Highlights
-
•
We studied the expression of Pitpnm1 in the developing mouse inner ear.
-
•
We covered several ages between E14.5 and P5, and also looked at adults.
-
•
Pitpnm1 is expressed in the inner hair cells from before birth to adulthood.
-
•
Pitpnm1 is expressed transiently in the outer hair cells at early postnatal stages.
-
•
Mice lacking Pitpnm1 display no obvious auditory defects.
Abbreviations: ABR, auditory brain stem response; ANOVA, analysis of variance; E, embryonic day; EDTA, ethylenediaminetetraacetic acid; P, postnatal day; PCR, polymerase chain reaction
Key words: Pitpnm1, inner ear, hair cell, mouse mutant, deafness, Mir96
Abstract
Deafness is a genetically complex disorder with many contributing genes still unknown. Here we describe the expression of Pitpnm1 in the inner ear. It is expressed in the inner hair cells of the organ of Corti from late embryonic stages until adulthood, and transiently in the outer hair cells during early postnatal stages. Despite this specific expression, Pitpnm1 null mice showed no hearing defects, possibly due to redundancy with the paralogous genes Pitpnm2 and Pitpnm3.
Introduction
Deafness is the most common sensorineural disorder in the human population and is a particular problem in the older population; 60% of people over the age of 70 have a hearing loss of 25 dB or more, and would benefit from a hearing aid (Davis, 1995). Many genes are known to cause deafness in humans, but there are many more loci for which the gene responsible remains unknown (http://hereditaryhearingloss.org). One way to discover new genes involved in hearing is to carry out an expression screen, looking for genes specifically expressed in the inner ear. However, expression of a gene in a specific organ does not mean that the gene is critical for the development or function of that organ; studies of lack-of-function mutants are a necessary complement to an interesting expression pattern. Here we present a functional study of and expression data for such a gene which was found to be specifically expressed in hair cells.
Pitpnm1, also known as Nir2 or RdgB, was first found to be expressed in the hair cells in a study of mice mutant for the microRNA Mir96, being one of several genes that were markedly downregulated in the mutant (Lewis et al., 2009). The gene encodes a phosphatidylinositol transfer protein bound to the cytosolic side of the Golgi apparatus membrane (Aikawa et al., 1997, 1999; Lev et al., 1999; Litvak et al., 2002a). It is a homologue of the Drosophila retinal degeneration B gene, as are the two other members of the same family, Pitpnm2 and Pitpnm3 (Aikawa et al., 1997; Lev et al., 1999; Lu et al., 1999). Flies lacking RdgB display rapid photoreceptor degeneration (Harris and Stark, 1977).
The Pitpnm1 protein has two major functions: to regulate diacylglycerol (DAG) levels in the Golgi apparatus membrane and to regulate RhoA function (Litvak et al., 2002b, 2005; Tian et al., 2002). Through its phosphatidylinositol transfer domain, Pitpnm1 upregulates DAG and thereby increases the recruitment of other proteins required for trans Golgi network to plasma membrane transport (Jamora et al., 1999; Baron and Malhotra, 2002; Litvak et al., 2005). Pitpnm1 also helps maintain the Golgi apparatus structure (Litvak et al., 2005). With regard to RhoA, Pitpnm1 has an amino-terminal Rho inhibitory domain, which allows it to bind GDP–RhoA and prevent it from exchanging GDP (Guanosine diphosphate) for GTP Guanosine triphosphate (Tian et al., 2002). This inhibition of RhoA promotes cell extension, and facilitates cytokinesis since RhoA activity prevents cell division (Etienne-Manneville and Hall, 2002; Litvak et al., 2002b, 2004; Tian et al., 2002; Servotte et al., 2006; Morin et al., 2009).
miR-96 is a master regulator of inner ear development and is thought to coordinate a network of genes (Kuhn et al., 2011); thus, Pitpnm1 was an excellent candidate for further study of its role in inner ear function. We therefore examined its expression at different time points during development and characterised the hearing of mice lacking the gene.
Experimental procedures
Sample collection and preparation
Wildtype mice from the C57BL/6J-Tyrc-Brd strain were used for the main expression study. Embryonic samples were collected at embryonic day (E) 14.5, E16.5, and E18.5; the day of plug discovery was deemed E0.5. Postnatal pups were collected at the day of birth (P0), P3, P4 (for the Pitpnm1 null mice) and P5. The 9-week-old mice were from a mixed background of C57BL/6J-Tyrc-Brd and C57BL/6N. The heads for all samples were bisected, fixed in 10% formalin for two days, dehydrated and embedded in paraffin wax. The half heads from the 9-week-old mice were decalcified for two weeks in 10% EDTA after washing and before dehydration.
