Abstract
During translation initiation in eukaryotes, the small ribosomal subunit binds mRNA at the 5′-end and scans in the 5′ to 3′ direction to locate the initiation codon, form the 80S initiation complex and start the protein synthesis. This simple, yet intricate, process is guided by multiple initiation factors. We determined the structures of three complexes of the small ribosomal subunit that represent distinct steps in mammalian translation initiation. These structures reveal the locations of eIF1, eIF1A, mRNA and initiator tRNA bound to the small ribosomal subunit and provide insights into the details of translation initiation specific to eukaryotes. Conformational changes associated with the captured functional states reveal the dynamics of the interactions in the P site of the ribosome. These results have functional implications for the mechanism of mRNA scanning.
In bacteria, translation initiation is controlled by initiation factors IF1, -2 and -31. In eukaryotes it is more complex: multiple initiation factors facilitate the recruitment and scanning of mRNA, selection of initiator tRNA, the large and small ribosomal subunits joining2–4. Eukaryotic initiator tRNA binds specifically to eIF2-GTP, forming a stable ternary complex (TC). The TC forms the 43S preinitiation complex (PIC) by binding to the small ribosomal subunit (40S). Formation of the 43S PIC is promoted by the eukaryotic initiation factors (eIF) eIF1, -1A, -3, -55. 43S PIC then binds to the capped 5′-end of mRNA through eIF4F and eIF3, and scans the mRNA up to the first AUG codon.
The scanning hypothesis (first AUG rule) was proposed by Kozak6 and subsequently refined to reflect that sequences surrounding AUG, particularly at the −3 and +4 positions with respect to the A, are critical for initiation in mammals7,8. Despite widespread acceptance of this model, its molecular mechanism remains unknown2,9. Scanning stops upon AUG recognition, triggering eIF1 dissociation. The release of phosphate from the TC makes this reaction irreversible and promotes dissociation of eIF2-GDP10,11. The 48S PIC that is now formed contains the initiator tRNA base paired with AUG in the P site of the 40S subunit. The subsequent attachment of the large (60S) ribosomal subunit is facilitated by GTP binding factor, eIF5B. eIF5B and eIF1A dissociate from the 40S subunit during or after 80S ribosome assembly, leaving the ribosome primed for elongation2.
We have determined the crystal structures of the rabbit 40S subunit in complex with: (i) eIF1 (PIC1), (ii) eIF1 and eIF1A (PIC2) and (iii) mRNA, tRNA and eIF1A (48S PIC). These structures establish the positions of eIF1, eIF1A, mRNA and tRNA and reveal the architectural principles governing the assembly of the pre-initiation complex on mRNA. Comparison of these structures shows that a conformational rearrangement in the P site and a rotation of the head of the 40S subunit accompany complex assembly. This provides the basis for understanding the functions of eIF1 and eIF1A during scanning.
Crystallization and structure determination
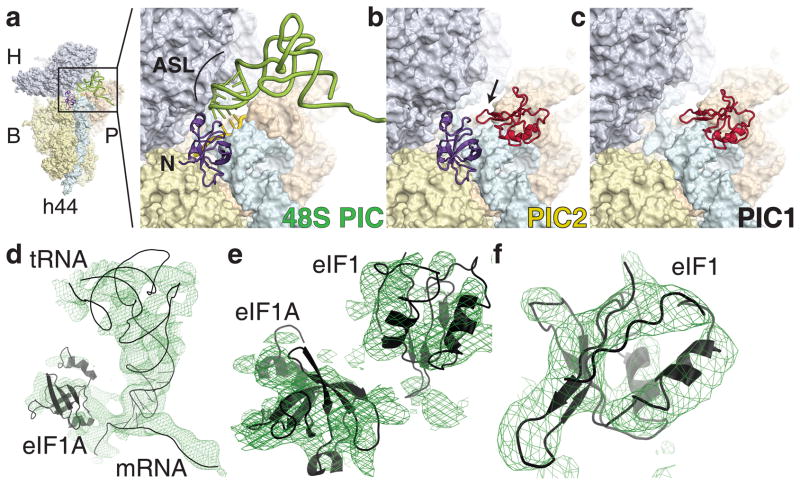
Rabbit 40S ribosomal subunit complexes were crystallized in the P3121 space group and contain one 40S subunit per asymmetric unit (Fig. 1, Supplementary Fig. 1). Complete data sets were collected to 7.9–9 Å resolution (Supplementary Table 1). The 48S PIC was formed by incubating 40S ribosomal subunits with mRNA, non-aminoacylated, in vitro transcribed initiator tRNA (tRNAi), and human eIF1A and eIF1. Although the presence of eIF1 in the reaction mixture is essential for crystallization, this factor is not seen in the 48S PIC structure. The PIC1 complex was formed by incubating 40S ribosomal subunits with eIF1, and the PIC2 complex was formed by soaking eIF1A in crystals of PIC1 (Methods).
