Abstract
Deficiency in human mitochondrial Complex-1 has been linked to a wide variety of neurological disorders. Homozygous deletion of the Complex-1 associated protein, Ndufaf2, leads to a severe juvenile onset encephalopathy involving degeneration of the substantia nigra and other sub-cortical regions resulting in adolescent lethality. To understand the precise role of Ndufaf2 in Complex-1 function and its links to neurologic disease, we studied the effects on Complex-1 assembly and function, as well as pathological consequences at the cellular level, in multiple in vitro models of Ndufaf2 deficiency. Using both Ndufaf2-deficient human neuroblastoma cells and primary fibroblasts cultured from Ndufaf2 knock-out mice we found that Ndufaf2-deficiency selectively reduces Complex-1 activity. While Ndufaf2 is traditionally referred to as an assembly factor of Complex-1, surprisingly, however, Ndufaf2-deficient cells were able to assemble a fully mature Complex-1 enzyme, albeit with reduced kinetics. Importantly, no evidence of intermediate or incomplete assembly was observed. Ndufaf2 deficiency resulted in significant increases in oxidative stress and mitochondrial DNA deletion, consistent with contemporary hypotheses regarding the pathophysiology of inherited mutations in Complex-1 disorders. These data suggest that Ndufaf2, unlike other Complex-1 assembly factors, may be more accurately described as a chaperone involved in proper folding during Complex-1 assembly, since it is dispensable for Complex-1 maturation but not its proper function.
Keywords: Mitochondria, Complex-1 assembly, Oxidative stress, Complex-1 disease, Mitochondrial DNA mutations
Introduction
NADH Dehydrogenase, or mitochondrial Complex-1, is a part of the electron transport chain and is a multi-protein enzyme thought to be comprised of about 45 subunits (Koopman et al., 2010). It is also one of the largest and most intricate protein complexes of the eukaryotic cell (Janssen et al., 2006). As the first enzyme in oxidative phosphorylation, Complex-1 catalyzes the abstraction of two electrons from NADH and transfers them to ubiquinone. At the same time, it pumps four protons across the mitochondrial inner membrane, contributing to the proton gradient used for ATP production (Mitchell, 1961). The L-shaped Complex-1 consists of a membrane bound portion (anchored to the inner mitochondrial membrane) and a large matrix-facing protrusion containing the NADH binding site (Sazanov et al., 2000). The assembly of Complex-1 is still poorly understood, but is thought to follow a stepwise process in which the subunits are first assembled in multiple, smaller independent sub-complexes. The membrane-bound sub-complexes consist predominantly of mitochondrially encoded subunits, whereas the matrix-facing sub-complexes are composed of largely nuclear encoded subunits (Ugalde et al., 2004). To date, an array of 7 assembly factors have been identified which are required to form mature Complex-1, which has a total molecular mass of ~1 MDa (Koopman et al., 2010).
Loss-of-function mutations within genes encoding mitochondrial Complex-1 subunits are among the most common cause of respiratory chain disorders, and can result in a wide variety of clinical phenotypes spanning from early onset fatal phenotypes to adult onset exercise intolerance (Loeffen et al., 2000). The tissues affected in these disorders can manifest a similarly diverse set of pathologies, but frequently involve neurodegeneration, such as in Leigh's syndrome, Leber's hereditary optic neuropathy, MELAS, or MERFF (McFarland et al., 2007). In addition to these primary genetic defects in Complex-1 associated genes, deficits in Complex-1 activity have also been implicated in neurodegenerative diseases such as idiopathic Parkinson's disease, Alzheimer's disease, Huntington's disease and Amyotrophic lateral sclerosis(Schon and Przedborski, 2011).
It has become clear in recent years that Complex-1 deficiency disorders do not arise solely from mutations in the structural or enzymatic subunits, but rather are increasingly linked to mutations in its assembly factors, as well. Ndufaf2 is a Complex-1 assembly factor and five patients with homozygous deletion of Ndufaf2 have been described thus far (Herzer et al., 2010). They developed severe encephalopathy, mostly involving the midbrain and brainstem, leading to premature death during childhood. A heterozygous Ndufaf2 deletion has been observed in a patient with attention-deficit/hyperactivity disorder, suggesting a link between Complex-1 deficits and Ndufaf2 in basal ganglia dysfunction, and perhaps consistent with the midbrain pathology in homozygous Ndufaf2 patients (Lesch et al., 2010). Ndufaf2 is a 24 kDa protein with a functional mitochondrial targeting sequence bearing significant homology to the Complex-1 subunit, Ndufa12 (Ogilvie et al., 2005; Tsuneoka et al., 2005). Functionally, Ndufaf2 has been shown to influence Complex-1 activity and assembly in patient fibroblasts (Hoefs et al., 2009; Janssen et al., 2009; Ogilvie et al., 2005; Vogel et al., 2007). It has been shown to stably associate with a novel ~830 kDa subcomplex of Complex-1 resulting from the deletion of NdufS4, NdufS6 or NdufV1 (Calvaruso et al.; Lazarou et al., 2007; Ogilvie et al., 2005), but not with the mature complex in normal cells. Based on these findings, Ndufaf2 has been proposed by many to be required in the final stages of Complex-1 assembly (Ogilvie et al., 2005; Vogel et al., 2007). Therefore, we examined the assembly and activity of multiple electron transport chain enzymes, and found selective deficits in Complex-1 activity in Ndufaf2-deficient cells. However, Ndufaf2 knockdown or knockout (KO) cells were able to produce a fully mature Complex-1 enzyme, albeit with reduced activity and assembly kinetics. In contrast to the traditional role of Ndufaf2, these data demonstrate that Ndufaf2 is not required for Complex-1 assembly, but nonetheless does play an important role in Complex-1 function.
Inefficient transfer of electrons and resulting oxidative stress are thought to play a pathogenic role in both Complex-1 deficiency diseases (Koopman et al., 2010) and Parkinson's disease (Abou-Sleiman et al., 2006), and the severity of the inherited Complex-1 diseases may correlate better with increases in leaked electrons from the mutant Complex-1 than with decreased ATP production (Verkaart et al., 2007a) (Verkaart et al., 2007b). In fact, our in vitro models of Ndufaf2 deficiency showed significant increases in numerous markers of oxidative stress, consistent with data from patient tissue and other models of Complex-1 dependent pathology (Janssen et al., 2009; Pitkanen and Robinson, 1996).
Material and Methods
Cell lines and stable transduction
Human SK-N-MC neuroblastoma cells (ATTC number: HTB-10™) were cultured in growth media consisting of Dulbecco's Modified Eagle's Media with 10% fetal bovine serum, 100 I.U./ml penicillin, 100 I.U./ml streptomycin and 0.2 mM L-Glutamine. SK-N-MC cells were stably transduced with plasmids containing Ndufaf2 shRNA (Sigma Mission clone IDs NM_174889.2-287, -117, -407, -192, -169) or scrambled shRNA (SHC002). Cells were selected for 48 h with 0.8 μg/ml puromycin, and then cultivated in growth media with 0.04 μg/ml puromycin. The -407 shRNA showed the greatest knockdown efficiency and was used for all experiments, unless noted otherwise. Ndufaf2 KO mice, generated by InGenious Targeting Laboratory, Inc. under our direction, were created using a targeting construct designed to conditionally ablate Ndufaf2 Exon 2 (detailed information on the cloning of the targeting construct, ES cell injections and breeding will be published elsewhere). Resultant F1 mice were crossed with CMV-Cre (Jackson), and bred to homozygosity. Genotyping was performed by a PCR-based strategy utilizing intronic primers that span Exon 2. Deletion of Ndufaf2 was confirmed at the genomic, mRNA and protein level. Mouse skin fibroblasts were isolated as described before (Takashima, 1998), cultured in growth media, and spontaneously immortalized. NdufS4 knockout and WT mouse embryonic fibroblasts were kindly provided by Richard Palmiter.
SDS-PAGE, BN-PAGE and Western Blot
SDS-PAGE
Total cell lysates were prepared in 1% NP-40/phosphate buffered saline (PBS) containing protease inhibitor (Sigma), sonicated, and protein normalized by BCA assay (Pierce). Samples were mixed with 4x Laemmli sample buffer with 20% β-mercaptoethanol, heated at 65°C for 10 minutes and loaded onto Novex (Invitrogen) 4-20% Tris-Glycine pre-cast gels. The proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore), probed with a rabbit polyclonal antibody directed against Ndufaf2 (Ogilvie et al., 2005), NdufS4 (sc-100567; Santa Cruz) or MnSOD (06-984 EMD Millipore) and stripped and reprobed for actin (ab6276; Abcam, Cambridge, MA, USA). Secondary antibodies and ECL-plus were purchased from GE Healthcare. Densitometry was performed using ImageJ.
