; Magidson et al. 2007). GFP signal is shown in green in all panels. A, E: Cells prior to ablation with the intact centrosomes (arrows). B, C: Success of the ablation was confirmed by collecting an image stack at a significant time after the operation. Arrows in B and F indicate the lack of centrosomes at the location indicated in A and E, respectively. C: The cell shown in A and B was fixed and immunostained for tubulin (red) and the Golgi marker G97 (cyan). D: A bystander cell in the same preparation as C with an intact centrosome (arrow). G: The cell shown in E and F was incubated on ice for 40 minutes resulting in complete MT depolymerization (Vinogradova et al. 2012), allowed to recover at room temperature for 5 minutes to initiate MT re-growth, and then fixed and immunostained for tubulin (red) and the Golgi marker GM130 (cyan). H: A bystander cell in the same preparation as G with an intact centrosome (arrow). White outlines show cell borders in G and H. I, J: The central regions of the cells shown in G, H, respectively. Yellow chevrons indicate Golgi-derived MTs. White arrow indicates the centrosome.
Thus, it is clear today that discovery of Golgi-derived MTs has dramatically changed our understanding of MT network organization in interphase cells. An emerging new field of research addresses the specific regulation and functions of this MT sub-population.
Molecular machinery underlying MT nucleation at the Golgi
Because Golgi-derived MTs are nucleated directly at the Golgi membrane, MT nucleation machinery must exist at this location. Not surprisingly, siRNA depletion of the major MT nucleating factor, γ-tubulin, results in elimination of Golgi-derived MTs along with centrosomal MTs (Efimov et al. 2007). Moreover, MT are organized at the Golgi membrane in vitro only when γ-tubulin is present in isolated Golgi preparations (Chabin-Brion et al. 2001; Ori-McKenney et al. 2012). A search for specific factors required for formation of MTs at the Golgi but not at the centrosome yielded two important results. First, we determined that MT-stabilizing proteins CLASPs, which are localized at the Golgi, are essential for Golgi-derived MT formation (Efimov et al. 2007). Second, a large multi-functional protein AKAP-9 (AKAP350, AKAP450, CG-NAP; will be referred to herein as “AKAP”) was found necessary for this phenomenon (Rivero et al. 2009). The specific mechanisms whereby these proteins facilitate MT nucleation are not completely elucidated.
It has been proposed that the role of AKAP in this process is in the recruitment of γ-TuRC to the Golgi membrane (Rivero et al. 2009). However, the direct proof for this appealing hypothesis remains to be elucidated. Though the N-terminal domain of AKAP has been described to bind γ-TuRC components GCP2 and GCP3 (Takahashi et al. 2002), this binding has not been confirmed despite careful evaluation (Hurtado et al. 2011); moreover, the same domain was identified as a membrane-binding domain of AKAP (Hurtado et al. 2011). γ-tubulin and GCP2 are found co-immunoprecipitated from cell lisates with endogenous AKAP (Rivero et al. 2009) within a protein complex, which also includes GM130 and PTTG1/securin (Moreno-Mateos et al. 2011). However, it is unclear whether AKAP or another protein within this complex is responsible for γ-TuRC binding. Clear co-localization of PTTG1/securin, AKAP and γ-TuRC at the centrosome interfere with the interpretation of these data even more. Besides, while γ-tubulin can be detected in vitro on vesiculated Golgi membranes (Chabin-Brion et al. 2001; Ori-McKenney et al. 2012), it is not enriched at the Golgi in cells as compared to the surrounding cytoplasm. However, taking into account the high content of γ-tubulin in the cytosol of interphase cells (>80%(Moudjou et al. 1996; Khodjakov and Rieder 1999)), enrichment of γ-TuRC at the Golgi may not be necessary for MT nucleation. If the nucleation success rate at this particular location is high it is possible that association of a low number of γ-tubulin molecules with the Golgi membrane is sufficient to maintain the MTOC activity at the Golgi. We have previously proposed a model which includes formation of unstable MT seeds at the γ-TuRCs in the cytoplasm regardless of their location, and stabilization of selected seeds precisely at the Golgi membrane; such stabilization is likely performed by CLASPs, which coat short MTs emerging from the Golgi (Efimov et al. 2007). Thus, whether the function of AKAP in MT formation at the Golgi involves recruitment of γ-TuRCs to the membrane is still an open question. However, a few other known properties of this large scaffolding protein can potentially play a role is this activity. Interestingly, AKAP interacts with p150Glued (Hurtado et al. 2011; Kim et al. 2007). Blocking dynein/dynactin was shown to interfere with acentrosomal MT nucleation at melanosomes (Malikov et al. 2004) and the Golgi (Rivero et al. 2009); thus AKAP might support MT nucleation and/or stabilization via a dynein/dynactin-dependent mechanism. Finally, AKAP also regulates MT dynamics (Larocca et al. 2006) and has direct MT-binding activity (Hurtado et al. 2011), which might support MT formation at the Golgi as an alternative (or additional) mechanism to γ-TuRC recruitment by this protein.
