Abstract
The cell cycle regulatory gene INK4A-ARF (CDKN2A) has two alternative transcripts that produce entirely different proteins, namely p14ARF and p16, which have complementary functions as regulators of p53 and pRB tumor suppressor pathways, respectively. The unusual organization of INK4A-ARF has long led to speculation of a need for coordinated regulation of p14ARF and p16. We now show that p14ARF (ARF) regulates the stability of p16 protein in human cancer cell lines, as well as in mouse embryonic fibroblasts (MEFs). In particular, ARF promotes rapid degradation of p16 protein, which is mediated by the proteasome and, more specifically, by interaction of ARF with one of its subunits, REGγ. Furthermore, this ARF-dependent destabilization of p16 can be abrogated by knock-down of REGγ or by pharmacological blockade of its nuclear export. Thus our findings have uncovered a novel crosstalk of two key tumor suppressors mediated by a REGγ-dependent mechanism.
Keywords: p14ARF, p19Arf, p16, REGγ, proteasome, nuclear export
Introduction
Of four INK4 genes that encode inhibitors of Cyclin D-dependent protein kinases, INK4A-ARF (CDKN2A in humans) is most frequently deregulated in human cancer (1, 2). INK4A-ARF expresses two overlapping transcripts that encode two distinct proteins, namely p14ARF (hereafter referred as ARF) and p16, which share no sequence homology (3, 4), but nonetheless have complementary functions as regulators of two major cell cycle control pathways, namely p53 and RB, respectively (4–6). Notably, p16, as well as p53 and RB, have a greater degree of evolutionary conservation in vertebrates than ARF, which evolved much later during amniote development (7). The organization of the INK4A-ARF locus with its two highly similar transcripts yielding unrelated proteins has led to the speculation that the organization of the locus reflects a need for the coordinated regulation of ARF and p16 (7).
Although ARF and p16 have no sequence similarity they share the unusual feature of having no (or, in the case of mouse Arf, only one) lysine residues (3, 4), which impacts their overall structure as well as their ability to undergo cellular degradation. Furthermore, while ARF and p16 govern complementary regulatory pathways and both function as regulators of aging, cellular senescence and tumorigenesis (6, 8), their functions are complex as they are sometimes overlapping (e.g., (9)) and in other contexts they are opposing (e.g., (10)). Moreover, the functions of ARF are inherently complex; although its primary role is to regulate p53 by interfering with its negative regulator MDM2, ARF also has activities that are not dependent on p53, particularly its ability to promote protein SUMOylation of its various binding partners (11–14).
Thus, the INK4A-ARF locus is characterized by the unusual organization of its transcripts, the unusual sequences of its encoded proteins, and the complex functions of its protein products. In the present study, we sought to further understand their relationship by investigating the status of ARF and p16 proteins in human cancer. We find an unexpected inverse relationship of ARF and p16 protein levels, which reflects the regulation of p16 protein stability by ARF.
Methods summary
The bladder cancer and prostate cancer tissue microarrays (TMAs) used in this study are described in Supplementary Table S1. Human cancer cell lines were obtained from American Type Culture Collection (ATCC) and their authenticity was verified by ATTC; mouse embryonic fibroblasts (MEFs) were made from 13.5 dpc mutant mouse embryos from the indicated genotypes. Exogenous gene expression or introduction of siRNA were introduced via retroviral gene transfer or transient transfection, respectively; sequences of siRNA are provided in Supplementary Table S2. A summary of antibodies used in this study is provided in Supplementary Table S3. Quantitative analyses of protein levels were done using ImageJ software and half-lives were estimated by drawing approximate reduction curves. Full details of material and methods are provided in Supplementary Information.
