Abstract
Objective
Trichomonas vaginalis infection is the most prevalent treatable sexually transmitted infection (STI) in the world. An accurate point-of-care (PoC) molecular test would enable patients to be tested and treated for T vaginalis in a single visit to the genitourinary medicine clinic, community STI clinic, pharmacy or doctor’s office. In this report, we describe a rapid prototype assay for T vaginalis designed for use in conjunction with the Atlas io PoC platform, and initial verification of its performance using 90 clinical samples.
Methods
A rapid prototype T vaginalis assay was designed. The test, featuring novel electrochemical endpoint detection, used a multi-copy region of the T vaginalis genome as the assay target. Ninety clinical vaginal swab samples were used to verify the performance of the prototype assay.
Results
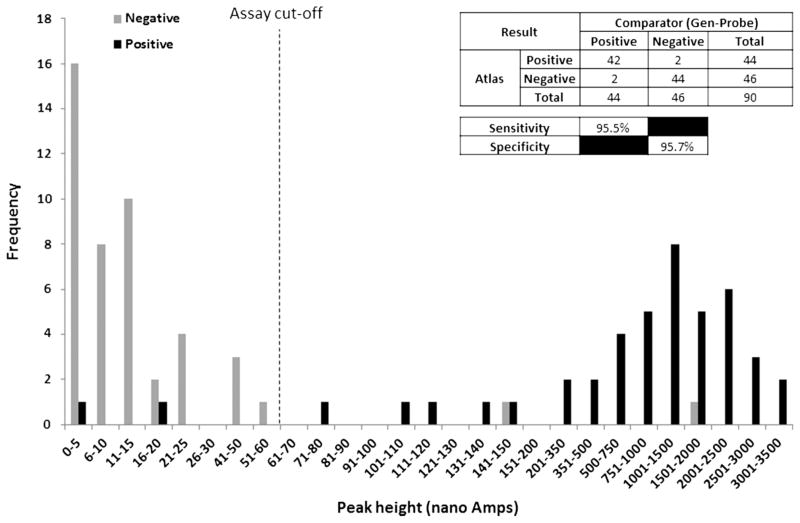
The assay demonstrated a sensitivity and specificity of 95.5% (42/44) and 95.7% (44/46), respectively, when tested using clinical samples. Assay inclusivity was demonstrated for a number of geographically diverse T vaginalis isolates, and the test showed no cross-reactivity with either human DNA or a panel of DNAs isolated from common cross-reactants.
Conclusions
The sensitivity and specificity achieved using this prototype assay is comparable with that achieved for existing central laboratory nucleic acid amplification tests used for screening patients for T vaginalis.
INTRODUCTION
Trichomoniasis is the most prevalent curable sexually transmitted infection (STI) in the world, causing an estimated 248 million new cases of genital Trichomonas vaginalis infection annually.1 The organism affects the urogenital tract of both sexes, where it can cause vaginitis in women, and non-gonococcal urethritis and prostatitis in men. Timely diagnosis and treatment is essential, but current STI screening tests suffer from lengthy turnaround times, exacerbating the spread of the disease through continued sexual activity during this period and the failure of patients to attend follow-up appointments where treatment can be provided. This is especially important in view of the established links between T vaginalis infection and an increased risk of HIV-1 acquisition,2 and between T vaginalis and low birth weight and preterm delivery of neonates.3 The treatment for trichomoniasis is a single oral dose of either metronidazole or tinidazole,4 which, in combination with a rapid test for T vaginalis, would allow a ‘test and treat’ approach to this infection at the point of care. Point-of-care (PoC) tests currently available for T vaginalis include wet mount microscopy (which is a subjective test and suffers from a lack of accuracy) and immunochomatographic assays that approach the sensitivities observed using molecular testing methods.5 To meet the need for rapid PoC diagnostics, Atlas is developing io: a platform technology capable of running molecular and immunoassays with a time-to-result of 30 min, which is within the optimum waiting period for a PoC diagnostic test result suggested in a recent study.6 The technology comprises a cartridge to which an unprocessed clinical sample is added. The sample is fully processed on the cartridge using on-cartridge reagents, a low-cost instrument that provides fluidic and temperature control, end-point electronics and software. We have previously reported a PoC test for Chlamydia trachomatis using this platform.7 In the present study, a T vaginalis assay was designed and tested with vaginal swab samples that had been pre-typed for T vaginalis using the Gen-Probe APTIMA T vaginalis test (Hologic Gen-Probe, San Diego, California, USA).8
MATERIALS AND METHODS