Immunohistochemistry
Embedded samples were cut into 8 μm thick sections along the sagittal plane. Immunohistochemistry was then carried out on slides using the Ventana Discovery machines (Ventana Roche, Tucson, AZ, USA) with the manufacturer’s reagents (CC1 (cat. no. 950-124), EZPrep (cat. no. 950-100), LCS (cat. no. 650-010), RiboWash (cat. no. 760-105), Reaction Buffer (cat. no. 95-300), and RiboCC (cat. no. 760-107)) according to the manufacturer’s instructions. The DABMap™ Kit (Ventana; cat. no. 760-124) with haematoxylin counterstain (cat. no. 760-2021) and bluing reagent (cat. no. 760-2037) was used. Slides covering the entire inner ear for three different mice at each age were stained, and the observed expression patterns were only considered reliable if present in all three samples. The primary antibody used was Abcam (Cambridge, Cambs, UK) goat polyclonal antibody to Nir2 (Pitpnm1) (ab22823), with the Jackson ImmunoResearch (West Grove, PA, USA) biotin-conjugated donkey anti-goat (705-065-147) secondary. All antibodies were diluted in ‘Antibody staining solution’: 10% foetal calf serum, 0.1% Triton, 2% bovine serum albumin (BSA) and 0.5% sodium azide in phosphate-buffered saline (PBS).
Controls were run for the secondary antibody, wherein the above immunohistochemistry protocol was used on slides at each age but the primary antibody was replaced with an equal volume of antibody staining solution. Some staining was observed in certain patches of brain tissue, but none at all in the cochlea or vestibular system.
Analysis
Stained slides were examined and images obtained using an AxioCam HRc camera mounted on a Zeiss microscope. Images were then processed in Photoshop CS2.
Generation and testing of knockout mice
The Pitpnm1 mutant allele (Pitpnm1tm1a(EUCOMM)Wtsi) is a “knockout-first” allele generated by the Sanger Institute Mouse Genetics Project as described in (Skarnes et al., 2011). The mutant was maintained on a C57BL/6N background. P4 pups were collected as described in Section “Sample collection and preparation” and tested with the same antibody used for the expression study. Genotyping was carried out by polymerase chain reaction (PCR) with the following primers: Wt_F: TCCCCTGAGGTAGTTGCTGAC; Wt_R: ATGTTGGCCCTGTGATGTTG; Mutant_F: TCCCCTGAGGTAGTTGCTGAC; Mutant_R: TCGTGGTATCGTTATGCGCC. The wildtype PCR gives a band of 355 bp in wildtypes and heterozygotes and the mutant PCR a band of 128 bp in heterozygotes and homozygotes.
Auditory brainstem response
In order to assess the hearing of mice homozygous for the Pitpnm1 mutation, mice were tested using auditory brainstem response measurements (ABR). Mice homozygous, heterozygous or wildtype for the targeted mutation Pitpnm1−/−, were anaesthetised and ABR recordings made by placing subcutaneous hook electrodes through the skin. Pure tone pips were presented in 5 ms bursts with a 1 ms rise/fall time, and responses to 6, 12, 18, 24 and 30 kHz were recorded, as were responses to multi-frequency 0.01 ms clicks. For some mice, additional frequencies were tested (3, 36 and 42 kHz). The responses to 256 repetitions were averaged using custom software, and the thresholds determined by identifying the lowest sound intensity at which any part of the ABR waveform was visually discernable (Ingham et al., 2011). Three ages were studied; 7, 14 and 24 weeks, and at least seven mice were tested for each genotype and age. A two-way analysis of variance (ANOVA) was used to determine whether there were significant differences between the genotypes at each frequency.
Real time PCR (RT-PCR) on Pitpnm genes
The organs of Corti of three P5 C57BL/6J-Tyrc-Brd mice were dissected out and stored at −20 °C in RNAlater stabilization reagent (cat. no. 76106, QIAGEN (Valencia, CA, USA)). RNA was extracted using QIAshredder columns (QIAgen, cat. no. 79654) and the RNeasy mini kit (QIAgen, cat. no. 74104), then treated with DNAse 1 (cat. no. AMP-D1, Sigma (St Louis, MO, USA)), all according to the manufacturer’s instructions. cDNA was generated from normalised quantities of RNA using the Superscript II Reverse Transcriptase kit (cat. no. 11904-018, Invitrogen (Carlsbad, CA, USA)). PCR was run with a touchdown protocol. Primer sequences were designed to cover at least one exon–exon junction:
All experiments were performed in accordance with the UK Home Office Animals (Scientific Procedures) Act 1986.