BN-PAGE of mitochondrial extracts
To enrich mitochondria, cells were homogenized for 10 passes on ice in a glass/Teflon power-driven Potter Elvehjem in isolation buffer (250 mM Sucrose, 2 mM HEPES pH 7.4 at 20°C, EGTA 0.1 mM). Nuclei and debris were removed by a 1000 × g spin 5 min 4°C, and mitochondria were pelleted at 10,000 × g for 10 min at 4°C (Wittig et al., 2006). Crude mitochondrial fractions were resuspended in 75 mM Bis-Tris, 1.5 M aminocaproic acid, pH 7 at 4°C, and a final concentration of 1% DDM, 0.5 or 1% Triton-X-100 or 1% Digitonin was added to extract respiratory complexes for 10 min on ice (McKenzie et al., 2011). Lysates were centrifuged at 21,000 × g for 30 min, supernatants were mixed with 4x BN-sample buffer (Invitrogen). Samples and high molecular weight native standards (GE Healthcare) were loaded on NativePage 3-12% gels (Invitrogen) and run according to manufacturer's instructions. Gels were soaked in 0.1% SDS for 10 min, and proteins were transferred in buffer containing 0.01% SDS onto PVDF membranes, as previously described (LaVoie et al., 2003). Membranes were dried, reactivated/destained with quick immersions in methanol, and probed with antibodies: Ndufa9 (MitoSciences, MS111), SDHA/FP70 (Abcam ab14715), Rieske-FeS (Santa Cruz sc 79037), MTCO1 (Abcam ab14705), ATP5A (Abcam ab14748), Ndufaf2 (Ogilvie et al., 2005). Stripping and reprobing of membranes were performed as described above.
Real-time PCR
Ndufaf2
Total RNA was isolated using the PureLink RNA kit (Invitrogen) following the manufacturer's instructions. One ug of RNA was transcribed to cDNA using iScript cDNA synthesis kit (BioRad) and random primers. Each real-time PCR reaction included 1 μl of cDNA solution, 2x SYBR green PCR master mix (Applied Biosystems), and primers (human Ndufaf2 forward: 5’-GGACCAATTCGGGAACAAAT-3’; human Ndufaf2 reverse: 5’-AAGCTTCCCATTCTGTTGGA-3’). Serial dilution of the samples and primers was conducted to evaluate primer efficiency and the appropriate cDNA concentration to yield linear changes. Quantification was performed using 7500 Fast Real Time System (Applied Biosystems). RNA expression was normalized to actin and 18SrRNA. Relative expression was calculated using the ΔΔCt method.
ND4/ND1
Extraction and quantification via quantitative PCR was performed as previously described (Bender et al., 2006).
Mitochondrial ETC complexes enzymatic activity
Crude mitochondrial preparations were obtained by homogenizing cells for 10 passes on ice in a glass/Teflon power-driven Potter Elvehjem in isolation buffer (250 mM Sucrose, 2 mM HEPES pH 7.4 at 20°C, EGTA 0.1 mM). Nuclei and debris were removed by a 1,000 × g spin 5 min 4°C, and mitochondria were pelleted at 10,000 × g for 10 min at 4°C (Wittig et al., 2006). The mitochondria-containing pellet was resuspended in hypotonic media (25 mM Potassium phosphate, 5 mM MgCl2), and 50 μg aliquots were stored at -80°C until measurement.
Complex-1 assay
Enzymatic activity was determined by following NADH absorbance at 340 nm, as described before (Birch-Machin and Turnbull, 2001). Briefly, 25 mM Potassium phosphate pH 7.4, 5 mM MgCl2, 2 mM KCN, 2.5 mg/ml bovine serum albumin Fraction V, 13 μM NADH, 0.65 mM Ubiquinone, 2 μg/ml Antimycin A were combined in a quartz cuvette and incubated for 3 min at 30°C. Absorbance was followed for 1 min at 340 nm, then 50 μg of freeze-thawed crude mitochondria were added. Absorbance was followed for 3 min, then Rotenone was added at 5 μg/ml, and absorbance was again followed for 3 min. Rotenone-sensitive activities were calculated using linear regression.
Complex-2 assay
Complex-2 activity was determined by following dichloroindophenol (DCPIP) absorbance at 600 nm, with 520 nm as a reference. 50 mM potassium phosphate, 20 mM Succinate, 2 μg/ml Antimycin A, 2 μg/ml Rotenone, 2 mM KCN, 50 μM DCPIP and 50 μg freeze-thawed, crude mitochondrial preparation were combined in a plastic cuvette. Blank rate was recorded for 1 min, then 50 μM decylubiquinone was added, and absorbance was recorded for 3 min. Complex-2 activities were calculated using linear regression.
Citrate synthase assay
0.25% Triton X-100, 0.31 mM Acetyl-CoA, 0.1 mM DTNB and 50 μg crude mitochondrial preparation were combined in a quartz cuvette. The reaction was started by adding 0.5M Oxaloacetate, and absorbance was followed at 412 nm for 2 min.
Cell death assays
Cell growth and trypan blue exclusion
Cells were seeded at a density of 1×105 per well in 3.5 cm 6-well dishes. Each of the following 4 days, one well was trypsinized and counted using a Neubauer chamber and trypan blue exclusion of dead cells. Cell growth rates were calculated (Roth V. 2006 <http://www.doubling-time.com/compute.php>). Percent of live and dead cells were calculated relative to the total number of counted cells.
Cleaved Caspase-3 assay
Cells were fixed with 3% PFA/PBS for 10 min at room temperature, permeabilized for 30 min on ice in 90% methanol/PBS, and blocked for 10 min in 0.5% BSA/PBS at room temperature. To detect cleaved Caspase-3, cells were incubated with primary antibody (Cell Signaling, #9661) for 1 h at room temperature, followed by Cy2 coupled secondary antibody (Jackson ImmunoResearch) for 30 min at room temperature. Green fluorescence was recorded on a FACS Calibur, and median fluorescence intensity was calculated using FlowJo 7.6.1 (Tree Star, Inc.). As a positive control, cells were treated prior to fixation with 1 μM staurosporine for 3 h at 37°C.
Cell cycle analysis
Cells were trypsinized, washed in PBS and fixed in 70% ethanol on ice for 15 min, washed with PBS, and resuspended in propidium iodide (PI) staining solution containing 50 μg/ml PI, 0.1 mg/ml RNase A, and 0.05% Triton-X 100, and incubated for 40 min at 37°C. Cell cycle profiles were recorded on a FACS Calibur (BD Biosciences) using the CellQuest Pro software. Cell cycle analysis was performed using FlowJo 7.6.1 software (Tree Star, Inc.)
ATP assay
ATP levels were measured using the CLS II Kit (Roche), according to the manufacturer's instructions.
Live cell oxygen consumption
Cells were seeded onto fibronectin-coated Seahorse 24 well plates at a density of 80,000 cells per well. The next day, media was changed 1h prior to measurement to unbuffered media containing 25 mM Glucose or 2 mM pyruvate. Oxygen consumption rates (OCR) were measured in using a Seahorse XF-24 analyzer according to the manufacturer's instructions during baseline, and in the presence of 10 μM oligomycin. In the final step, 1 μM rotenone was added to determine non-mitochondrial respiration. To account for any variations in density and cell growth, the DNA concentrations of each well were determined by gently washing the cells once with PBS, resuspending 90 μl of 50 mM NaOH, adding 10 μl of 50 mM Tris-HCl pH 6.8 and measuring DNA content via spectrophotometer. The data from each well were normalized against their DNA content. Basal respiration was calculated by subtracting OCR after rotenone injection from baseline OCR. ATP turnover was determined by subtracting OCR after oligomycin injection from baseline OCR. (Brand and Nicholls, 2011)
Determination of membrane potential
Cells were grown to 70% confluence in regular growth media then changed to DMEM containing 25 mM glucose or 1 mM pyruvate for 16h. After trypsinisation, 2×106 cells were used for each staining condition, centrifuged at 300 × g at room temperature, washed once with HBSS-/-, and resuspended in HBSS-/- (no dye control) or staining solution (HBSS-/- with 10 nM TMRM (Invitrogen)), and incubated for 15 min at 37°C. After 10 min, 50 μM FCCP was added to the staining control. Fluorescence was recorded in the FL-2 channel of a FACS Calibur (BD Biosciences). Data analysis, using the FlowJo 7.6.1 (Tree Star, Inc.) software, encompassed gating for live cells in FSC and SSC scatter plots, and calculation of median fluorescence intensities in FL-2.
Assays for oxidative stress
Mitochondrial superoxide
For quantification of mitochondrial reactive oxygen species, cells were trypsinized, washed in Hank's balanced salt solution without calcium and magnesium (HBSS -/-) and stained with 5 μM MitoSox (Molecular Probes) for 20 min at 37°C. As positive controls, cells were treated with 10 μM Antimycin A for 30 min at 37°C, and for rescue experiments cells were pre-treated with 100 μM Vitamin E for 18h prior to analysis. Fluorescence was recorded on an LSR II (BD Biosciences), gating for live cells and median PE-H intensities were determined using FlowJo 7.6.1 software (Tree Star, Inc.). For imaging, cells were cultured on glass bottom dishes (MakTek) coated with fibronectin. The next day, cells were simultaneously stained with 0.5 μM Mitotracker green, 5 μM MitoSox and 0.5 μg/ml CellMask (all Invitrogen), mounted in warm buffer and imaged on an inverted LSM 510 (Zeiss) confocal microscope.