The published T vaginalis draft genome sequence has identified a superabundance of regions with repeats and transposable elements comprising up to two-thirds of the genetic material in this organism.9 A bench-top (laboratory) version of the T vaginalis assay was developed using a genetic biomarker identified as being both unique to the parasite and present in multi-copy. The assay procedure was developed in a similar manner to that described for Atlas’s C trachomatis assay.7 Molded plastic ‘subcircuits’, each representative of one of the three stages of the assay (target DNA extraction, amplification and electrochemical detection) were employed. Following extraction in the presence of an internal assay control, genetic material from T vaginalis was amplified using PCR. Amplicons generated were subsequently detected using electrochemical detection.10 The output of the assay is an electrochemical signal obtained using differential pulse voltammetry.11 A high peak indicated the presence of T vaginalis, whereas a low (or no) peak indicated absence of T vaginalis in a clinical sample. Assay inclusivity was tested using a panel of 13 T vaginalis isolates obtained from worldwide geographical locations (see web appendix). Assay exclusivity was tested using a panel of common cross-reactants comprising genomic material from organisms generally associated with the genitourinary tract and human DNA, plus the DNA from two protists that are closely genetically related to T vaginalis: Trichomonas tenax and Pentatrichomonas hominis (see web appendix). Analytical sensitivity of the T vaginalis assay was determined by adding serial 10-fold dilutions of T vaginalis cells to aliquots of swab transport medium. Clinical sample testing was performed by similarly testing 90 self-collected vaginal swab residual samples (44 positive, 46 negative) that were available for use at the Johns Hopkins International STD Laboratory. The samples had previously been typed using the Gen-Probe APTIMA T vaginalis assay before being stored frozen for this evaluation. The swab used for collection was that supplied in the Gen-Probe vaginal specimen collection kit.
RESULTS
The detection limit of the T vaginalis assay was shown to be five T vaginalis cells. No cross-reactivity with the nucleic acids from organisms commonly associated with the genitourinary tract, human DNA, T tenax or P hominis was detected, indicated by electrochemical signals below cut-off being observed for these sample types (see web appendix). All 13 T vaginalis isolates obtained from a range of geographical locations were successfully detected, with electrochemical peak heights above cut-off being obtained for each isolate (see web appendix). The clinical sample testing data (figure 1) demonstrated a clinical sensitivity and specificity of 95.5% (42/44, 95% CI: 83.2% to 99.2%) and 95.7% (44/46, 95% CI: 84.0% to 99.2%), respectively. The cut-off value of 51.48 nA was selected by taking the mean peak height values of the negative clinical sample plus 3 SD.
Figure 1.
Clinical sample data.
DISCUSSION
We have designed a highly sensitive assay for T vaginalis that detects all T vaginalis isolates tested and excludes common cross-reactants. A qualitative study involving STI clinic directors, clinicians and public health professionals in the USA suggests the sensitivity and specificity demonstrated by this assay is in line with expectations for a nucleic acid amplification test that could be used for detecting STIs in the PoC setting.6,12 Combined with an overall duration of under 30 min, this PoC test would allow a considerable reduction in time-to-result to be realised compared to laboratory-based molecular screening tests. The speed, simplicity and robustness of the electrochemical end-point detection method are well suited to a PoC diagnostic and offer a number of benefits over fluorescence detection methods. The instrumentation involved in electrochemical analysis is simpler and more robust compared to fluorescence analysis, making the instrument easier to miniaturise, simpler to maintain and produce at a cost compatible with PoC settings. Multiple labels have been synthesised that will permit multiplex assays to be developed, meeting the requirements of clinicians and laboratory directors.12 In this study, there was insufficient clinical material to allow the resolution of discrepant samples. This was a pilot study and in a future prospective evaluation, we would consider using a third molecular method to resolve such samples. The testing conducted in this study was achieved using moulded plastic subcircuits, each performing a single stage of the assay process. The next stage of this work will be to adapt the T vaginalis assay onto a fully integrated cartridge capable of being run on the Atlas io instrument. We will then complete a more detailed performance evaluation of the final PoC assay using a greater number of clinical samples.