Figure 1. The crystal structure of rabbit preinitiation complexes.
a. 48S PIC, b. PIC2, c. PIC1. H, head; B, body; P, platform; h44, helix 44. eIF1A (violet), eIF1 (red), tRNAi (green) and mRNA (yellow) are shown. The basic loop of eIF1 is marked by an arrow; the N-terminus of eIF1A is marked by N. Initial unbiased electron density maps Fo-Fc contoured at σ=2.0 (green mesh) for 48S PIC (d), PIC2 (e), PIC1 (f). Docked structures are shown in black.
The presence of additional electron densities in the maps allows us to unambiguously identify the locations of eIF1, eIF1A, tRNA, mRNA (Fig. 1, Supplementary Fig. 2). We provide E. coli nomenclature for ribosomal proteins and rRNA in parentheses. We focus predominantly on the details related to translation initiation, which include the interactions of initiation factors, tRNA and mRNA with the 40S ribosomal subunit and their implications for the mechanistic understanding of the scanning process.
Initiator tRNA in the P site
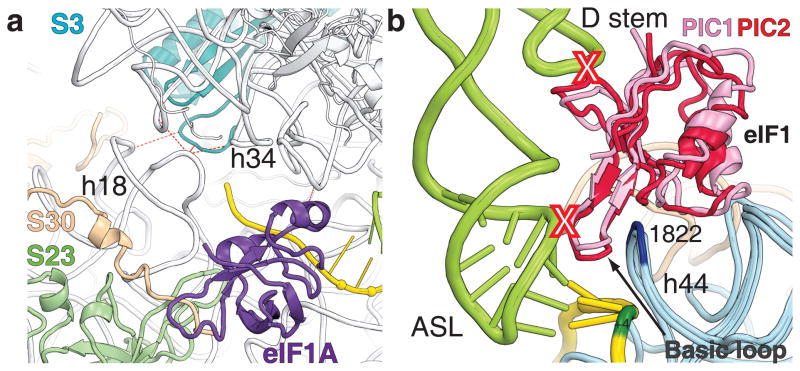
The P site of the 40S subunit contains a binding pocket, occupied by the anticodon stem-loop (ASL) of tRNAi. This pocket is formed by the 18S rRNA helices h28, h44 of the body, h24 of the platform, h28, h29, h30, h31 of the head domains and by three ribosomal proteins (rpS) located in the head of 40S subunit - rpS15(19), rpS16(9) and rpS18(13) (Fig. 2, Supplementary Fig. 7). The regions of tRNAi that interact with rRNA are similar to those seen for the P site tRNA bound to the 70S ribosome12,13. However, the rRNA is arranged differently in the tRNAi binding pockets of PIC1, PIC2 and 48S PIC, reflecting the differences between the scanning-competent (PIC1, 2) and scanning-incompetent (48S PIC) conformations. Movement of the tRNAi toward the E site is blocked in the 48S PIC by helices h24 and h29, which contact the ASL from the platform and head side of the 40S subunit, respectively (Supplementary Fig. 3). The position of h29 is different in the PIC1 and PIC2 structures, compared to that in 48S PIC, and allows the P site bound tRNA to move toward the E site (Fig. 2b). The corresponding region of h29 has shifted ~4Å toward the E site in PIC1 and ~2Å in PIC2. The binding of tRNAi on the opposite side of the pocket in the 48S PIC might be stabilized by the C-terminus of rpS15(19) and by the N-terminal tail (NTT) of eIF1A, as evidenced by the presence of additional electron density near the ASL of the tRNAi in the corresponding map (Supplementary Fig. 4). The suggested orientation of the C-terminus of rpS15(19) in the 48S PIC can explain the crosslink reported between rabbit rpS15(19) and the +4 position on mRNA in the 48S complex14. Positioning of the NTT of eIF1A near the ASL of tRNAi in 48S PIC is consistent with biochemical data demonstrating that NTT contributes to eIF1A binding to the 40S subunit15 and may interact with the ASL16. The NTT of eIF1A remains unstructured in PIC2. We propose that the architecture of the P site in PIC1 and PIC2 represents the scanning-competent conformation of the 40S subunit, with tRNAi in the Pout mode2. In contrast, the P site in 48S PIC represents a scanning-incompetent conformation of the 40S subunit, with the ASL of tRNAi in Pin mode - locked in position by base-pairing with AUG and interactions with the binding pocket, possibly with rpS15(19) and the NTT of eIF1A2,17. This position of tRNAi is similar to the P/I state in the bacterial 30S IC18,19 (Supplementary Fig. 6).
Figure 2. Interactions of tRNAi and eIF1A with the 40S subunit.