DNA-damage
To detect oxidative DNA damage, cells were trypsinized, washed in PBS and fixed in 70% ethanol on ice for 15 min. 8-Oxoguanine staining was performed according to manufacturer's instructions (OxyDNA assay kit, Merck). Fluorescence was recorded in the FL-1 channel containing a 530/30 bandpass filter on a FACS Calibur (BD Biosciences) with the CellQuest Pro software. Data analysis was performed as described above, using the FlowJo 7.6.1 (Tree Star, Inc.) software. To keep cells under non-glycolytic conditions, media was changed to DMEM containing 0 mM glucose, 5 mM galactose, 50 μg/ml Uridine, 1mM pyruvate 24h prior to measurement.
Lipid peroxidation
To analyze the content of lipid peroxides, cells were incubated with 5 μM of the lipid peroxidation sensor Bodipy 581/591 C11 (Molecular Probes) in Optimem (Invitrogen) for 30 min at 37°C, trypsinized, and rinsed with HBSS-/-. As positive controls, cells were treated with 80 μM FeCl2 for 30 min at 37°C prior to collection and staining. The shift from red to green fluorescence upon oxidation was detected on a FACS Calibur (BD Biosciences) in the FL-1 channel containing a 530/30 bandpass filter. Analysis was performed as described for the 8-Oxoguanine staining. For rescue experiments, cells were pre-treated with 100 μM Vitamin E for 18 h.
Statistics
Each experiment was performed in at least three independent experiments. Using GraphPad Prism software (v5.0), Student's t-test was used for pairwise comparison, and one-way ANOVA with Tukey's, Dunnett's or Holm-Sidak's post-hoc test was used to compare multiple groups.
Results
Ndufaf2 knockdown alters Complex-1 assembly kinetics and enzymatic activity
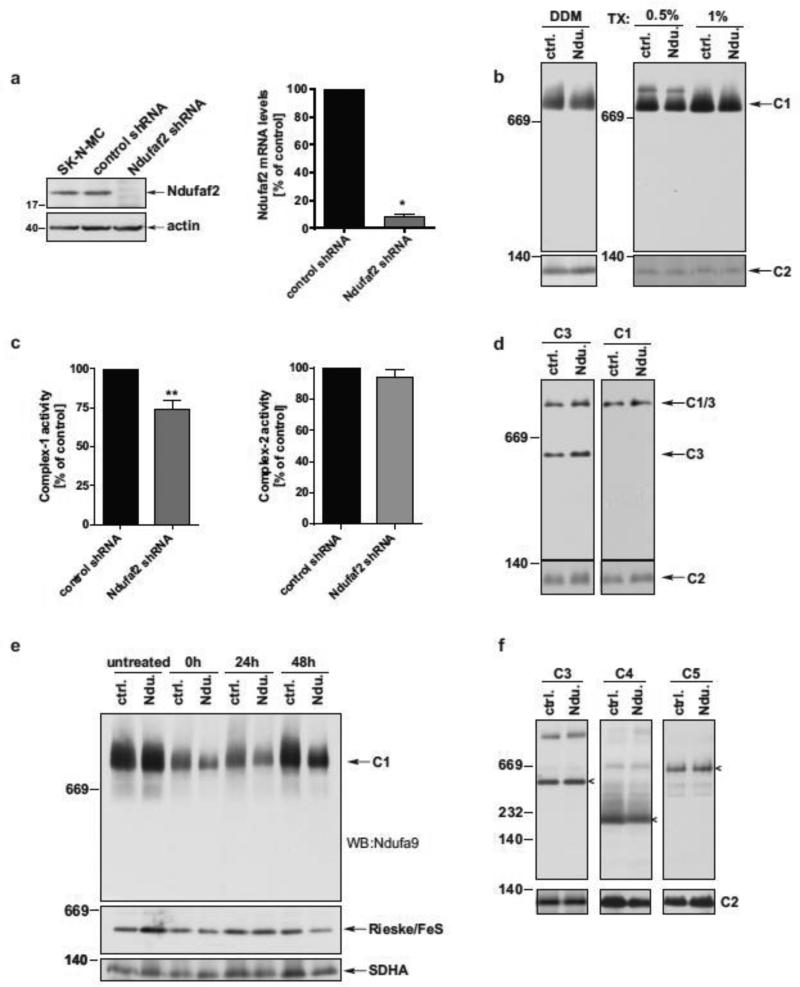
To target Complex-1 function in cell culture, an Ndufaf2-deficient human neuroblastoma cell line was established. SK-N-MC cells were transduced with lentiviral particles each encoding for one of five shRNAs directed against human Ndufaf2, or a scrambled shRNA sequence as a control. Transduced cells were selected using puromycin to derive stably transduced lines. Out of the five shRNAs tested, shRNA -407 demonstrated the greatest knock down of Ndufaf2 levels, and this line will be referred throughout as Ndufaf2 knockdown cells. Parental SK-N-MC cells were transduced with the -407 shRNA in multiple iterations to confirm our results. Using this shRNA, Ndufaf2 protein was below the detection limit in whole cell extracts (Fig. 1a, left panel) and reduced by 92% at the mRNA level (Fig. 1a, right panel), compared to the scrambled shRNA control and to parental SK-N-MC cells. The literature suggests that Ndufaf2 is a critical assembly factor for Complex-1, but this has not been systematically studied outside of the limited analysis from patient samples. Therefore, we sought to examine Complex-1 assembly in this well-controlled knockdown system using the standard Blue Native PAGE and SDS-PAGE Western-blot analyses for mature Complex-1. We probed isolated mitochondria for the Ndufa9 subunit and found that the native molecular weight of Complex-1 was not affected by Ndufaf2 deficiency (Fig. 1b). Importantly, we analyzed Complex-1 assembly under a wide variety of detergent extraction conditions known to result in differential electrophoretic mobilities of Complex-1, and the molecular weight of Complex-1 was consistently identical in the control and Ndufaf2 knockdown cells (Fig. 1b,d).
Figure 1.
Ndufaf2 downregulation leads to mitochondrial impairment in human neuroblastoma cells. a) Left: Stable transduction of SK-N-MC cells with Ndufaf2 shRNA -407 reduces protein levels of Ndufaf2 as determined by SDS-PAGE and Western blot. Actin was used as a loading control. Right: Total Ndufaf2 mRNA levels were decreased after stable transduction with Ndufaf2 shRNA. Actin, 18SrRNA and TBP were used as housekeeping genes in the ΔΔCT method (n=3, mean+SEM, Student's T-Test, *, p<0.0001). b) Mitochondrial preparations were extracted with 1% DDM, 0.5% or 1% Triton-X100 Complex-1 was analyzed on BN-PAGE and Western blot and probing for Complex-1 subunit Ndufa9 (C1: Complex-1), and stripped and reprobed for SDHA (C2: Complex-2) c) Protein-normalized crude mitochondrial preparations were analyzed for enzymatic activities of Complex-1 and Complex-2 (n=5, Student's T-Test, **, p<0.005). Citrate synthase activities of the corresponding preparations were equal (not shown). d) Mitochondrial preparations were extracted with 1% Digitonin. Complex-1 and Complex-3 were analyzed on BN-PAGE and Western blot and probing for Complex-1 subunit Ndufa9 (C1/3: superomplex-1/3) and Complex-3 subunit Rieske-FeS (C3: Complex-3). e) Cells were kept in 1 μg/ml doxycycline for 5 days, changed back to normal growth media, and collected at the times indicated, after media change. DDM-extracts of mitochondrial fractions were analyzed on BN-PAGE for Complex-1 subunit Ndufa9, Complex-3 subunit Rieske-FeS, and Complex-2 subunit SDHA (Ctrl. = control shRNA, Ndu. = Ndufaf2 shRNA). f) Mitochondrial preparations of control and Ndufaf2 shRNA cells were extracted with 1% Triton-X, and analyzed by BN-PAGE and Western blot, using antibodies directed against Complex-2 subunit SDHA (C2), Complex-3 subunit Rieske-FeS (C3), Complex-4 subunit CO1 (C4) and Complex-5 subunit ATP5A (C5). Complexes are indicated by arrowheads.
Electron transport chain activities were measured using enzymatic assays on isolated mitochondria. Results showed a significant 26.5% decrease in Complex-1 activity in Ndufaf2 knockdown cells compared to controls. This effect appeared to be specific for Complex-1, as citrate synthase activity (data not shown) and Complex-2 activity of the same mitochondrial preparations, analyzed in parallel, were not affected (Fig. 1c).