Results
Expression in wildtypes
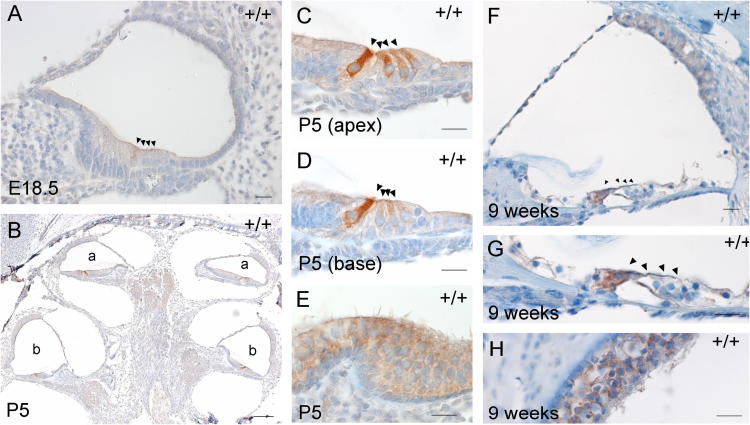
Specific expression of Pitpnm1 was visible from E18.5, when inner hair cells showed faint staining (Fig. 1A). Expression at P0 was generally identical to that seen at E18.5 but was more clear and well established (data not shown). By P3, all hair cells showed strong expression of Pitpnm1 (data not shown). Strikingly, at P5 all inner hair cells still maintained heavy expression, but outer hair cells showed a steep gradient: at the base outer hair cells showed essentially no Pitpnm1 expression, while at the apex expression in outer hair cells was similar to P3 levels (Fig. 1B–D). In the vestibular system, moderate levels of Pitpnm1 expression could be seen in both hair cells and supporting cells from E18.5, and were maintained at a fairly constant level up to P5 (Fig. 1E). At 9 weeks old, Pitpnm1 staining was still faintly visible in the inner hair cells and the vestibular system (Fig. 1F–H).
Fig. 1.
Expression of Pitpnm1. Pitpnm1 expression in wildtype mice at E18.5 (A), P5 (B–E) and 9 weeks old (F–H). (A) Only inner hair cells show expression at E18.5. (B) The organ of Corti at P5, with staining in all hair cells at the apex (a) but in the inner hair cell only at the base (b). (C) and (D) are close-ups of the apical turn and basal turn respectively. (E) Fainter staining in vestibular hair cells and supporting cells of the crista at P5. (F) Staining in the inner hair cells only at 9 weeks old; (G) is a close-up of (F). (H) Staining in the macula at 9 weeks old. Positive staining is in brown; tissue is counterstained blue. Cochlear hair cells are indicated by arrowheads. (A, C–H) Scale bar = 20 μm, (B) Scale bar = 100 μm.
Pitpnm1−/− mouse phenotyping
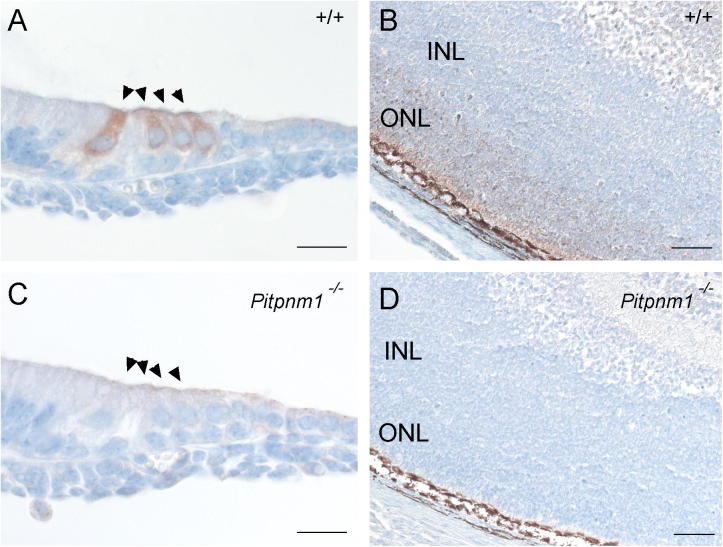
Pitpnm1 knockout mice are viable and fertile. The absence of Pitpnm1 protein in the knockout animals was confirmed by immunohistochemistry; Pitpnm1−/− animals had no expression in the hair cells or the retina (Fig. 2). The homozygous Pitpnm1 mutants were subjected to a broad phenotypic screen through the Sanger Institute Mouse Genetic Programme (http://www.sanger.ac.uk/mouseportal/). The only tests where Pitpnm1 null mice show a difference from the wildtype are the plasma chemistry tests, where male homozygotes showed a decreased circulating cholesterol level and hypocalcaemia (http://www.sanger.ac.uk/mouseportal/phenotyping/MCTJ/plasma-chemistry/), and the haematology test, where female homozygotes show a decrease in leucocyte cell number (http://www.sanger.ac.uk/mouseportal/phenotyping/MCTJ/haematology-cbc/). No obvious histopathological defects were observed in the eyes, and no consistent eye phenotype was visible in 13 mutants examined using a slit lamp, ophthalmoscope and endoscope (data not shown; http://www.sanger.ac.uk/mouseportal/, http://www.sanger.ac.uk/mouseportal/phenotyping/MCTJ/eye-morphology/).