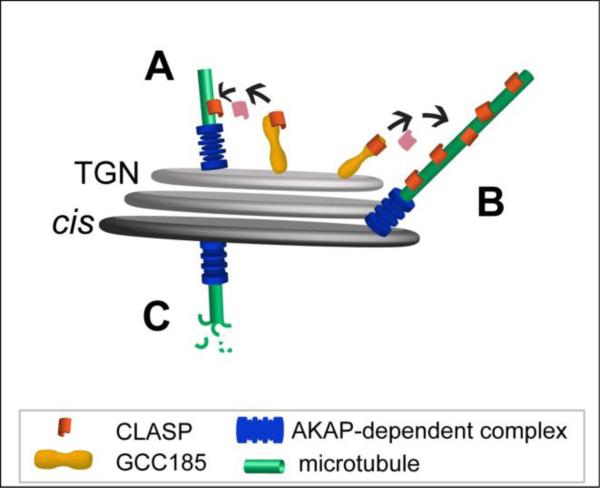
Another puzzling question that arises when a model of MT nucleation at the Golgi is built is the location of the nucleation sites within a Golgi stack. AKAP is a cis-Golgi protein recruited to the membranes by a major cis-Golgi organizer GM130 (Rivero et al. 2009). CLASPs, on the contrary, bind to the TGN membrane via a large coiled-coil golgin GCC185 (Efimov et al. 2007). How can these proteins work in concert to promote MT nucleation? If the role of AKAP is the recruitment of γ-TuRC, MTs must first be nucleated at AKAP-rich cis-Golgi cisternae and then stabilized by CLASPs as they grow (Efimov et al. 2007). This can occur if MT seeds relocate to the TGN from the cis-Golgi and stabilized there by CLASPs ((Stephens 2012), Fig. 2A). It has also been proposed by others and us (Vinogradova et al. 2009; Sutterlin and Colanzi 2010) that CLASPs might stabilize only those MTs which extend toward the TGN from their nucleation sites at cis-Golgi (Fig. 2B), while all other MT seeds are unstable and depolymerize (Fig. 2C). Such selective MT outgrowth is consistent with our finding that the majority of Golgi-derived MTs are directed toward the front of motile cells (Efimov et al. 2007). Due to steric constrains, the extension of emerging MTs toward the TGN must occur at the sides of a Golgi stack (Fig. 2B). Of note, if AKAP facilitates formation of Golgi-derived MTs by an alternative mechanism that does not involve γ-TuRC recruitment, it is still likely that MTs are nucleated at the edges of Golgi stacks where dilated edges of opposite cisternae could come into contact, so that essential molecules could work in concert.
Figure 2. Model of potential MT nucleation sites at a Golgi stack.