Electron transport can be impaired not only by structural defects of individual complexes, but also by a disorganization of the respiratory chain supercomplexes (McKenzie et al., 2011). Analysis of the Complex 1/3 supercomplex by extracting mitochondrial preparations with 1% digitonin and BN-PAGE separation did not reveal any indication for defects in 1/3 supercomplex assembly in Ndufaf2 knockdown cells compared to controls (Fig. 1d). Similarly, the assembly of other respiratory complexes were unaffected by Ndufaf2 knockdown (Fig. 1f), consistent with our enzymatic data suggesting a specific role for Ndufaf2 on Complex-1 activity. To further investigate the potential role of Ndufaf2 as a Complex-1 chaperone, we studied the kinetics of Complex-1 assembly. We treated control and knockdown cells with doxycycline, an inhibitor of mitochondrial protein synthesis that inhibits the formation of mitochondrially encoded subunits of Complex-1 (and Complex-3). Upon removal of doxycycline, mitochondrially encoded proteins are synthesized and Complex-1 and Complex-3 assembly can begin, where the kinetics of this assembly can be studied (Ugalde et al., 2004; Yadava et al., 2004). After 5 days of doxycycline treatment, the total levels of mature Complex-1 were substantially reduced in both control and Ndufaf2 shRNA cells (Fig. 1e, 0h). However, 24h after doxycycline removal, there was a substantial increase in Complex-1 levels in the control cells while Complex-1 levels in the Ndufaf2 knockdown cells remained decreased. Similar to Complex-1, Complex-3 is composed of nuclear and mitochondrial subunits and is likewise affected by doxycycline treatment, but not by Ndufaf2 knockdown (Fig. 1e, second panel), indicating a specific function of Ndufaf2 on Complex-1 assembly kinetics. As both a loading and treatment control, membranes were stripped and re-probed for Complex-2 (Fig. 1e, bottom panel), which contains only nuclear encoded subunits and should not be affected by doxycycline. At no time did we observe intermediate or meta-stable isoforms of immature Complex-1 in the Ndufaf2 knockdown cells. This was examined using a variety of detergent conditions (Fig. 1d and data not shown), antibodies and on 2D BN-PAGE/SDS-PAGE, to capture labile species (data not shown). Therefore, Ndufaf2 is dispensable for assembly of Complex-1, while our data suggests it does play a chaperone-like function accelerating assembly and facilitating proper activity.
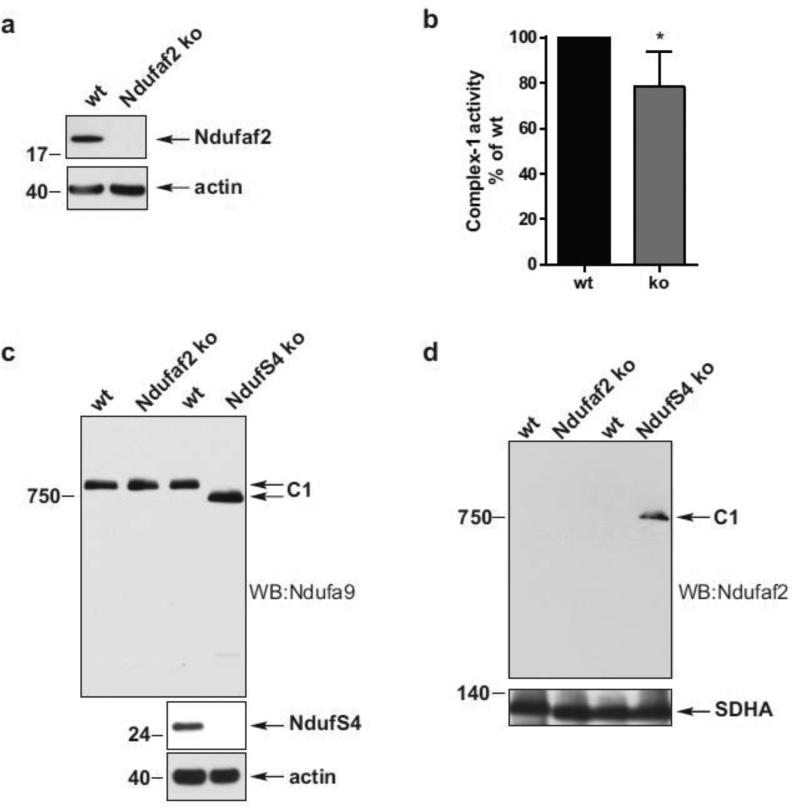
Ndufaf2 is not essential for Complex-1 assembly in mouse cells
The surprising observation that Ndufaf2 knockdown impaired Complex-1 activity without a more dramatic effect on its assembly prompted us to consider the possibility that the incomplete knockdown of Ndufaf2 still allowed a near physiological level of assembly. To address this potential confound, we generated a mouse skin fibroblast cell line cultivated from Ndufaf2 KO mice and wild-type (WT) controls (Fig. 2a). Mitochondria isolated from these cells confirmed a significant 21.7% decrease of Complex-1 activity, comparable to that observed in the knockdown cells (Fig 1c) and consistent with the previously published data from patient skin fibroblast cell lines (Barghuti et al., 2008; Herzer et al., 2010; Hoefs et al., 2009). However, as in the knockdown cells, Complex-1 assembled to its full, mature molecular weight and the total levels of mature Complex-1 protein were not affected by Ndufaf2 deletion (Fig. 2c. lanes 1 and 2). To confirm the resolution of our BN-PAGE system to detect Complex-1 assembly defects, we analyzed WT and NdufS4 KO mouse embryonic fibroblasts (kindly provided by Richard Palmiter). NdufS4 deletion has been previously shown to cause a Complex-1 assembly defect, reduced Complex-1 activity (Kruse et al., 2008; Petruzzella et al., 2001), and the association of Ndufaf2 with a Complex-1 intermediate of approximately 830 kDa (Lazarou et al., 2007; Ogilvie et al., 2005). NdufS4 KO cells exhibited the predicted stalled assembly of Complex-1 (Fig. 2c), demonstrating our ability to resolve this incomplete Complex-1 assembly intermediate despite its absence in multiple Ndufa2 deficient cell models. Interestingly, we also observed the association of endogenous Ndufaf2 with this incomplete Complex-1 intermediate in NdufS4 KO cells (Fig. 2d), whereas we did not detect Ndufaf2 associated with WT Complex-1. Thus, Ndufaf2 is not essential for the complete assembly of Complex-1, but may act as a chaperone modulating the kinetics of assembly and/or efficiency of the mature enzyme Complex-1, possibly by assisting in proper folding of specific subunits.
Figure 2.
Normal Complex-1 maturation in Ndufaf2 KO fibroblasts. a) SDS-PAGE and Western blot analysis of Ndufaf2 KO in mouse skin fibroblasts. Actin was used as a loading control. b) Protein-normalized crude mitochondrial preparations of WT and Ndufaf2 KO skin fibroblasts were analyzed for enzymatic activities of Complex-1. c) Mitochondrial preparations of WT, Ndufaf2 KO and NdufS4 KO fibroblasts were extracted with 0.5% DDM. Complex-1 maturation was analyzed on BN-PAGE and Western blot, by probing for Complex-1 subunit Ndufa9. d) Mitochondrial preparations of WT, Ndufaf2 KO and NdufS4 KO fibroblasts were extracted with 0.5% DDM, and analyzed on BN-PAGE and Western blot by probing for Ndufaf2.