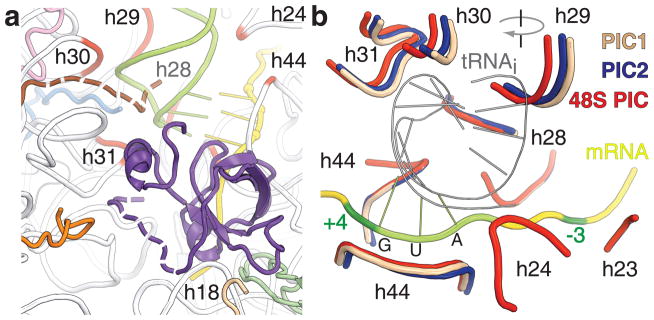
a. eIF1A (violet) binds to h44, rpS30e (wheat) and rpS23 (green). Possible positions of the NTT of eIF1A and the C-terminus of rpS15 (brown) are shown as dashed lines. Conserved regions of 18S rRNA, which form tRNAi binding pocket, are in red. rpS16 (blue), rpS18 (pink), rpS31 (orange) are shown. b. Superposition of 48S PIC, PIC1 and PIC2. Only regions of rRNA that are important for the interaction with the tRNAi are shown. Clockwise rotation of the head domain is indicated.
Rearrangements of the P site in the PICs are accompanied by a 3° clockwise rotation of the head of the 40S subunit after the binding of eIF1A to PIC1, and an additional 3° upon formation of the 48S PIC. Rotations of the head shift part of the tRNAi binding pocket toward the A site by ~2Å with each rotation. At the same time, h28, which comprises the neck that connects the head and body of the 40S subunit, does not change its position (Fig. 2b, Supplementary Fig. 5). The role of h28 as a pivot point for head rotation suggests that mutations in the neck affect rotation of the head and repress the conformational rearrangements of the P site that are required during scanning. This likely explains the leaky scanning phenotype seen in mutational studies of h28 in the yeast 18S rRNA20.
The mRNA channel and the mRNA path
The electron density map of the 48S PIC reveals the position of the mRNA on the 40S subunit from nucleotides −6 to +7 (Fig. 3a). Its conformation does not have the sharp kink between the A and P site codons that is seen in the bacterial 70S ribosomal complexes. This kink allows the simultaneous pairing of the A and P site tRNA anticodons with the mRNA21 (Fig. 3a), that defines the reading frame and prevents slippage of the mRNA13. The conformation of the mRNA in the scanning complex should promote the slippage of mRNA. eIF1A assists the slippage of the 40S subunit along the mRNA by occupying the A site, which prevents possible interactions between tRNAs and mRNA that might otherwise fix the mRNA on the ribosome (Fig. 3a, Supplementary Fig. 7).
Figure 3. Path of the mRNA in the 48S PIC.
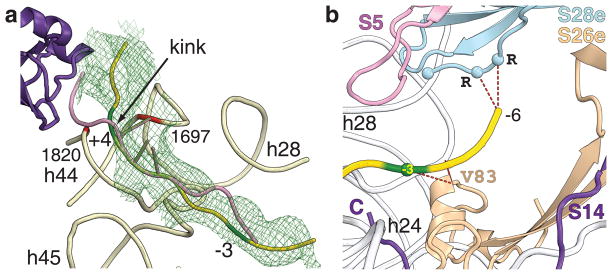
a. Initial unbiased electron density map Fo-Fc contoured at σ=2.0 (green mesh). eIF1A (violet), mRNA in eukaryotic 48S PIC (yellow), mRNA from bacterial 70S translating ribosome (pink) are shown. Nucleotides +4 and −3 of the mRNA are in dark green. b. rpS26e is ~5Å away from nucleotide −3 of the mRNA. The C-terminus of rpS28e is ~7Å from the −5, −6 region of the mRNA.
The mRNA path in the 48S PIC is similar to that in 70S ribosomal complexes14,22. Eukaryote specific features are localized in the mRNA channel near nucleotides −3 and +4, which are the most sensitive positions in the consensus sequence around the initiation codon, and also upstream of the E site, because eukaryotes lack the Shine-Dalgarno sequence (SD)22,23. Two ribosomal proteins, rpS26e and rpS28e, which do not have homologs in eubacteria are present in these regions. rpS26e has an α helix (C74-V83) that is located ~5Å away from positions −4 and −3 on the mRNA as well as from h28 (Fig. 3b). This helix may change the orientation of the −3 nucleotide during scanning, thus affecting its interaction with the α-subunit of eIF2, implicated in the discrimination of the AUG context at position −314. While rpS26e is bound to the platform of the 40S subunit, its N-terminus interacts with the neck of the 40S subunit, possibly facilitating communication between position −3 on the mRNA, the head domain, and the platform.
mRNA position −6 is located in the vicinity of rpS28e. This region of mRNA is of particular interest because it is involved in the SD interaction in prokaryotes and modulates translation initiation on non-AUG initiation codons in eukaryotes24. The C-terminus of rpS28e contains three conserved arginine residues, located ~7Å from nucleotide −6, that may interact with it (Fig. 3b). We suggest that these eukaryote-specific interactions may stabilize the position of mRNA during initiation. The initiation codon and nucleotide +4 are located in a gorge formed by the top of h44 (Fig. 3a, Supplementary Fig. 7). We propose that nucleotides 1697(1401) and 1820(1494) of h44 of 18S rRNA, together with the NTT of eIF1A, modulate the orientation of nucleotide +4 of the mRNA, which may pause mRNA sliding during scanning (Fig. 3a). This pause would allow the AUG codon or ASL of tRNAi to be accommodated for codon-anticodon base-pairing. The presence of electron density attributed to the NTT of eIF1A in the 48S PIC (Supplementary Fig. 4) suggests that upon base-pairing with the initiation codon, the NTT might stabilize the Pin conformation of the tRNAi17 by interacting with the mRNA’s +4 nucleotide and/or with the ASL of tRNAi (Fig. 2, Supplementary Fig. 7).