A, MT seeds nucleated at the cis-Golgi by an AKAP-dependent protein complex (possibly including PTTG1/securin and γ-TuRC) might relocate to the TGN where they are stabilized by CLASPs and elongate. B, Molecules located at the sides of both cis-Golgi cisternae and the TGN act in concert for nucleation and stabilization of MTs. C, MTs that are nucleated by an AKAP-dependent complex at the cis-Golgi regions distant from the TGN are unstable in the absence of CLASP-stabilization.
Because our knowledge of the molecular machinery that drives MT nucleation at the Golgi is insufficient, signaling pathways that regulate this process remain largely unclear. It is possible that a small GTPase Arl4a is involved in this regulation because it modulates recruitment of CLASPs to GCC185 at the TGN membrane (Lin et al. 2011). Also, MT formation at the Golgi is tightly regulated by the cell cycle signaling: extensive nucleation throughout interphase and prophase is sharply down-regulated when a cell enters prometaphase, and starts to increase again in telophase (Maia et al. 2013). Interestingly, Cdk1-dependent phosphorylation of PTTG1/securin reduces binding of this protein to the Golgi membrane and delays MT regrowth after nocodazole washout (Moreno-Mateos et al. 2011). It is possible that this mechanism contributes to down-regulation of Golgi-derived MT nucleation at the onset of mitosis. Another contributing factor might involve mitotic phosphorylation of CLASP2 (Maia et al. 2012). Overall, the observed cell cycle-dependent modulation of MT nucleation at the Golgi suggests that this MT subpopulation likely performs specific functions as a component of the interphase/prophase MT network rather than the mitotic spindle.
Functions of Golgi-derived MTs
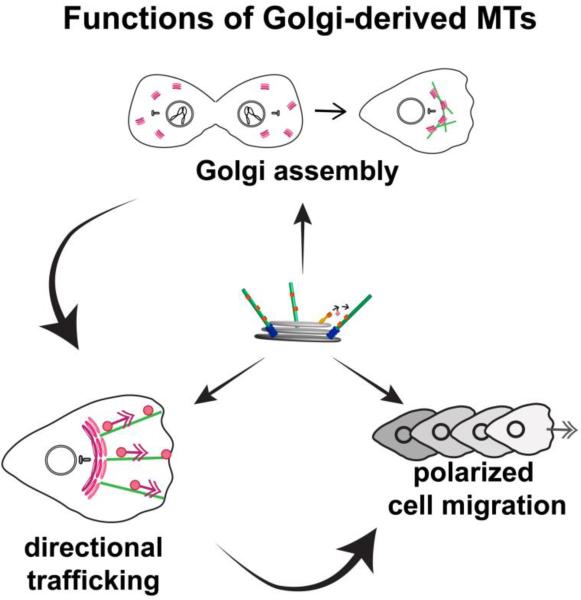
It is well known that Golgi organization is supported by MTs and MT-dependent molecular motors, which are responsible for transport of vesicular carriers involved in the Golgi trafficking and of Golgi stacks themselves (reviewed in (Brownhill et al. 2009; Stephens 2012; Watson and Stephens 2005). However, the functional distinction between MT sub-populations of distinct origins has been addressed only in a few recent studies. It is logical to suggest Golgi-derived MTs serve as preferred tracks for MT-dependent transport of the Golgi membranes because their nucleation occurs at Golgi membranes, and MT minus-ends remain connected with their sites of origin (Efimov et al. 2007). Indeed, existing data indicate that a number of specific MT functions in Golgi organization and/or Golgi trafficking can be accomplished only by Golgi-derived MTs (Fig. 3).
Figure 3. Functions of Golgi-derived MTs and their interdependence.
Golgi-derived MTs drive the correct assembly of polarized Golgi complex at the cell center, which is essential for directional post-Golgi trafficking toward the front of a motile cell; this is, in turn, critical for cell polarity and polarized cell migration.