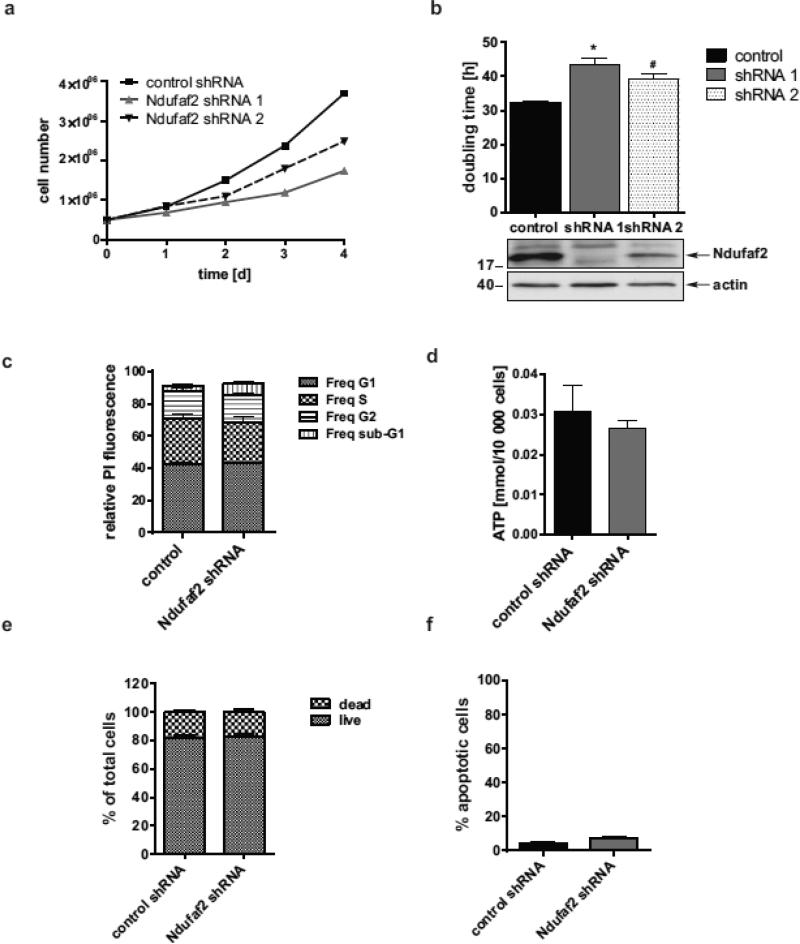
Delayed cell growth in Ndufaf2 knockdown cells
To study the pathological consequences of reduced levels of Ndufaf2, we determined cell growth by counting live cells using trypan blue exclusion and found the doubling time of Ndufaf2 knockdown cells to be increased by 35% compared to control (Fig. 3 a and b). We also tested an shRNA with intermediate knockdown efficiency (shRNA 2), and found a 22% increase in doubling time, suggesting a dose-dependent influence of Ndufaf2 deficiency on cell growth. We next tested several pathways that might explain the delayed growth. No obvious changes in cell cycle were apparent (Fig. 3c), and indices of cell death and apoptosis were the same in control and Ndufaf2 knockdown conditions (Fig. 3e and f). Since the respiratory chain was mildly disturbed in Ndufaf2 knockdown cells, we measured ATP levels, which were not significantly affected (Fig. 3d) following the modest decrease in Complex-1 activity (Fig. 1c). These data likely exclude cell death, cell cycle and energy deficiency as explanations for the delayed cell growth in this cell culture model after Ndufaf2 knockdown.
Figure 3.
Delayed cell growth in Ndufaf2 knockdown cells. a) Representative growth curves of control and two Ndufaf2 shRNA stable cell lines. b) SK-N-MC control and Ndufaf2 shRNA cells were seeded at 5×105 cells per well, collected at various time points, and doubling times were calculated based on the number of live cells using trypan blue exclusion. Shown are control and Ndufaf2 shRNAs -407 (1) and -169 (2) of different knockdown efficiency (n=4, mean +SEM, 1way ANOVA/Dunnett's, *,p<0.001, #,p<0.01).c) Control and Ndufaf2 cells were fixed and permeabilized at 70% confluency during linear growth, and stained with propidium iodide to label DNA. Cell cycle phases were recorded by flow cytometry and analyzed with Flow-Jo (n=6, mean+SEM, one-way ANOVA/Tukey's multiple comparison test, not significant (ns)). d) ATP levels in total cell lysates of control and Ndufaf2 shRNA SK-N-MC cells were determined by luminometry (n=7, mean+SEM, Student's T-Test, ns). e) To analyze overall rate of cell death, live and dead cell number was determined by trypan blue exclusion, and shown as percent live and dead of total counted cells (n=3, mean+SEM, Student's T-Test, ns). f) Apoptosis rates of control and Ndufaf2 shRNA SK-N-MC cells were quantified by Caspase-3 staining and flow cytometry (n=3, mean+SEM,Student's T-Test, ns).
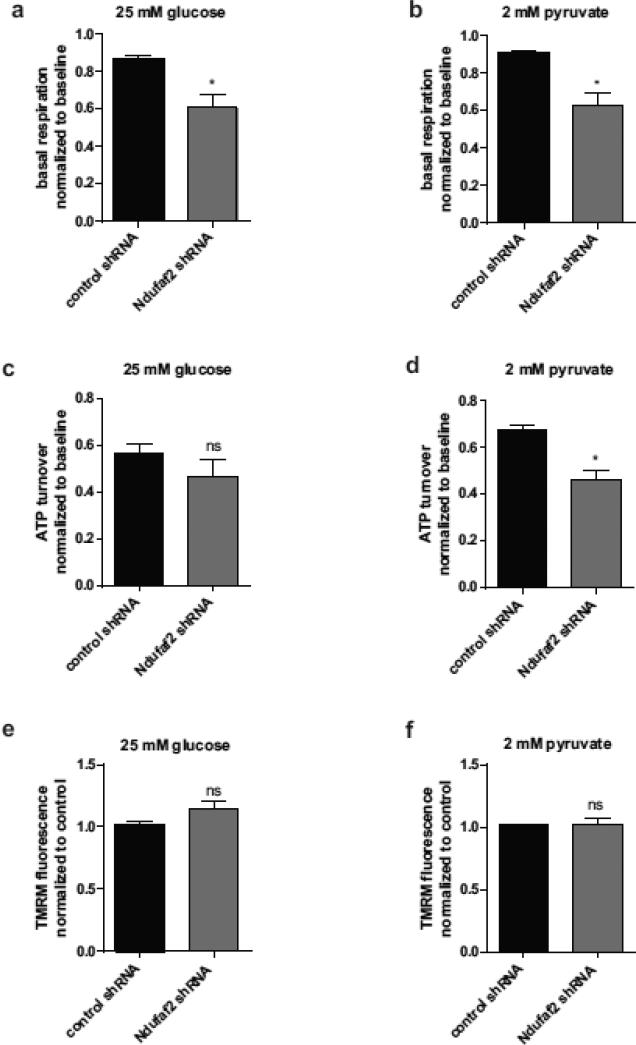
Impaired cell respiration and ATP turnover in Ndufaf2 knockdown cells
To examine mitochondrial function in an intact cellular environment, oxygen consumption rates of live Ndufaf2 knockdown and control cells were determined using a Seahorse XF analyzer. Basal respiration was decreased by 70.4% in glycolytic (25 mM glucose) and 69.6% in oxidative (2 mM pyruvate) conditions in Ndufaf2 knockdown cells compared to control (Fig. 4a, b), demonstrating a defect consistent with the decreased Complex-1 enzymatic activity (Fig. 1c). ATP turnover was not significantly altered under glycolytic conditions (Fig. 4c), consistent with the ATP content measurements (Fig. 3d), but showed a tendency to decrease in the Ndufaf2 knockdown cells. Under oxidative conditions, we found a significant decrease in ATP turnover (Fig. 4d) in Ndufaf2 knockdown cells compared to control cells, confirming the reliance of the cell lines on glucose for ATP production. Membrane potential was assessed by measuring TMRM fluorescence and found to be not substantially altered in Ndufaf2 knockdown cells compared to control, in glycolytic (Fig. 4e), or oxidative conditions (Fig. 4f).
Figure 4.
Altered bioenergetics and membrane potential after Ndufaf2 knockdown. a) and b) Basal respiration was determined under glycolytic (a) and oxidative (b) conditions, by subtracting rotenone-inhibited OCR from baseline OCR. ATP turnover was determined by subtracting oligomycin-inhibited OCR from baseline OCR under glycolytic (c) and oxidative (d) conditions (n=4, mean+SEM, student's T-Test.* p<0.005, ns). Mitochondrial membrane potential was analyzed using TMRM in live cells and quantified by flow cytometry under glycolytic (e) and oxidative (f) conditions (n=3, mean+SEM, one-way ANOVA, ns).