Initiation factor eIF1A and the latch
The universally conserved eIF1A stimulates binding of the TC to the 40S subunit and is involved in the scanning and selection of the initiation codon2. Its bacterial ortholog IF1 is bound to the A site of the 30S ribosomal subunit, where it sterically prevents the binding of tRNA25. The location of the binding site of eIF1A on the 40S ribosomal subunit was previously mapped to the A site16. An NMR structure of eIF1A revealed that its main domain has an oligonucleotide/oligosaccharide binding (OB) fold that is similar to IF1, an additional C-terminal subdomain with two α-helices, and long unstructured N- and C-terminal tails (CTT)26. While the CTT is required for stringent AUG selection by promoting the scanning-competent conformation of the initiation complex, mutations in the NTT suppress the elevated level of UUG initiation15,27.
Electron density maps of 48S PIC and PIC2 show clear density for eIF1A (Fig. 1)26. While the CTT of eIF1A remains disordered, the NTT is partially visible in the 48S PIC. The additional density in the difference map (Fo-Fc) of the 48S PIC can be attributed to the NTT of eIF1A. In this case, it is located 7–8Å from nucleotide +4 of the mRNA and the ASL of the tRNAi. (Fig. 2, Supplementary Fig. 4). eIF1A binds to the 40S subunit at the top of h44, which forms a part of the intersubunit bridge B2a28, and to h18 of the 18S rRNA through its OB domain, similar to that of IF125. eIF1A interacts with the N-terminus of rpS30e and with the rpS23(12) (Fig. 2a, Supplementary Fig. 7). This position of eIF1A ensures that the A site on the 40S subunit remains inaccessible to tRNAs during initiation.
eIF1A forms a bridge between the head and the body of the 40S subunit through its helical subdomain (PIC2 and 48S PIC) and NTT (48S PIC) (Figs. 2a, 4). These eukaryote specific interactions are consistent with hydroxyl radical cleavage experiments16. Bridging together the head and the body of the 40S subunit ensures the closure of the mRNA channel around the A site, which promotes scanning of the mRNA.
Figure 4.
a. Closed conformation of the latch. Interactions between body and head domains are connected by dashed lines. b. The P site of the 40S subunit. eIF1 from PIC1 (pink) or PIC2 (red) would sterically clash (red X) with the ASL and the D stem of tRNAi (green) that is positioned in the 48S PIC.
The “latch” of mRNA entry channel remains closed in the structures of all three PICs presented (Fig. 4a). It is formed by interactions between h18 of the body with h34 and ribosomal protein rpS3(3) of the head, which clamp around the incoming mRNA29,30. It was proposed that the closed conformation of the latch is a feature of ribosomes in the active conformation, which “provides geometry that guarantees processivity and ensures maximum fidelity”30. We propose that closure of the latch is necessary for the scanning function of the 40S ribosomal subunit. Closing of the latch and eIF1A binding alter the environment of the mRNA binding channel and ensure that the mRNA remains unstructured, correctly oriented for scanning and free to move toward the exit.
The closed conformation of the latch in our structure of PIC2 is different from the open conformation of the latch in the cryo-EM reconstruction of yeast 40S subunit complex with eIF1 and eIF1A29. It is possible that eIF3 may be involved in the latch opening in mammals, because unlike in yeast, eIF3 is required for TC to bind to the 40S subunit. The latch closure might be influenced by crystallization, although the crystal packing does not affect rotation of the head domain of the 40S subunit. Furthermore, the latch could sample different conformational states during initiation. The open conformation of the latch allows direct loading of the mRNA in the mRNA binding channel, instead of threading it through a tunnel29. We propose that the cryo-EM structure of the yeast 40S subunit with bound eIF1 and eIF1A represents the mRNA binding state of the ribosome, while the rabbit PIC2 represents its scanning mode.
eIF1 is poised to destabilize tRNAi binding
eIF1 is a functional homologue of the bacterial IF331,32. During scanning, eIF1 allows proof-reading and sensing a non-optimal context of AUG codons, while promoting the dissociation of aberrantly assembled complexes33,34. eIF1 binds to the top of h44 and to helices h24 and h45 of the 18S rRNA in the vicinity of the P site (Figs. 1, 4b), consistent with previously published studies31,35,36. The region of interaction between eIF1 and 18S rRNA consists of the intersubunit bridge B2a and part of the bridge B2b28. Superposition of the 48S PIC onto PIC1 and PIC2 reveals moderate sterical clashes between eIF1 and tRNAi, as was predicted earlier by comparison of structures of 40S subunit complex with eIF1 from T. thermophila and bacterial 70S ribosomal complexes35. The basic loop of eIF1 (R38-K42) in both PIC1 and PIC2 is positioned between the ASL of tRNAi and nucleotides 1821–1823(1495–1497) of h44, which stabilizes the interaction between AUG and the anticodon in the P site of the 48S PIC. This results in steric clashes between the basic loop of eIF1 and the ASL of tRNAi and between the region around residues P77-G80 of eIF1 and the D-stem of tRNAi (Fig. 4b).