Golgi assembly
One important function is that Golgi-derived MTs are indispensable for correct post-mitotic assembly of the Golgi complex. Every time a cell enters mitosis, the Golgi disassembles to assure equal inheritance of the Golgi membrane into the two daughter cells (reviewed in (Shorter and Warren 2002; Wei and Seemann 2009)). Accordingly, every mitotic exit requires rapid, coherent re-establishment of the integral polarized Golgi complex assembled anew from the vesiculated membranes. Golgi assembly occurs through several stages. First, scattered mini-stacks of Golgi cisternae are formed throughout the cell by membrane fusion and stacking (Meyer 2005; Shorter and Warren 2002; Tang et al. 2008). Then, a single Golgi complex is assembled by MT-dependent transport driven by dynein (Corthesy-Theulaz et al. 1992). Such minus end-directed transport along Golgi-derived MTs moves the Golgi stacks toward each other. Finally, the resulting Golgi stack clusters are collected toward the cell center by dynein moving along the radial centrosomal MTs (Miller et al. 2009). In the absence of Golgi-derived MTs, the Golgi complex accumulates in the pericentrosomal region but remains fragmented suggesting that centrosomal MTs are unable to support Golgi stack fusion (Miller et al. 2009; Vinogradova et al. 2012). Moreover, rates of Golgi assembly using in silico modeling only fit the experimentally determined rates under the assumption that Golgi-derived MTs support Golgi stack fusion, while centrosomal MTs do not (Vinogradova et al. 2012). This indicates that Golgi-derived MTs are necessary and sufficient for Golgi stack fusion.
Trafficking
Observation of cells, in which the centrosome has been ablated by laser microsurgery, indicate that the Golgi-derived MT subset alone is sufficient to maintain Golgi integrity and shape in acentrosomal cells over a significant period of time (Vinogradova et al. 2012). Thus, Golgi-derived MTs are capable of supporting all MT-dependent processes necessary for Golgi homeostasis, which requires balanced membrane exchange with the ER, exocytic and endocytic membrane pools (Szul and Sztul 2011; Burd 2011; Anitei et al. 2010). It is well known that MTs and MT-dependent molecular motors play a significant role in transportation of vesicular carriers to and from the Golgi (Stephens 2012; Brownhill et al. 2009), and it is possible that Golgi-derived MTs provide specialized MT tracks for this transportation. It is interesting in this regard that in motile cells, Golgi-derived MTs extend asymmetrically from the centrally located Golgi complex toward the cell front thereby serving as tracks for directional post-Golgi trafficking (Miller et al. 2009; Vinogradova et al. 2012).
Cell polarity and architecture
MTs are known to determine cell polarity in a variety of contexts. Not surprisingly, the asymmetric Golgi-derived MT array has been found to be essential for polarized cell organization in diverse cell systems. Several lines of evidence indicate that motile cells lacking Golgi-derived MTs are incapable of polarized migration (Rivero et al. 2009; Miller et al. 2009; Drabek et al. 2006). The likely reason is that a fragmented disorganized Golgi, in combination with the lack of directional MT tracks, is unable to provide directional trafficking of components toward the cell front (Fig. 3). This is in agreement with accumulating evidence that motile cells require a polarized Golgi complex in proximity to the centrosome for directional post-Golgi trafficking and directional cell migration (Yadav et al. 2009; Prigozhina and Waterman-Storer 2004).