Ndufaf2 deficiency leads to increased production of reactive oxygen species (ROS) and oxidative damage
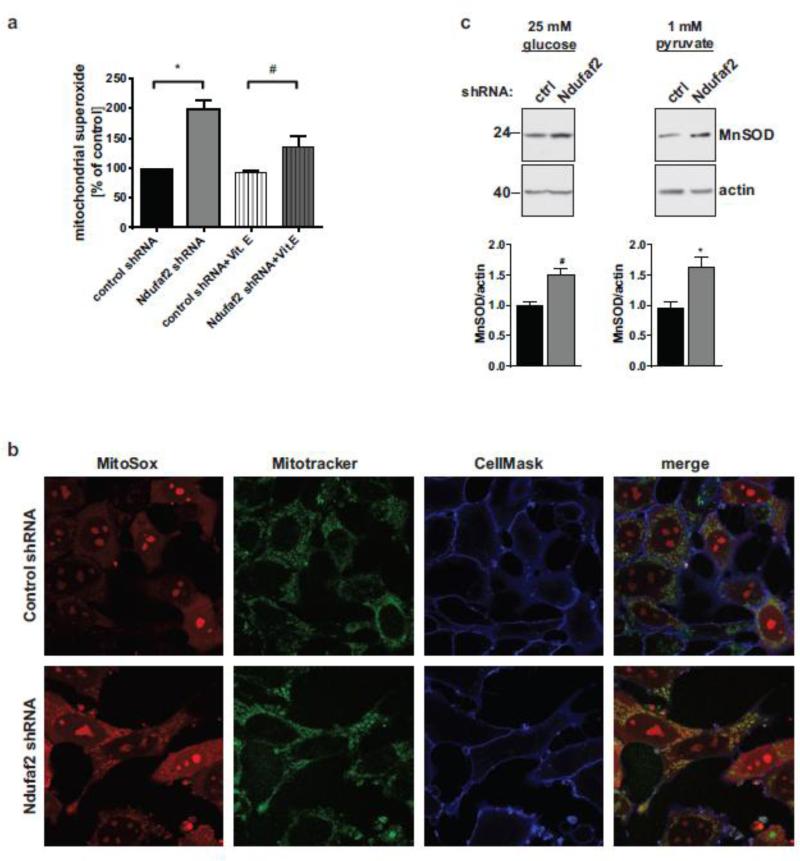
Genetic and toxin-induced insults to Complex-1 have been reported to induce oxidative stress by faulty transport of electrons derived from NADH reduction (Testa et al., 2005; Turrens, 2003). These electrons quickly react with molecular oxygen to form superoxide, instead of being transferred to coenzyme Q (Koopman et al., 2010). Therefore, we analyzed the Ndufaf2 knockdown cells for the levels of mitochondrial ROS. We employed the mitochondria-specific fluorescent dye MitoSox, which is taken up by mitochondria in live cells and fluoresces upon reaction with superoxide. Quantification of the fluorescence intensities by flow cytometry revealed that mitochondrial superoxide was elevated 2.1-fold in Ndufaf2 knockdown cells compared to controls (Fig. 5a). This effect could be reversed by pretreating the cells with the antioxidant vitamin E (Fig. 5a), confirming a bona fide increase in ROS. In an independent measure, higher MitoSOX fluorescence was also observed by live cell confocal imaging (Fig. 5b). Additionally, we determined the degree of activation of the mitochondrial antioxidant system by probing for levels of the mitochondrial superoxide dismutase, MnSOD, under glycolytic and oxidative conditions. MnSOD levels were increased in Ndufaf2 knockdown cells compared to control (Fig. 5c) in both cases, supporting an increased load of mitochondrial ROS.
Figure 5.
Ndufaf2 knockdown increases mitochondrial superoxide. a) Mitochondrial superoxide was labeled with MitoSOX, and fluorescence was quantified using flow cytometry (n=7). For rescue experiments, cells were pretreated with 100 μM Vitamin E for 18 h (n=3). Shown are median fluorescence intensities (mean+SEM, 1way ANOVA/Tukey's multiple comparison test.*, p<0.001, #, p<0.01). b) Control and Ndufaf2 shRNA SK-N-MC cells were grown on fibronectin-coated glass bottom dishes, stained in parallel with Mitotracker (green), MitoSOX (red) and CellMask (blue), and imaged live on a confocal microscope. Shown are representative images of three independent experiments. c) SDS-PAGE and Western-blot analysis of MnSOD in glycolytic (left panels) and oxidative (right panels) condition. Actin was used as a loading control. Quantifications were performed using densitometry (n=6, mean+SEM, unpaired student's T-Test.*,p<0.005, #, p<0.05)
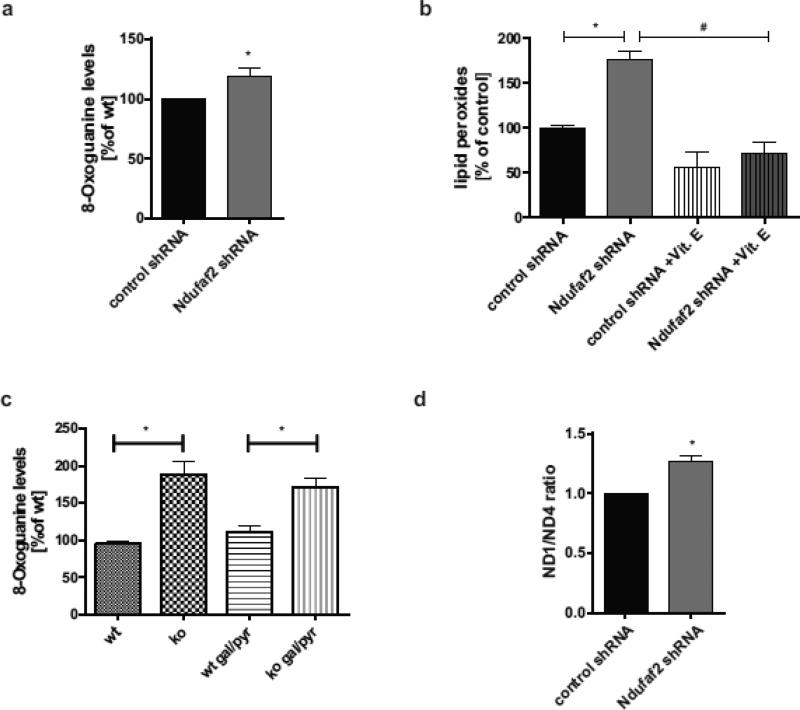
To further explore the biological significance of this increase in ROS, we analyzed several cellular consequences of oxidative stress in Ndufaf2 knockdown and Ndufaf2 KO cells. First, we quantified damage to DNA by measuring the levels of 8-Oxoguanine, an oxidative metabolite of guanine which can cause C to A and G to T substitutions during DNA replication (Cheng et al., 1992). Using a FITC-coupled antibody directed against 8-Oxoguanine, a significant 19% increase in oxidative DNA damage in Ndufaf2 shRNA cells was observed (Fig. 6a), providing evidence that Ndufaf2 deficient cells accumulate DNA damage due to an increased ROS load. Second, we observed a 1.8-fold increase in lipid peroxides in the Ndufaf2 knockdown cells compared to the controls (Fig. 6b), as detected with a Bodipy lipid probe. Again, this effect could be reversed by antioxidant pretreatment with Vitamin E (Fig. 6b).
Figure 6.
Increased oxidative damage after Ndufaf2 downregulation. a) Oxidative damage was detected using a FITC-labeled antibody directed against 8-Oxoguanine on fixed cells. Median fluorescence was quantified by flow cytometry (n=4, mean+SEM, Student's T-Test, *, p<0.05). b) Levels of lipid peroxides were determined using a Bodipy 581/591 probe (n=4). For rescue experiments, cells were pretreated with 100 μM Vitamin E for 24 h (n=3, mean+SEM, 1way ANOVA/Tukey's multiple comparison test , *, p<0.001, #, p<0.01). c) Ndufaf2 ko mouse skin fibroblasts were analyzed as in a). In addition, cells were cultured under non-glycolytic conditions (5 mM galacotse, 50 μg/ml pyruvate, 1 mM pyruvate; n=3, mean+SEM, 1way ANOVA/Holm-Sidak's multiple comparison test, *<0.005). d) Elevated mitochondrial DNA deletion after Ndufaf2 knockdown. Total DNA was isolated from control and Ndufaf2 shRNA SK-N-MC cells, and real-time PCR were run for the mitochondrial ND1 and ND4 genes. Shown are ND1/ND4 ratios relative to control (n=3, mean+SEM, Student's T-Test, *, p<0.01).
To exclude a non-specific increase in ROS due to the tumor-genic origin of the neuroblastoma cells and their maintenance in high glucose media, we studied the consequences of Ndufaf2 deficiency on cellular ROS in Ndufaf2 KO mouse skin fibroblasts, under various metabolic conditions. We found a 2-fold increase in basal 8-Oxoguanine staining in Ndufaf2 knockout fibroblasts, which was consistent under non-glycolytic conditions by maintaining them in DMEM containing 5 mM galactose, 50 μg/ml Uridine, and 1 mM pyruvate 24h prior to measurement (Fig. 6c). Together, these results demonstrate a significant increase in oxidative stress at several cellular levels as a consequence of Ndufaf2 deficiency.