It has been proposed that eIF1 modulates recognition of the start codon by restricting tRNAi binding to AUG only in an optimal context33,34. Fluorescent resonance energy transfer experiments (FRET) with eIF1 and eIF1A demonstrated that AUG recognition triggers the dissociation of eIF1 from 48S PIC, which allows the release of the phosphate from the eIF2-GDP-Pi complex10,11. Our data provide an explanation by revealing that eIF1 spatially interferes with the adjustment of the ASL of the initiator tRNA in the P site during scanning. Establishing the interaction between the AUG codon and the anticodon results in locking of the P site tRNA, which can prevent the basic loop of eIF1 from re-entering the P site. Kinetic experiments show that the dissociation constant of eIF1 from the 40S subunit is around 1.4 nM in the presence of the TC, while the high affinity of the TC to the 40S subunit results from base-pairing with mRNA11,29. This affinity is low when no mRNA is bound to the 40S subunit or when only one nucleotide in the P site is base-paired. It is increased with the ability of the initiator tRNA to base pair with two nucleotides and becomes very strong upon base pairing with the AUG (Kd <1 nM)11,37. At this point, upon the correct codon-anticodon base pairing eIF1 cannot compete with the ASL of tRNAi for the P site and must dissociate.
The basic loop of eIF1 is located within 5Å of h44 and 10Å of the +4 position on mRNA (Fig. 4b). That may allow eIF1 to “inspect” the important region downstream of AUG. The same region of h44 undergoes small conformational changes upon formation of 48S PIC, which can weaken its interaction with eIF1 and contribute to the dissociation of eIF1 from the 40S subunit. Consequently, our structures of rabbit PICs provide mechanistic insights into the current model of eukaryotic translation initiation.
Scanning model
The observed conformations of the preinitiation complexes described in this study together with the prior biochemical data2 allow us to propose a model for scanning. Briefly, the mRNA slides through the tunnel, formed by the body and the head of the 40S subunit (the latch), eIF1A and eIF1. The tunnel ensures scanning processivity by keeping the mRNA unstructured and properly oriented for the “inspection” of the nucleotide sequence in the P site by tRNAi. The tRNAi “inspects” or scans the mRNA by attempting to establish Watson-Crick base-pairing between its anticodon and a nucleotide triplet of mRNA moving through the P site. The tRNAi is in Pout state because the orientation of the head domain of the 40S subunit allows a minor movement of the tRNAi in order to avoid a steric clash with eIF1. The affinity of the basic loop of eIF1 to the P site is sufficient to displace the ASL of tRNAi and prevent the locking of the tRNAi in the P site (Fig. 5).
Figure 5. Scanning model.
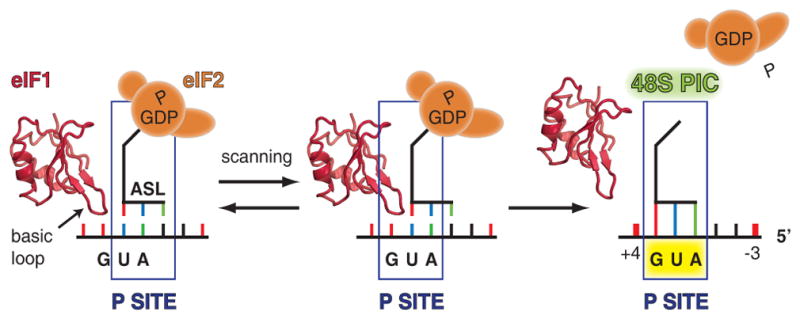
Basic loop of eIF1 competes for the P site of the 40S subunit with the ASL of tRNAi, which results in displacement of each other during mRNA scanning.
When the AUG codon is in the P site, it becomes base-paired with all three nucleotides of the anticodon of the tRNAi, thereby stabilizing the conformation of the tRNAi, while allowing it to displace the basic loop of eIF1. Subsequent rotation of the head domain locks tRNAi in the Pin state and completes the scanning (Fig. 5). The transition from Pout to Pin mode of tRNAi is likely promoted by the C-terminus of rpS15(19) and the NTT of eIF1A17, which together might stabilize the base-paired conformation of tRNAi in the P site. Nucleotides +4 and −3 on the mRNA may cause a similar effect by their interaction with h44, eIF1A, eIF1 and h23, rpS5(7), rpS26e respectively. The dissociation of eIF1 and eIF2 from the 40S subunit results in a stable 48S PIC similar to the one observed in this study. eIF5B then binds to this complex and orients the acceptor stem of tRNAi towards the P site on the 60S subunit to facilitate subunit joining in a manner that is similar to its bacterial analog IF2 (Supplementary Fig. 6).