Results and Discussion
ARF regulates p16 protein levels in human cancer
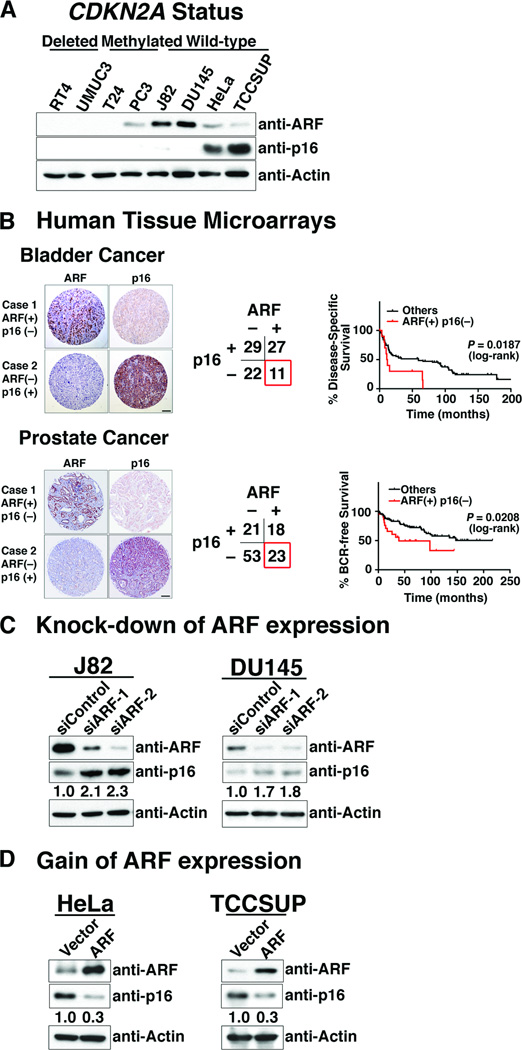
In many human cancers, CDKN2A is either deleted or methylated; however, in cases when CDKN2A is intact the corresponding protein products are often expressed at elevated levels (12). We examined a panel of representative human cancer cell lines, in which CDKN2A was alternatively homozygously deleted (RT4 and UMUC3), epigenetically silenced (T24 and PC3) or intact (J82, DU145, HeLa, TCCSUP) (Fig. 1A). We found that cells lines having intact CDKN2A (i.e., neither deleted nor silenced) had either high levels of ARF protein expression (J82 and DU145) or high levels of p16 protein expression (HeLa and TCCSUP), but not both (Fig. 1A).
Figure 1.
ARF regulates p16 protein levels in human cancer cells. (A) Inverse expression of ARF and p16 in human cancer cells. Western blot analyses of showing the expression levels of ARF and p16 proteins in the indicated human cancer cell lines, in which the CDNK2A gene is either deleted, methylated, or intact, as indicated. (B) Association of ARF and p16 expression with clinical outcome in bladder and prostate cancer. Representative images and categorical results of ARF and p16 immunostaining of tissue microarrays of human bladder and prostate cancer. Kaplan-Meier analyses show disease-specific survival of bladder cancer patients, and biochemical relapse (BCR)-free survival of prostate cancer patients. (C) Consequences of ARF knock-down for expression of p16 protein in J82 and DU145 cells using two independent ARF siRNA (or a scrambled siRNA as a control). (D) Consequences of expressing exogenous ARF in HeLa and TCCSUP cells following transfection with an ARF cDNA (or the empty vector as a control). In C and D the relative expression levels of p16 are indicated as determined using ImageJ software.
To assess the potential clinical relevance of these observations, we evaluated the expression of ARF and p16 on human cancer tissue microarrays. We used two representative tissue microarrays, one comprised of invasive bladder tumors (n = 89) and another of prostate tumors (n = 128) (Supplementary Table S1). Considering the prevalence of CDKN2A loss in human cancer (1, 2), many of these primary tumors express neither ARF nor p16 (bladder = 22/89 and prostate = 53/128) (Fig. 1B), while some express both ARF and p16 (bladder = 27/89 and prostate = 18/128) (Fig. 1B), and are therefore presumably unaffected at this locus. Notably, however, a subset of tumors express ARF but not p16 (bladder = 11/89 and prostate = 23/128) and, conversely, p16 but not ARF (bladder = 29/89 and prostate = 21/128) (Fig. 1B). Furthermore, as evident by Kaplan Meier analyses the ARF(+)/p16(−) sub-group had a significantly worse outcome compared with the population as a whole in both the bladder and prostate cancer cohorts (log rank p-value = 0.0187 and 0.0208, respectively) (Fig. 1B). These findings suggest that tumors with elevated ARF expression but low p16 expression may be associated with poorer outcome.