Supplementary Material
Acknowledgments
The authors thank the Johns Hopkins International STD Laboratory for help in processing the T vaginalis clinical samples and for assistance with this project, Dr Jane Carlton and Dr Barbara Van Der Pol for providing T vaginalis isolates, and Dr Wayne Wilson and Dr Andrew Brittingham for providing genomic DNA from T tenax.
Funding This work was supported by Atlas and the US National Institutes of Health under grant U54 EB007958.
Footnotes
Contributors DMP: Experimental conception and design, data acquisition, data analysis and interpretation of the results, drafting and approving the final version of the manuscript. DNS: Experimental conception and design, data acquisition, data analysis and interpretation of the results, approving the final version of the manuscript. JPH: Sample acquisition, data analysis and interpretation of the results, approving the final version of the manuscript. CAG: Data analysis and interpretation of the results, approving the final version of the manuscript.
Competing interests DMP and DNS receive salaries and share options from Atlas. JPH and CAG have no competing interests to declare.
Ethics approval This study was approved by the Johns Hopkins Institutional Review Board.
Provenance and peer review Not commissioned, externally peer reviewed.
References
- 1.World Health Organisation (WHO) Global prevalence and incidence of selected sexually transmitted infections Chlamydia trachomatis, Neisseria gonorrhoeae, syphilis and Trichomonas vaginalis: methods and results used by WHO to generate 2005 estimates. Geneva: WHO; 2011. [Google Scholar]
- 2.McClelland RS, Sangaré L, Hassan WM, et al. Infection with Trichomonas vaginalis increases the risk of HIV-1 Acquisition. J Inf Dis. 2007;195:698–702. doi: 10.1086/511278. [DOI] [PubMed] [Google Scholar]
- 3.Cotch MF, Pastorek JG, Nugent RP, et al. Trichomonas vaginalis associated with low birth weight and preterm delivery. The Vaginal Infections and Prematurity Study Group. Sex Transm Dis. 1997;24:353–60. doi: 10.1097/00007435-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recomm Rep. 2010;59:1–110. [PubMed] [Google Scholar]
- 5.Huppert JS, Mortensen JE, Reed JL, et al. Rapid antigen testing compares favorably with transcription-mediated amplification assay for the detection of Trichomonas vaginalis in young women. Clin Inf Dis. 2007;45:194–8. doi: 10.1086/518851. [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Gaydos CA, Barnes MR, et al. Comparative effectiveness of a rapid point-of-care test for detection of Chlamydia trachomatis among women in a clinical setting. Sex Transm Inf. 2013;89:108–14. doi: 10.1136/sextrans-2011-050355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce DM, Shenton DS, Holden J, et al. Evaluation of a novel electrochemical detection method for Chlamydia trachomatis: application for point-of-care diagnostics. IEEE Trans Biomed Eng. 2011;58:755–8. doi: 10.1109/TBME.2010.2095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrea SB, Chapin KC. Comparison of Aptima Trichomonas vaginalis transcription-mediated amplification assay and BD Affirm VPIII for detection of T vaginalis in symptomatic women: performance parameters and epidemiological implications. J Clin Microbiol. 2011;49:866–9. doi: 10.1128/JCM.02367-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlton JM, Hirt RP, Silva JC, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–12. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillier SC, Flower SE, Frost CG, et al. An electrochemical gene detection assay utilizing T7 exonuclease activity on complementary probe-target oligonucleotide sequences. Electrochem Commun. 2004;6:1227–32. [Google Scholar]
- 11.Bard AJ, Faulkner LR. Electrochemical methods: fundamentals and applications. 2. Hoboken, NJ: Wiley; 2001. [Google Scholar]
- 12.Hsieh Y-H, Hogan MT, Barnes M, et al. Perceptions of an ideal point-of-care test for sexually transmitted infections—a qualitative study of focus group discussions with medical providers. PLoS ONE. 2010;5:e14144. doi: 10.1371/journal.pone.0014144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.