Conformational changes in the P site may affect the interaction of eIF1with the top of h44 to which it binds. This likely provides synchronization between the rotations of the head domain of the 40S subunit, which rearrange the P site, and the presence of eIF1 on the ribosome. Interaction of eIF1A with the h44 may cause the same effect and explain the cooperative binding of eIF1A and eIF1 to the 40S subunit29,38. Future high-resolution structures of the 40S ribosomal subunit complexes with the initiation factors, mRNA and tRNA coupled with biochemical studies are needed to refine and complete the model presented here.
METHODS
Cloning and Protein Expression
The DNAs encoding full-length human eIF1 and eIF1A with additional N-terminal affinity tag and TEV protease cleavage site sequences (MGHHHHHHDYDIQTTENLYFQ↓G) were synthesized by PCR and cloned into pET28b expression vector (Novagen, Madison, WI). Recombinant initiation factors were expressed in E. coli Rosetta™2(DE3)pLysS cells (Novagen).
Protein Purification
Recombinant eIF1 and eIF1A were purified using a HisTrap™ HP column (GE Healthcare, Piscataway, NJ), essentially as previously described40. The proteins were then treated with AcTEV protease (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The affinity tag was removed by chromatography on a HisTrap™ HP column (GE Healthcare). Tag-free eIF1 and eIF1A were further purified on Mono S and Mono Q columns (GE Healthcare), respectively. Finally, both proteins were purified on a Superdex 75 column (GE Healthcare) equilibrated with buffer H300 (5 mM HEPES-KOH, 300 mM KCl, 5% glycerol, pH 7.1). eIF1 and eIF1A were frozen in liquid nitrogen in small aliquots and stored at −80°C.
Initiator tRNA
Mammalian initiator tRNA41 was transcribed using T7 RNA polymerase and chemically synthesized DNA olygonucleotides as suggested in ref. 42. tRNA was extracted by a phenol chloroform mixture (5:1, pH 4.3) and precipitated by ethanol. tRNA was dissolved in the buffer (10 mM Tris-HCl, 90 mM KCl, 3.0 mM Mg acetate, 0.5 mM EDTA, pH 7.4) and purified on a Superdex 75 column (GE Healthcare), equilibrated with the same buffer. tRNA was not aminoacylated. tRNA was concentrated to 2.9 mg/ml, frozen in liquid nitrogen in small aliquots and stored at −80°C.
mRNA
A 24-nucleotide long mRNA with an AUG start codon was chemically synthesized by Thermo Fisher Scientific, Inc. (Waltham, MA); the sequence was 5′-CAACAACAACAACAAAUGUUUCAA-3′.
Purification of 40S ribosomal subunits from rabbit reticulocyte lysate (RRL)
Frozen RRL was purchased from Green Hectares (Oregon, WI). The following components were added to 1 liter of thawed RRL: 5 mM HEPES-KOH (pH 7.1), 15 mM KCl, 11 mM Mg acetate, 1.0 mM EDTA (pH 8.0), 2 mM DTT, 10 μM MG132, 500 mg heparin, 150 mg Pefabloc, 12 tablets of Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN,). Ribosomes were pelleted by centrifugation at 40,000 rpm for 10.5 h in a Beckman Type 45 Ti rotor at 4°C through a sucrose cushion (7 ml/tube; 30% (w/v) sucrose, 20 mM Bis-Tris (pH 5.9), 200 mM NH4 acetate, 300 mM KCl, 10 mM Mg acetate, 5 mM DTT). The pellet was resuspended in 25 ml of buffer A (20 mM HEPES-KOH, 30 mM KCl, 11 mM Mg acetate, 1 mM EDTA, 2 mM DTT, 10 μM MG132, 1.0 mg/ml heparin, 0.2 mg/ml Pefablock, 1 tablet/100ml Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN,), 200 U/ml SUPERaseIN (Ambion®), pH 7.1). Subsequently, NH4Cl was added to the suspension to a final concentration of 0.4 M and the suspension was mixed on ice for at least 0.3 h. One third of the suspension was then clarified by centrifugation in an Eppendorf microcentrifuge for 10 min at 13,200 rpm at 4°C and loaded on a Sephacryl S-400 HR column (GE Healthcare) equilibrated with buffer B (20 mM Tris-HCl, 500 mM KCl,10 mM MgCl2, 5% (w/v) glycerol, 2 mM DTT, pH 7.4). This step was repeated for the rest of the ribosomal suspension. Fractions (8 ml) were collected and analyzed by SDS PAGE (4–12%, MES SDS running buffer, NuPAGE®). Those containing 80S ribosomes were combined, brought to a final concentration of 2.5 mM MgCl2 and 1% glycerol and concentrated to a final volume of 25 ml. Optical density of concentrated ribosomes (OD260) should be 200–350 U/ml. Ribosomes were subsequently supplemented with 1 mM puromycin and incubated for 20 min at 37°C followed by 20 min incubation at room temperature and then chilled on ice for at least 10 min.