We next asked whether the inverse correlation of p16 and ARF protein expression in human cancer cells might reflect their reciprocal regulation. In the cell lines tested, we found that the expression levels of ARF affected those of p16 protein expression, but not the reverse. Specifically, knock-down of ARF in cells that normally expressed ARF (i.e., J82 and DU145) resulted in increased levels of p16 protein (Fig. 1C) and, conversely, forced expression of ARF in cells that normally have low levels of ARF (i.e., HeLa and TCCSUP) resulted in reduced levels of p16 protein (Fig. 1D); however, in neither case did manipulating ARF expression affect p16 mRNA levels (Supplementary Fig. S1). On the other hand, reciprocal experiments in which p16 expression levels were manipulated either by its knock-down in cells that normally express p16 (i.e., HeLa and TCCSUP) or by its forced expression in cells that normally do not express p16 (i.e., J82 and DU145) had virtually no effect on the expression of ARF protein or mRNA (Supplementary Fig. S2, and data not shown). Taken together, these findings suggest that ARF is a posttranscriptional regulator of p16.
Arf regulates the stability of p16 protein via REGγ-dependent proteasome degradation
To further evaluate the consequences of Arf expression for p16 protein levels, as well as to study the underlying mechanism(s), we used mouse embryonic fibroblasts (MEFs), which have been widely used to evaluate Arf expression and function (5, 11). Consistent with previous reports, Arf protein levels are relatively low in early passage wild-type MEFs (Arf+/+; p53+/+; Pten+/+); nonetheless, deletion of Arf in otherwise wild-type MEFs (Arff/f; p53+/+; Pten+/+) resulted in increased levels of p16 protein (Supplementary Fig. S3A, Lanes 1, 2). Furthermore, MEFs lacking p53 and Pten (Arf+/+; p53f/f; Ptenf/f) express robust levels of Arf protein but very low levels of p16 (Supplementary Fig. S3A, Lane 7). Deletion of Arf in this context (Arff/f; p53f/f; Ptenf/f) resulted in high levels of p16 protein (Supplementary Fig. S3A, Lane 8). Importantly, cell cycle analyses revealed that the Arf-null (Arff/f; p53f/f; Ptenf/f) MEFs, which have elevated p16 protein levels, were increased in G1 phase compared to the Arf-positive (Arf+/+; p53f/f; Ptenf/f) MEFs (56.2% versus 36.9%), indicating that p16 is functionally active in these Arf-null MEFs (Supplementary Fig. S3B). Therefore these Arf-positive and Arf-null MEFs provide a model for studying the consequences of Arf for expression of p16.
Indeed, as we had observed in the human cancer cells (see Fig. 1C, D), knockdown of Arf in the Arf-positive MEFs resulted in increased levels of p16 protein while, conversely, forced expression of Arf in the Arf-null MEFs resulted in reduced levels of p16 protein (Supplementary Fig. S3C); in neither case, did manipulation of Arf expression affect p16 mRNA levels (Supplementary Fig. S3D). Consistent with the apparent post-transcriptional consequences of Arf for p16 protein expression, we found that Arf status was well-correlated with p16 protein stability. Specifically, following treatment with the protein synthesis inhibitor, cycloheximide, p16 protein was significantly less stable in the Arf-positive MEFs as compared to the Arf-null MEFs (T1/2 = 2.4 versus 8.6 hours, respectively) (Fig. 2A). Furthermore, reduced stability of p16 in Arf-positive MEFs could be overcome by inclusion of bortezomib, a proteasome inhibitor, while bortezomib had no effect on p16 in the Arf-null MEFs (Fig. 2B). These findings suggest that Arf regulates p16 protein stability in a proteasome-dependent manner.