Ndufaf2 knockdown leads to mitochondrial DNA damage
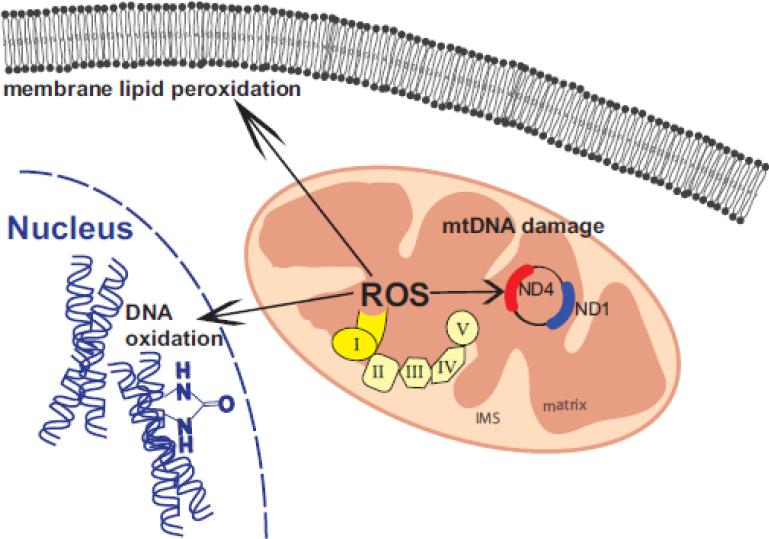
Mitochondrial DNA (mtDNA) mutations tend to accumulate in the aging substantia nigra in humans, and are further increased in PD compared to normal age-matched controls (Bender et al., 2006; Corral-Debrinski et al., 1992; Kraytsberg et al., 2006). The minor arc of the mitochondrial genome is more stable towards the most common deletions and contains the ND1 gene. However, the major arc of the mitochondrial genome is the area in which most deletions occur, and that segment contains the ND4 gene (MITOMAP, (Kogelnik et al., 1996). Therefore, changes in the ratio of ND4/ND1 can be used to quantify the selective loss of major arc DNA (Bender et al., 2006; He et al., 2002). A semi-quantitative real-time PCR assay for ND1 and ND4 levels revealed a significant 27% increase in ND4/ND1 ratio in the Ndufaf2 knockdown cells compared to control (Fig. 6d), indicating a greater number of ND4 deletion events per mitochondrial genome. These data implicate a free radical-mediated induction of mitochondrial DNA damage and genomic deletion following impairment of Complex-1 assembly in live neural cells (Fig. 7).
Figure 7.
Schematic model of pathological consequences of Ndufaf2 downregulation. Using an shRNA and KO approach, we observed indices of increased oxidative stress on several levels, as well as mtDNA damage.
Discussion
Ndufaf2 was first identified in a screen for myc-regulated genes, it was named “mimitin”, an abbreviation of ‘mitochondrial protein induced by myc’ due to its mitochondrial localization (Tsuneoka et al., 2005). Other initial names included Ndufa12L and B17.2L, given its high homology to the B17.2 (Ndufa12) subunit of Complex-1. Interestingly, the regulated nature of Ndufaf2 expression has since been extended to include a responsiveness to pro-inflammatory cytokines (Wegrzyn et al., 2009). Soon after the initial discovery of Ndufaf2, patients with a severe juvenile-onset encephalopathy accompanied by reduced Complex-1 levels were genetically traced to compound heterozygous loss-of-function mutations in Ndufaf2 (Ogilvie et al., 2005) with several cases reported since (Barghuti et al., 2008; Herzer et al., 2010; Hoefs et al., 2009). The fact that Ndufaf2 deletion resulted in reduced Complex-1 activity from patient tissue but did not stably associate with the mature Complex-1 in controls led to the current prevailing hypothesis that Ndufaf2 is an essential assembly factor, and was thus renamed ‘Ndufaf2’. In this study, we sought to examine the physiological function of this disease-linked mitochondrial protein and investigate further the biochemical and pathological effects of its deletion in a well-controlled neural cell system.
While incredibly powerful and possessing advantages to in vitro model systems, the biochemical analyses of patient-derived tissues can suffer from confounds including clonal variation in the cultivation of human fibroblasts, the lack of replicates from limited tissue availability, and the unknown contribution of other genetic and environmental factors among the patients studied. In addition, there have been a very limited number of Ndufaf2 patients thus far identified to overcome the heterogeneity of patient-derived tissue analysis. To study the consequences of Ndufaf2 deficiency in vitro, we developed novel, well-controlled cell culture systems where Ndufaf2 was selectively down-regulated by shRNA in human neuroblastoma cells, or eliminated by knocking out Exon 2 in mouse fibroblasts, and analyzed the mitochondrial and biochemical consequences of a selective reduction in Ndufaf2 expression.
We show here that Ndufaf2 deficiency in multiple cell lines leads to a mild and selective functional impairments of Complex-1 of the respiratory chain. The magnitude of these changes was consistent with that reported from patient fibroblasts in multiple reports, but less substantial than observed in muscle tissue (Hoefs et al., 2009; Ogilvie et al., 2005). Additionally, we observed that the rate of cell division was significantly decreased by Ndufaf2 deficiency, a finding consistent with a previous report on transient siRNA-mediated knockdown of Ndufaf2 in immortalized cell lines (Tsuneoka et al., 2005). This attenuated cell growth under glycolytic conditions was not attributable to cell-wide energy crisis, as ATP levels and turnover were not significantly altered by Ndufaf2 deficiency. A likely explanation for this discrepancy is that ATP production in proliferating cells is largely glycolytic, despite the cells being cultured under aerobic conditions (Gatenby and Gillies, 2004), an interpretation supported by our finding of decreased ATP turnover under glucose-free conditions.
NdufS4 is a critical member of Complex-1 and is part of the lambda subcomplex, within the matrix-facing arm of Complex-1 (Petruzzella et al., 2001). Autosomal recessive mutations in NdufS4 result in a severe encephalopathy, however, one that is phenotypically distinct from that seen in Ndufaf2 patients. Studies of NdufS4 patient tissue have shown that NdufS4 deficient cells cannot fully assembly Complex-1, but rather show a stalled intermediate of ~830 kDa (Breuer et al.). Interestingly, Ndufaf2 can be found stably associated with this assembly intermediate (Ogilvie et al., 2005), as we show in Figure 2d. Since Ndufa2 is not found tightly associated with mature, fully assembled Complex-1 in normal cells it was concluded that Ndufaf2 serves as a necessary late-stage assembly factor (Lazarou et al., 2007; Vogel et al., 2007). However, this has not been rigorously examined across multiple Ndufaf2 patient samples, and the impact on Ndufaf2 deficiency on Complex-1 assembly versus activity has not been fully addressed. BN-PAGE analysis of Ndufaf2 patient samples performed by multiple groups has shown full assembly of Complex-1 (Ogilvie et al., 2005; Zurita Rendon and Shoubridge, 2012), consistent with our data on Complex-1 and Complex-1/3 supercomplex assembly. Nonetheless, prior interpretations of these data entertained the hypothesis that Ndufaf2 deficiency results in assembly defects. Therefore, we conclude that Ndufaf2 is not required for full maturation of Complex-1 (Fig 1d and 2c). Rather, we found that the assembly kinetics were impaired in Ndufaf2 knockdown cells compared to control, indicating that while the size of the assembled Complex-1 was unaffected, the process occurs at a slower rate.
The fact that Complex-1 is able to achieve full assembly in spite of genetic deletion of the Ndufaf2 gene prompted us to consider the hypothesis that this protein serves more as a chaperone to assist in the proper folding of proteins within Complex-1, as opposed to serving as conventional assembly factor. This model would fit better to the mild deficits in enzymatic Complex-1 activity we and others have reported (Barghuti et al., 2008; Hoefs et al., 2009), as well as the cytokine-induced regulation of its expression (Wegrzyn et al., 2009). Furthermore, a recent study on all known Complex-1 assembly factors demonstrated profound effects of each of the other Complex-1 assembly factors on the stability of the mitochondrially-encoded ND1 subunit, but no effect by Ndufaf2 deletion (Zurita Rendon and Shoubridge, 2012). Again, these data differentiate the actual phenotype of Ndufaf2 deletion from the prior predicted model as a necessary assembly factor. This current proposal of a chaperone-like activity of Ndufaf2 would explain its influence on the kinetics of assembly, the efficiency of the fully assembly Complex-1, and the association of Ndufaf2 with an Ndufs4-deficient intermediate. It will be important to examine in future studies whether metabolic or mitochondrial insults induce the up-regulation of Ndufaf2 to assist in Complex-1 folding or assembly under conditions of stress, consistent with a complementary perspective on Ndufaf2 function (Wegrzyn et al., 2009).
It has been proposed that a toxic gain of function through ROS production observed in Complex-1 disease patient-derived tissues correlates well with the overall severity of disease (Koopman et al., 2010; Verkaart et al., 2007b), however, not all respiratory chain dysfunction leads to evidence of increased oxidative stress (Kujoth et al., 2005; Trifunovic et al., 2005; Wang et al., 2001). In the case of Ndufaf2 related pathology, the only study that examined patient-based material used fibroblasts harboring a contiguous deletion of the neighboring genes ELOVL7, ERCC8 and NDUFAF2. There, it was reported that patient cells displayed increased ROS production as measured by hydroethidine oxidation products (Janssen et al., 2009). However, this large-scale deletion encompasses genes whose products are responsible for elongation of very long-chain fatty acids, DNA repair, and Complex-1 assembly, respectively, and the precise contribution of Ndufaf2 to the phenotype was not addressed. Our data following a discrete targeting of Nduaf2 alone, in multiple cell types, show that Ndufaf2 deletion is sufficient to produce increased ROS production and suggest that the prior observation of oxidative injury in this larger deletion may be ascribed to Ndufaf2, as was initially proposed (Janssen et al., 2009).