Additional exciting evidence for a specialized function of Golgi-derived MTs was recently found in sensory neurons in Drosophila, a cell type where polarized trafficking is a key determinant of proper cellular function (Ori-McKenney et al. 2012). These cells are characterized by highly branched dendrites where MTs equally extend to each process. The total number of MTs at all of the distal dendritic branches is significantly higher than the number of MTs at the base of a dendrite. Ori-McKenny et al found that small Golgi outposts located at every dendritic branchpoint serve as organizing centers for MTs extending into the processes, and that without MT nucleation at the Golgi (that is, in AKAP mutants), dendrite branching is severely disturbed. Strikingly, MTs always emanated from the Golgi outposts in the anterograde direction, establishing directional MT arrays within the distal branches. This observation indicates specific polarity in the MT nucleation machinery at neural Golgi outposts, similar to that in motile cells. In both scenarios, asymmetric Golgi-derived MTs facilitate polarized cell morphology, either by promoting cell protrusion, or by the extension of dendrites. It is possible that the role of Golgi-derived MTs in cell polarity is abundant in other cell types yet unexamined in this regard. For example, depletion of either AKAP or CLASPs in hepatocytes strongly diminishes structural and biochemical polarization of these cells during bile canaliculi morphogenesis (Mattaloni et al. 2012).
Overall, all these studies indicate that Golgi-derived MT subset has distinct functions and cannot be substituted by the centrosomal array. It is possible that the unique capacity of Golgi-derived MTs to support Golgi membrane trafficking and organization relies on the distinct biochemical properties of this MT subset. Such biochemical specificity may arise from enrichment of CLASPs (Efimov et al. 2007) or another yet unknown MT-binding protein at the MT lattice or from preferential accumulation of acetylated tubulin reported for Golgi-derived MTs (Chabin-Brion et al. 2001).
The majority of Golgi-derived MT functions discussed above have been already demonstrated by multiple approaches. Similar phenotypes found in cells where either AKAP or CLASPs were depleted or mis-placed from the Golgi, indicate that Golgi-derived MTs themselves, and not one of these molecular players, are required for such functions. Moreover, it is very likely that the role of this MT subset is not restricted to these already identified functions. For example, the abundance of Golgi derived MTs in prophase (Maia et al. 2013) suggests a yet unknown contribution to the MT network in cells preparing for mitosis.
Acknowledgements
We thank Drs. V. Magidson and A. Khodjakov (Wadsworth Center, Albany, NY) for the permission to use images obtained from our collaboration and presented in Fig. 1. This work was supported by National Institutes of Health grant R01-GM078373 (to I.K.).
Bibliography
- Anitei M, Wassmer T, Stange C, Hoflack B. Bidirectional transport between the trans-Golgi network and the endosomal system. Mol Membr Biol. 2010;27:443–456. doi: 10.3109/09687688.2010.522601. [DOI] [PubMed] [Google Scholar]
- Baas PW. The role of motor proteins in establishing the microtubule arrays of axons and dendrites. J Chem Neuroanat. 1998;14:175–180. doi: 10.1016/s0891-0618(98)00012-x. [DOI] [PubMed] [Google Scholar]
- Brownhill K, Wood L, Allan V. Molecular motors and the Golgi complex: staying put and moving through. Semin Cell Dev Biol. 2009;20:784–792. doi: 10.1016/j.semcdb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Burd CG. Physiology and pathology of endosome-to-Golgi retrograde sorting. Traffic. 2011;12:948–955. doi: 10.1111/j.1600-0854.2011.01188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, Ten Hagen T, Smits R, Fodde R, Grosveld F, Galjart N. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR, 3rd, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Priess JR. A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr Biol. 2012;22:575–582. doi: 10.1016/j.cub.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L, Caballero C, Gavilan MP, Cardenas J, Bornens M, Rios RM. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J Cell Biol. 2011;193:917–933. doi: 10.1083/jcb.201011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Takahashi M, Matsuo K, Ono Y. Recruitment of CG-NAP to the Golgi apparatus through interaction with dynein-dynactin complex. Genes Cells. 2007;12:421–434. doi: 10.1111/j.1365-2443.2007.01055.x. [DOI] [PubMed] [Google Scholar]