Figure 2.
Arf regulates the stability of p16 protein via REGγ-dependent proteasome degradation in mouse embryonic fibroblasts. (A) Arf regulates p16 protein stability. Arf(+) and Arf(−) MEFs were treated with cycloheximide (50 µg/ml) for indicated time in hours. (Left) Western blot analyses showing relative protein expression levels. (Right) Relative change in p16 expression as a function of time showing the half-life (T1/2) was calculated from approximation curves. Note that in all approximation curves shown, the change in p16 expression is presented relative to the normalized expression levels (so it takes into account the change in basal levels in the cells). (B) Arf-mediated destablilization of p16 protein is counteracted by proteasome inhibitor. Arf(+) and Arf(−) MEFs were untreated or treated with cycloheximide (50 µg/ml) in the presence or absence of bortezomib (5 µM) and analyzed by Western blot analyses. (C) Arf interacts with REGγ in MEFs. (Left) Co-immunoprecipitation of endogenous Arf with endogenous REGγ using an anti-Arf antibody. (Right) Co-immunoprecipitation of exogenous HA-tagged REGγ with endogenous Arf using an anti-HA antibody. (D) REGγ is required for Arf-mediated destablization of p16 protein levels. Arf(+) MEFs were treated with treated two independent REGγ siRNA (or a scrambled siRNA as a control) followed by cycloheximide (50 µg/ml) for indicated time in hours. (Left) Western blot analyses showing relative protein expression levels. (Right) Relative change in p16 expression as a function of time showing the half-life (T1/2) was calculated from approximation curves. In A, B, and D the relative expression levels of p16 are indicated as determined using ImageJ software.
Since p16 does not have lysine residues, it is not subject to ubiquitin-mediated degradation; instead, it may be targeted for degradation by ubiquitin-independent components of the proteasome and particularly by REGγ (also known as PSME3 or PA28γ), which is an ubiquitin-independent proteasome activator (15). We, therefore, asked whether REGγ contributes to the Arf-dependent destabilization of p16 protein. Indeed, we found that Arf interacts with endogenous as well as exogenous REGγ as evident by co-immunoprecipitation analyses (Fig. 2C). Furthermore, knock-down of REGγ resulted in increased p16 protein levels in the Arf-positive MEFs but did not further increase p16 protein levels in the Arf-null MEFs (Supplementary Fig. S4A). Additionally, knock-down of REGγ abrogated the Arf-dependent destabilization of p16 protein in Arf-positive MEFs (T1/2 = 9.2 hours with siREGγ versus 2.3 hours with siControl) (Fig. 2D). Taken together, these findings indicate that the Arf-dependent destabilization of p16 protein is mediated, at least in part, by the interaction of Arf with the proteasome subunit, REGγ.