Chilled ribosomes were clarified by centrifugation in an Eppendorf microcentrifuge for 10 min at 13,200 rpm at 4°C and loaded on a 10–40% sucrose density gradient, prepared in tubes for Beckman SW 32 Ti rotor (10–40% (w/v) sucrose, 20 mM Tris-HCl, 500 mM KCl, 5 mM Mg acetate, 2 mM DTT, pH 7.4). Sucrose density gradient centrifugation was performed at 25,000 rpm for 14.5 h in a Beckman SW 32 Ti rotor. Fractions (1 ml) were collected and analyzed by SDS-PAGE (4–12%, MES SDS running buffer, NuPAGE®). Fractions containing 40S subunits were combined, diluted five times with buffer B without KCl and loaded on a DEAE column equilibrated in buffer B with 50 mM KCl. Elution was performed by a 0.05–1.0 M KCl gradient in buffer B. Fractions containing 40S subunits were combined, concentrated to 391 U/ml (OD260) and dialyzed against buffer C (10 mM HEPES-KOH, 30 mM KCl, 5 mM Mg acetate, 5% (w/v) glycerol, 2 mM DTT, pH 7.1). Small aliquots were frozen in liquid nitrogen and stored at −80°C.
Activity test
Purified 40S ribosomal subunits were tested by toe printing analysis for their ability to form initiation complexes with mRNA containing CrPV IRES, as described earlier43.
Complex formation
Prior to crystallization, PIC1 complex was formed by mixing ribosomes and eIF1 to final concentrations of 2.6 μM and 9.1 μM, respectively, with subsequent dialysis against buffer DB (2.0 mM HEPES-KOH, 100 mM NH4Cl, 15 mM KCl, 3 mM Mg acetate, 2 mM TCEP, pH 7.1) for 2 h at room temperature. The complex was incubated at 37 °C for 15 min and kept at room temperature for crystallization setup. Complex PIC2 was formed by soaking eIF1A (final concentration 40 μM) during freezing crystals of the PIC1. The 48S PIC was formed similarly to PIC1 but with addition of eIF1A (final concentration 7.5 μM), mRNA (10 μM) and tRNAi (10 μM). Although the presence of eIF1 in the reaction mixture is essential for crystallization, this factor is not seen in the 48S PIC structure. We suggest that it promotes the homogeneity of the complex, which is required for crystal growth, by controlling the correct recognition of the initiation codon by tRNAi and by dissociating aberrantly assembled ribosomal complexes33,34.
Crystallization and cryoprotection
Crystals were grown in 24-well siting drop plates using vapor diffusion technique. 4 μl of the PIC1 were mixed with 4 μl of reservoir solution (50 mM Tris-HCl, 100 mM NH4 acetate, 5 mM Mg acetate, 50 mM sarcosine, 5 mM urea, 2.5% PEG 20000, 5% MPD, 2 mM TCEP, pH 8.5). Plates were incubated at 19 °C for 6 – 9 days. Crystals were stabilized by stepwise increase of MPD to the final concentration of 35%. After stabilization, crystals were frozen in a cryo stream. For PIC2 complex formation, crystals were stabilized in the same buffer containing 40 μM of eIF1A.
48S PIC (4 μl) was mixed with 7.4 μl of reservoir solution (50 mM Tris-HCl, 100 mM NH4 acetate, 5 mM Mg acetate, 50 μM Zn acetate, 2 mM putrescine, 2.5% PEG 20000, 5% 1,4-butanediol, 2 mM TCEP, pH 8.5). Plates were incubated at 19 °C for 6 – 13 days. Crystals were stabilized at 4°C by stepwise increase of 1,4-butanediol and PEG 20000 to the final concentration of 30% and 3.0%, respectively. After stabilization, crystals were frozen in liquid nitrogen.
Data collection and processing
Data collection was carried out at 100 °K at beamlines X25 and X29 at Brookhaven National Laboratory (Upton, NY) and 24ID at the Advanced Photon Source (Argonne, IL). A complete dataset was collected from a single crystal (PIC1, PIC2) or from two crystals (48S PIC, Supplementary Table 1). Data were integrated and scaled with the XDS program package44. All crystals belong to the hexagonal space group P3121 (Supplementary Table 1).
The structure was solved by molecular replacement using PHASER from the CCP4 program suite45. The search model was generated from the previously published structure of yeast 80S ribosome (ref. 20, PDB entries 3U5B, 3U5C). Both the 60S subunit and the suppressor protein STM1 were excluded from the search model. Before refinement all B factors in the model were set to an isotropic B of 80. The initial molecular replacement solution was refined by rigid body refinement with the 40S subunit split into multiple domains (Supplementary Table 2), followed by five cycles of grouped TLS and grouped B-factor refinement using PHENIX46. Bulk solvent correction was applied as recommended47. After initial refinement, the difference electron density corresponding to each initiation factor, mRNA and tRNA became clearly visible in the difference electron density map (Fobs – Fcalc, Fig. 1). The NMR structures of human eIF1 and eIF1A (refs 29, 39, PDB entries 2IF1, 1D7Q) without their flexible tails were docked into the corresponding electron density, followed by refinement using PHENIX46. Structures of the mRNA and P site tRNA were taken from the previously published structure of the 70S ribosome with bound mRNA and tRNAs (ref 13, PDB entry 2J00) were docked in the corresponding electron density in 48S PIC map and refined as above. The position of the mRNA (except AUG) was manually adjusted to fit the electron density before their refinement in PHENIX46. mRNA and tRNA were treated as one ridged body during refinement.