- Larocca MC, Jin M, Goldenring JR. AKAP350 modulates microtubule dynamics. Eur J Cell Biol. 2006;85:611–619. doi: 10.1016/j.ejcb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Lin YC, Chiang TC, Liu YT, Tsai YT, Jang LT, Lee FJ. ARL4A acts with GCC185 to modulate Golgi complex organization. J Cell Sci. 2011;124:4014–4026. doi: 10.1242/jcs.086892. [DOI] [PubMed] [Google Scholar]
- Magidson V, Loncarek J, Hergert P, Rieder CL, Khodjakov A. Laser microsurgery in the GFP era: a cell biologist's perspective. Methods Cell Biol. 2007;82:239–266. doi: 10.1016/S0091-679X(06)82007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia AR, Garcia Z, Kabeche L, Barisic M, Maffini S, Macedo-Ribeiro S, Cheeseman IM, Compton DA, Kaverina I, Maiato H. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J Cell Biol. 2012;199:285–301. doi: 10.1083/jcb.201203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia AR, Zhu X, Miller P, Gu G, Maiato H, Kaverina I. Modulation of Golgi-associated microtubule nucleation throughout the cell cycle. Cytoskeleton (Hoboken) 2013;70:32–43. doi: 10.1002/cm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikov V, Kashina A, Rodionov V. Cytoplasmic dynein nucleates microtubules to organize them into radial arrays in vivo. Mol Biol Cell. 2004;15:2742–2749. doi: 10.1091/mbc.E03-10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaloni SM, Kolobova E, Favre C, Marinelli RA, Goldenring JR, Larocca MC. AKAP350 Is involved in the development of apical “canalicular” structures in hepatic cells HepG2. J Cell Physiol. 2012;227:160–171. doi: 10.1002/jcp.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH. Golgi reassembly after mitosis: the AAA family meets the ubiquitin family. Biochim Biophys Acta. 2005;1744:481–492. [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113(Pt 17):3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Espina AG, Torres B, Gamez del Estal MM, Romero-Franco A, Rios RM, Pintor-Toro JA. PTTG1/securin modulates microtubule nucleation and cell migration. Mol Biol Cell. 2011;22:4302–4311. doi: 10.1091/mbc.E10-10-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109(Pt 4):875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Moynihan KL, Pooley R, Miller PM, Kaverina I, Bader DM. Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. Mol Biol Cell. 2009;20(22):4790–4803. doi: 10.1091/mbc.E09-07-0560. doi:10.1091/mbc.E09-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina NL, Waterman-Storer CM. Protein kinase D-mediated anterograde membrane trafficking is required for fibroblast motility. Curr Biol. 2004;14:88–98. doi: 10.1016/j.cub.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Stephens DJ. Functional coupling of microtubules to membranes - implications for membrane structure and dynamics. J Cell Sci. 2012;125:2795–2804. doi: 10.1242/jcs.097675. [DOI] [PubMed] [Google Scholar]
- Sutterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szul T, Sztul E. COPII and COPI traffic at the ER-Golgi interface. Physiology. 2011;26:348–364. doi: 10.1152/physiol.00017.2011. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y. Centrosomal proteins CGNAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell. 2002;13:3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova T, Miller PM, Kaverina I. Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle. 2009;8:2168–2174. doi: 10.4161/cc.8.14.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova T, Paul R, Grimaldi AD, Loncarek J, Miller PM, Yampolsky D, Magidson V, Khodjakov A, Mogilner A, Kaverina I. Concerted effort of centrosomal and Golgi-derived microtubules is required for proper Golgi complex assembly but not for maintenance. Mol Biol Cell. 2012;23(5):820–833. doi: 10.1091/mbc.E11-06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Stephens DJ. ER-to-Golgi transport: form and formation of vesicular and tubular carriers. Biochim Biophys Acta. 2005;1744:304–315. doi: 10.1016/j.bbamcr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Mitotic division of the mammalian Golgi apparatus. Semin Cell Dev Biol. 2009;20:810–816. doi: 10.1016/j.semcdb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Kaverina I. Quantification of asymmetric microtubule nucleation at subcellular structures. Methods Mol Biol. 2011;777:235–244. doi: 10.1007/978-1-61779-252-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]