Arf-mediated destablization of p16 protein is associated with nuclear export of REGγ
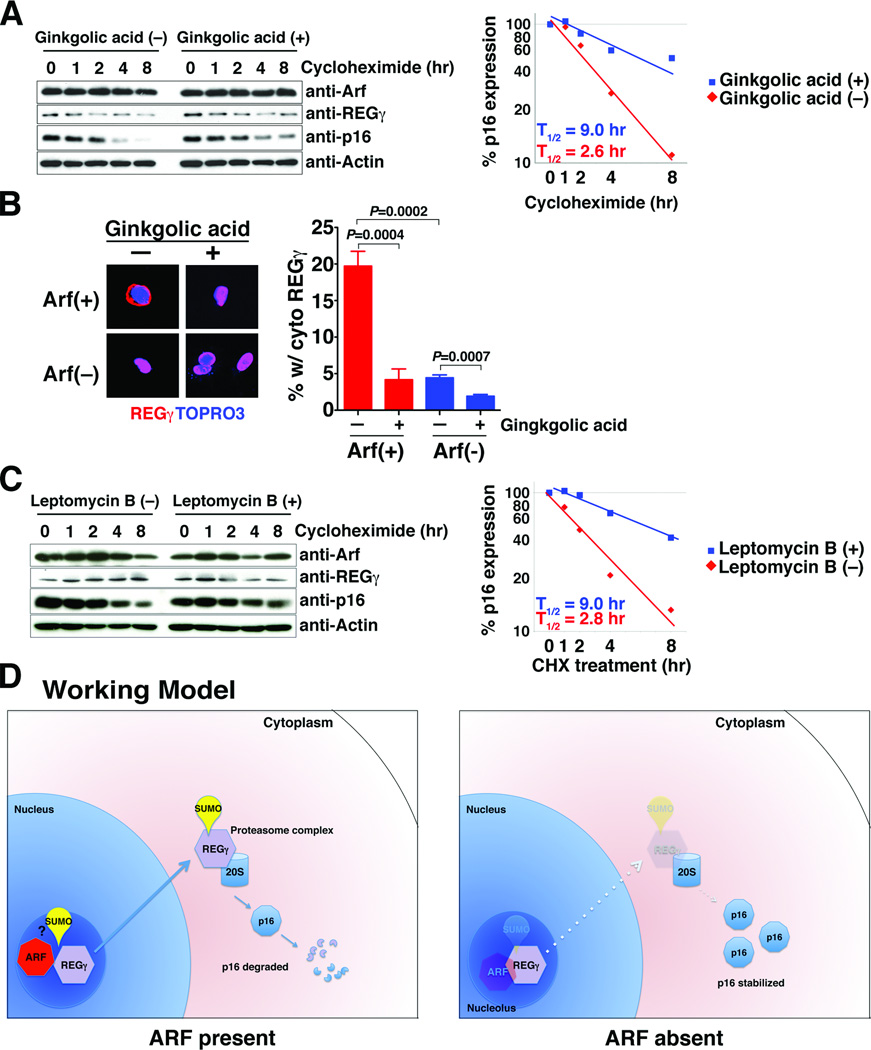
One of the main functions of Arf, and particularly one of its major p53-independent functions, is to promote SUMOylation of targets to which it is bound (12–14). Since REGγ is itself known to be SUMOylated (16), we asked whether SUMOylation contributes to the Arf-dependent REGγ-mediated regulation of p16 protein stability. We found that treatment of cells with a small molecular inhibitor of SUMOylation, namely ginkgolic acid (17), abrogated the rapid degradation of p16 in Arf-positive cells in a dose-dependent manner (Supplementary Fig. S4B) and resulted in a prolonged half-life of p16, similar to that observed following knock-down of REGγ (T1/2 = 9.0 versus 2.6 hours) (Fig. 3A).
Figure 3.
Arf-mediated destablization of p16 protein is mediated by nuclear export of REGγ. (A) Inhibition of SUMOylation stabilizes p16 expression in Arf-positive MEFs. Arf(+) MEFs were treated or untreated with ginkgolic acid (5 µM) for 4 hr followed by treatment with cycloheximide (50 µg/ml) for the indicated time in hours. (Left) Western blot analyses showing relative protein expression levels. (Right) Relative change in p16 expression as a function of time showing the half-life (T1/2) calculated from approximation curves. (B) Inhibition of SUMOylation reduces cytoplasmic localization of REGγ. Arf(+) and Arf(−) MEFs were transfected with an expression plasmid encoding HA-REGγ and treated with bortezomib (5 µM) and ginkgolic acid (5 µM) for 8 hr. (Left) Immunofluorescence images showing HA-REGγ localization in Arf(+) and Arf(−) MEFs detected using anti-HA antibody or detection of the nuclear marker TOPRO3. (Right) Percentage of cells in each condition having cytoplasmic expression of the REGγ. The chart summarize the results from 3 independent assays, each counting a minimum of 100 cells per variable. (C) Block of nuclear export of REGγ stabilizes p16 expression in Arf-positive MEFs. Arf(+) MEFs were treated or untreated with Leptomycin B (50 ng/ml) for 4 hr followed by treatment with cycloheximide (50 µg/ml) for the indicated time in hours. (Left) Western blot analyses showing relative protein expression levels. (Right) Relative change in p16 expression as a function of time showing the half-life (T1/2) calculated from approximation curves. (D) Working model. Discussed in the text. In A, and C the relative expression levels of p16 are indicated as determined using ImageJ software.