Somatic mtDNA mutations have been shown to be associated with both neurodegeneration and the normal aging process (Corral-Debrinski et al., 1992; Kraytsberg et al., 2006), presumably due to the lack of protective histones and DNA repair mechanisms in mitochondria. A recently published mouse model highlights the potential involvement of mtDNA damage in neurodegeneration by the targeted overexpression of a mitochondrially targeted restriction enzyme that induces mtDNA cleavage (Pickrell et al., 2011). The human substantia nigra, a target in both PD and Ndufaf2-associated neuropathology, has been shown to contain particularly high levels of mtDNA deletions compared to other brain regions (Bender et al., 2006; Kraytsberg et al., 2006). Therefore, we examined control and Ndufaf2 deficient neuroblastoma cells for the relative genomic levels if ND1 and ND4, an established marker of mtDNA deletion (He et al., 2002), and found that Ndufaf2 deficiency promotes the mtDNA deletion (Fig. 5d).
This study demonstrates that downregulation of the mitochondrial Complex-1 chaperone, Ndufaf2, leads to pathological consequences for Complex-1 function throughout the cell, as illustrated in the model in Fig. 7. However, Complex-1 and supercomplex-1/3 assembly was not dramatically affected by knockdown or by genetic deletion of Ndufaf2, and its enzymatic activity was modestly reduced, suggesting that it is dispensable for many aspects of Complex-1 assembly. However, in spite of its modest effect on Complex-1 assembly, Ndufaf2 deficiency resulted in dramatic oxidative damage throughout the cell, consistent with known mechanisms of neurodegeneration. This work supports the utility of these in vitro models to analyze cellular pathological consequences that mirror those observed in Complex-1 deficiency-related encephalopathies, and indicates that further work will be required to fully understand the contribution of Ndufaf2 to Complex-1 function.
Highlights.
Ndufaf2 is important to Complex-1 assembly, but is not required
Mitochondrial ROS and DNA and lipid damage are increased by Ndufaf2 deficiency
Increased mitochondrial DNA deletions occur in Ndufaf2 deficient cells
Acknowledgements
We thank Eric A. Shoubridge for kindly providing the Ndufaf2 polyclonal antibody, Richard Palmiter for the control and NdufS4 MEFs, Tetyana Labunska for technical assistance, and Jason Schapansky and Jonathan Nardozzi for critical review of the manuscript. This study was supported by NIH grants NS069949 and AG023049 (MJL) and the German Research Foundation (DFG; JSS). JSS and MJL designed research; JSS, MSMJ, KPT, KDA, MJL performed research; JSS, MSMJ, KPT, KDA, analyzed data; JSS, MJL wrote the paper.
Abbreviations
- PBS
phosphate buffered saline
- PVDF
polyvinylidene fluoride
- WT
wildtype
- KO
knockout
- OCR
oxygen consumption rate
- ROS
reactive oxygen species
- mtDNA
mitochondrial DNA
- ns
not significant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interest to declare.
References
- Abou-Sleiman PM, et al. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat Rev Neurosci. 2006;7:207–19. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- Barghuti F, et al. The unique neuroradiology of complex I deficiency due to NDUFA12L defect. Mol Genet Metab. 2008;94:78–82. doi: 10.1016/j.ymgme.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer ME, et al. Cellular and animal models for mitochondrial complex I deficiency: A focus on the NDUFS4 subunit. IUBMB Life. doi: 10.1002/iub.1127. [DOI] [PubMed] [Google Scholar]
- Calvaruso MA, et al. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum Mol Genet. 21:115–20. doi: 10.1093/hmg/ddr446. [DOI] [PubMed] [Google Scholar]
- Cheng KC, et al. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem. 1992;267:166–72. [PubMed] [Google Scholar]
- Corral-Debrinski M, et al. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324–9. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- He L, et al. Detection and quantification of mitochondrial DNA deletions in individual cells by real-time PCR. Nucleic Acids Res. 2002;30:e68. doi: 10.1093/nar/gnf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer M, et al. Leigh disease with brainstem involvement in complex I deficiency due to assembly factor NDUFAF2 defect. Neuropediatrics. 2010;41:30–4. doi: 10.1055/s-0030-1255062. [DOI] [PubMed] [Google Scholar]
- Hoefs SJ, et al. Baculovirus complementation restores a novel NDUFAF2 mutation causing complex I deficiency. Hum Mutat. 2009;30:E728–36. doi: 10.1002/humu.21037. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, et al. Contiguous gene deletion of ELOVL7, ERCC8 and NDUFAF2 in a patient with a fatal multisystem disorder. Hum Mol Genet. 2009;18:3365–74. doi: 10.1093/hmg/ddp276. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, et al. Mitochondrial complex I: structure, function and pathology. J Inherit Metab Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Kogelnik AM, et al. MITOMAP: a human mitochondrial genome database. Nucleic Acids Res. 1996;24:177–9. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, et al. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid Redox Signal. 2010;12:1431–70. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–20. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Kruse SE, et al. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 2008;7:312–20. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–4. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, et al. Assembly of the gamma-secretase complex involves early formation of an intermediate subcomplex of Aph-1 and nicastrin. J Biol Chem. 2003;278:37213–22. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- Lazarou M, et al. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–37. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, et al. Genome-wide copy number variation analysis in attention-deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Mol Psychiatry. 2010;16:491–503. doi: 10.1038/mp.2010.29. [DOI] [PubMed] [Google Scholar]
- Loeffen JL, et al. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum Mutat. 2000;15:123–34. doi: 10.1002/(SICI)1098-1004(200002)15:2<123::AID-HUMU1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- McFarland R, et al. Mitochondrial disease--its impact, etiology, and pathology. Curr Top Dev Biol. 2007;77:113–55. doi: 10.1016/S0070-2153(06)77005-3. [DOI] [PubMed] [Google Scholar]
- McKenzie M, et al. Mutations in the gene encoding C8orf38 block complex I assembly by inhibiting production of the mitochondria-encoded subunit ND1. J Mol Biol. 2011;414:413–26. doi: 10.1016/j.jmb.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–8. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, et al. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest. 2005;115:2784–92. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzella V, et al. A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Hum Mol Genet. 2001;10:529–35. doi: 10.1093/hmg/10.5.529. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, et al. Striatal dysfunctions associated with mitochondrial DNA damage in dopaminergic neurons in a mouse model of Parkinson's disease. J Neurosci. 2011;31:17649–58. doi: 10.1523/JNEUROSCI.4871-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkanen S, Robinson BH. Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J Clin Invest. 1996;98:345–51. doi: 10.1172/JCI118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA, et al. Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry. 2000;39:7229–35. doi: 10.1021/bi000335t. [DOI] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–53. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima A. Establishment of Fibroblast Cultures. Current Protocols in Cell Biology. 1998:2.1.1–2.1.12. doi: 10.1002/0471143030.cb0201s00. [DOI] [PubMed] [Google Scholar]
- Testa CM, et al. Rotenone induces oxidative stress and dopaminergic neuron damage in organotypic substantia nigra cultures. Brain Res Mol Brain Res. 2005;134:109–18. doi: 10.1016/j.molbrainres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, et al. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci U S A. 2005;102:17993–8. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneoka M, et al. A novel Myc-target gene, mimitin, that is involved in cell proliferation of esophageal squamous cell carcinoma. J Biol Chem. 2005;280:19977–85. doi: 10.1074/jbc.M501231200. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–44. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde C, et al. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum Mol Genet. 2004;13:2461–72. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- Verkaart S, et al. Mitochondrial and cytosolic thiol redox state are not detectably altered in isolated human NADH:ubiquinone oxidoreductase deficiency. Biochim Biophys Acta. 2007a;1772:1041–51. doi: 10.1016/j.bbadis.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Verkaart S, et al. Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochim Biophys Acta. 2007b;1772:373–81. doi: 10.1016/j.bbadis.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Vogel RO, et al. Investigation of the complex I assembly chaperones B17.2L and NDUFAF1 in a cohort of CI deficient patients. Mol Genet Metab. 2007;91:176–82. doi: 10.1016/j.ymgme.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A. 2001;98:4038–43. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn P, et al. Mimitin - a novel cytokine-regulated mitochondrial protein. BMC Cell Biol. 2009;10:23. doi: 10.1186/1471-2121-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, et al. Blue native PAGE. Nat Protoc. 2006;1:418–28. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Yadava N, et al. Development and characterization of a conditional mitochondrial complex I assembly system. J Biol Chem. 2004;279:12406–13. doi: 10.1074/jbc.M313588200. [DOI] [PubMed] [Google Scholar]
- Zurita Rendon O, Shoubridge EA. Early complex I assembly defects result in rapid turnover of the ND1 subunit. Hum Mol Genet. 2012;21:3815–24. doi: 10.1093/hmg/dds209. [DOI] [PMC free article] [PubMed] [Google Scholar]