Despite that at resolution of about 8–11Å the structures cannot be build de novo, the availability of the high resolution models allows to detect the movement of the structural domains of the ribosome, locate positions of the bound protein factors and even antibiotics48–50. The conformational changes of the 40S subunit described in our manuscript are reproducible during refinement independently of the starting model: for example, refining the 40S structure from PIC1 against the data of PIC2 results in conformation of PIC2 while refining the 40S structure from PIC2 against the data of PIC1 results in conformation of PIC1. That assures us that movements of the domains of the 40S subunit that were observed are not biased to the starting models.
When the manuscript was under review the high resolution cryo-EM structure of the human 80S ribosome was published39. Because of the high identity between rabbit and human ribosomal proteins and 18S rRNA (Supplementary Table 3) we used the structure of the human 40S subunit for the final refinement.
Figures
Figures showing electron densities and atomic models were generated using PYMOL (DeLano, W.L. The PyMOL Molecular Graphics System (2002), http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank the members of the Steitz lab for useful suggestions and discussions, the staff of the Advanced Photon Source beamline 24-ID, the National Synchrotron Light Source beamlines X25 and X29 and the Richards Center facility at Yale University for support. Special thanks to Y. Polikanov, J. Wang and Y. Xiong for advice with crystallographic software. This work was supported by the National Institutes of Health (NIH) grant GM022778 (to T.A.S.).
Footnotes
Supplementary Information (Supplementary Figures, Supplementary Tables 1–3, Notes and Supplementary References) is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
I.B.L. designed and performed experiments, analyzed data and wrote the paper; T.A.S. analyzed data, wrote the paper and directed research.
The structural coordinates of PIC1, PIC2 and 48S PIC have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) under accession codes 4KZX, 4KZY, 4KZZ, respectively.
Reprints and permissions information is available at www.nature.com/reprints. T.A.S. owns stock in Rib-X Pharmaceuticals, which does structure-based design of drugs that target the bacterial ribosome.
References
- 1.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Current opinion in structural biology. 2009;19:300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiology and molecular biology reviews : MMBR. 2011;75:434–467, first page of table of contents. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/Nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 4.Voigts-Hoffmann F, Klinge S, Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Current opinion in structural biology. 2012 doi: 10.1016/j.sbi.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Lorsch JR, Dever TE. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. The Journal of biological chemistry. 2010;285:21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 7.Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic acids research. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic acids research. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews. Molecular cell biology. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Molecular cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Molecular cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 12.Noller HF, Hoang L, Fredrick K. The 30S ribosomal P site: a function of 16S rRNA. FEBS letters. 2005;579:855–858. doi: 10.1016/j.febslet.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 14.Pisarev AV, et al. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes & development. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fekete CA, et al. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. The EMBO journal. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic acids research. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes & development. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julian P, et al. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS biology. 2011;9:e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonetti A, et al. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 20.Dong J, et al. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes & development. 2008;22:2242–2255. doi: 10.1101/gad.1696608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 22.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. The EMBO journal. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 24.Wegrzyn JL, Drudge TM, Valafar F, Hook V. Bioinformatic analyses of mammalian 5′-UTR sequence properties of mRNAs predicts alternative translation initiation sites. BMC bioinformatics. 2008;9:232. doi: 10.1186/1471-2105-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter AP, et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 26.Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Molecular cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 27.Fekete CA, et al. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. The EMBO journal. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 29.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Molecular cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Schluenzen F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 31.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & development. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. The EMBO journal. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 34.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & development. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 36.Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. The EMBO journal. 1999;18:2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolitz SE, Takacs JE, Lorsch JR. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA. 2009;15:138–152. doi: 10.1261/rna.1318509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. Journal of molecular biology. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- 39.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
Methods References
- 40.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova TV, Hellen CU. Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Meth Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 43.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, et al. Overview of the CCP4 suite and current developmentsActa crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rees B, Jenner L, Yusupov M. Bulk-solvent correction in large macromolecular structures. Acta crystallographica Section D, Biological crystallography. 2005;61:1299–1301. doi: 10.1107/S0907444905019591. [DOI] [PubMed] [Google Scholar]
- 48.Vila-Sanjurjo A, et al. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc Natl Acad Sci USA. 2003;100:8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lancaster L, et al. The location of protein S8 and surrounding elements of 16S rRNA in the 70S ribosome from combined use of directed hydroxyl radical probing and X-ray crystallography. RNA. 2000;6:717–729. doi: 10.1017/s1355838200000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.