Although REGγ is preferentially localized to the nucleus, the SUMOylated form is located in the cytoplasm (16) and p16 is also located primarily in the cytoplasm. Therefore, we examined the localization of REGγ in Arf-positive versus Arf-null MEFs. We found that REGγ was located in the cytoplasm in a significant percentage (20%) of Arf-positive MEFs, but only 5% of the Arf-null MEFs (p= 0.004, Fig. 3B). However, treatment of Arf-positive MEFs with ginkgolic acid reduced the cytoplasmic REGγ to ~5%, similar to that seen in the Arf-null MEFs (p= 0.0007; Fig. 3B). Furthermore, we looked more directly at whether nuclear export of REGγ might contribute to the Arf-dependent REGγ-mediated regulation of p16 protein stability using a small molecular inhibitor of nuclear export, namely leptomycin B which inhibits CRM1, a protein required for nuclear export of proteins containing a nuclear export sequence (18, 19). We found that leptomycin B resulted in a prolonged half-life of p16 in Arf-expressing cells (T1/2 = 9.0 versus 2.8 hours) but not in Arf-null cells (Fig. 3C and data not shown).
These findings indicate that blocking nuclear export of REGγ inhibits Arf-dependent p16 turnover. Interestingly, REGγ is rapidly degraded in the Arf-positive MEFs while it is highly stable in Arf-null MEFs (T1/2 = 8.7 versus 95 hour, respectively), which was completely abrogated by treatment with ginkgolic acid (T1/2 = 9.4 hours with ginkgolic acid versus 77 hours without ginkgolic acid) (Supplementary Fig. S5), indicating that REGγ is itself degraded in an Arf- and SUMOylation-dependent manner. Taken together, these findings suggest that both SUMOylation and the nuclear export of REGγ are required for the Arf-dependent destabilization of p16.
ARF regulates stability of p16 protein via REGγ-dependent proteasome degradation in human cancer cells
Finally, we asked whether these observations regarding the regulation of p16 protein stability by Arf from analyses of MEFs were also relevant for human cancer cells. Indeed, we found that in J82 human cancer cells, which normally express ARF (see Fig. 1A), the increased expression of p16 protein observed following knock-down of ARF was independent of bortezomib (Supplementary Fig. S6A) and reflective of increased p16 stability (T1/2 = 6.2 hours with siARF versus 2.5 hours with siControl) (Supplementary Fig. S6B). Conversely, in HeLa cancer cells, which normally do not express ARF (see Fig. 1A), we found that the reduced p16 protein levels observed following ARF gain of expression was partially abrogated by bortezomib (Supplementary Fig. S6E), and reflective of reduced p16 stability (T1/2 = 3.5 hours in ARF-expressing cells versus 8.0 hours in vector-expressing cells) (Supplementary Fig. S6F). Furthermore, the ARF-dependent destabilization of p16 observed in these human cancer cells was abrogated either by knock-down of REGγ (Supplementary Fig. S6C, G) or by pharmacological inhibition of SUMOylation (Supplementary Fig. S6D, H). Thus, these findings demonstrate that in human cancer cells ARF regulates p16 protein stability via a REGγ mediated mechanism.
Conclusions
Our findings address a long-standing issue regarding the potential coordinate regulation of the two distinct proteins encoded by the INK4A-ARF gene, namely ARF and p16. Thus, we find that expression of ARF and p16 are often inversely correlated in cancer, wherein tumors having high levels of ARF and low levels of p16 tend to have poorer outcomes. Furthermore, their inverse expression reflects the ability of ARF to regulate p16 protein stability, which is mediated by REGγ, an ubiquitin-independent activator of the proteasome. Notably, REGγ is dysregulated in a variety of cancers, and its targets for degradation include various tumor regulators, such as p53 and p21 (15, 20). Thus, the involvement of REGγ as an ARF-dependent regulator of p16 stability further highlights its significance, as well as that of the proteasome, as a key regulator of growth control. We propose a model (Fig. 3D) in which ARF interacts with REGγ to promote its SUMOylation as well as its nuclear export, which in turn promotes degradation of p16 as well as itself. In the absence of ARF, REGγ is stabilized, either by limiting its SUMOylation or inhibiting its de-SUMOylation (13), and as a consequence p16 is also stabilized.
Finally, our findings showing that ARF regulates p16, but not the reverse, are notable given that ARF evolved significantly later than p16 (6, 7). Thus, we speculate that the unusual organization of the INK4A-ARF genes reflects an additional level of regulatory control that occurred during evolution. We envision ARF as having evolved to provide a fail-safe mechanism to maintain the appropriate levels of expression of the cell cycle regulator, p16. Notably, since this regulatory relationship occurs post-transcriptionally it might have been overlooked if we had focused exclusively on expression profiling. This emphasizes the importance of evaluating key regulators at multiple, independent levels in order to get a complete picture of the mechanisms that control their functions.
Supplementary Material
Acknowledgements
We acknowledge the support of the Herbert Irving Comprehensive Cancer Center Shared Resource in Molecular Pathology for the generation of the prostate cancer tissue microarray. We are grateful to Drs. Richard Baer, Riccardo Dalla-Favera, Wei Gu, Michael Shen, and Charles Sherr for their helpful comments and discussion. We also thank Dr. Sherr for generously providing the Arf mutant mice. This work was supported in part by NIH grants to CAS (CA084294) and funding from the Alexander and Margaret Stewart Trust provided to the Institute for Cancer Genetics. TK was supported by post-doctoral training grants from the American Urological Association Foundation and the American Association for Cancer Research, as well as funding from the, and Uehara Memorial Foundation. CAS is an American Cancer Society Research Professor supported in part by a generous gift from the F.M. Kirby Foundation.
Footnotes
Disclosure: The authors disclose no potential conflicts of interest.
References
- 1.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 2.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 4.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ. The INK4a/ARF network in tumour suppression. Nature reviews Molecular cell biology. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 6.Sherr CJ. Ink4-Arf Locus in Cancer and Aging. Wiley interdisciplinary reviews. Developmental biology. 2012;1:731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nature reviews Molecular cell biology. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 8.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Carnero A, Hudson JD, Price CM, Beach DH. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nature cell biology. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- 10.Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nature cell biology. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ, Bertwistle D, W DENB, Kuo ML, Sugimoto M, Tago K, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harbor symposia on quantitative biology. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 13.Kuo ML, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378–3387. doi: 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- 14.Tago K, Chiocca S, Sherr CJ. Sumoylation induced by the Arf tumor suppressor: a p53-independent function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7689–7694. doi: 10.1073/pnas.0502978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Molecular cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Wang L, Zhou P, Wang G, Zeng Y, Wang Y, et al. Regulation of REGgamma cellular distribution and function by SUMO modification. Cell research. 2011;21:807–816. doi: 10.1038/cr.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chemistry & biology. 2009;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Experimental cell research. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 20.Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond? Cellular and molecular life sciences : CMLS. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.