Fig. 2.
Pitpnm1 expression in homozygote knockout mice at P4. Pitpnm1 expression is present at P4 in wildtype mice (A, B) but not in a homozygote littermate (C, D), in both hair cells (A, C; the middle turn of the cochlea is shown) and the retina (B, D). Positive staining is indicated by brown; tissue is counterstained blue (the brown cells in D are melanocytes in the retinal pigment epithelium). Hair cells are indicated by arrowheads. INL = inner nuclear layer; ONL = outer nuclear layer. Scale bar in A and C = 20 μm, bar in B and D = 50 μm.
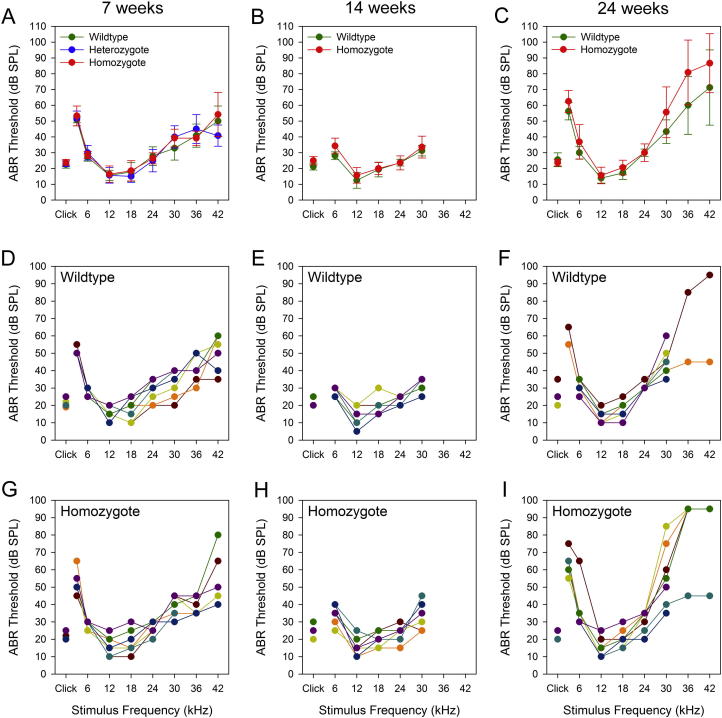
All Pitpnm1−/− homozygote animals had normal waveforms, wave amplitudes and latencies at all ages tested (data not shown). There was no significant difference in ABR threshold between wildtype and homozygote null animals at 7 weeks (ANOVA, p = 0.322, Fig. 3A). At 14 weeks there is a slight increase in threshold in homozygotes at 6 kHz (Fig. 3B) but it is not sufficient to be biologically meaningful. At 24 weeks, some individual mice showed increased thresholds at high frequencies, but this occurred in homozygotes and wildtypes and is probably just a reflection of variable age-related hearing loss on the C57BL/6N background (Fig. 3E, F, H, I).
Fig. 3.
ABR results of Pitpnm1 null mice. (A–C) ABR results showing average hearing thresholds from 3 to 42 kHz for wildtype (green), heterozygous (blue) and homozygous (red) mutant Pitpnm1 mice at 7 weeks (A), 14 weeks (B) and 24 weeks (C). Thresholds for click stimuli are shown as a single data point on the left of the graph. No difference is visible between wildtypes and mice carrying the null allele at 7 weeks (ANOVA, p = 0.322), and although there is a significant difference in 14-week-old mice at 6 kHz (ANOVA, p = 0.009), the difference is too small to be biologically meaningful. There is also a significant difference in 24-week-old mice at 30, 36 and 42 kHz (ANOVA, p = 0.022, 0.004 and 0.03 respectively), but in this case examination of the individual traces (F, I) show that the wildtypes are just as variable. Error bars are standard deviation. D–I) Thresholds for individual mice (one colour per mouse) at the three ages (D, G – 7 weeks; E, H – 14 weeks; F, I – 24 weeks), grouped by genotype; D–F are wildtypes and G–I homozygotes.
Expression of Pitpnm1, Pitpnm2 and Pitpnm3 in wildtypes
cDNA made from RNA from the organ of Corti of P5 wildtype mice was used as the template for PCR with primers against the spliced mRNA sequences of Pitpnm1, Pitpnm2 and Pitpnm3. All three genes produced a clear band in the samples tested, indicating that they are all expressed in the organ of Corti (Fig. 4).
Fig. 4.
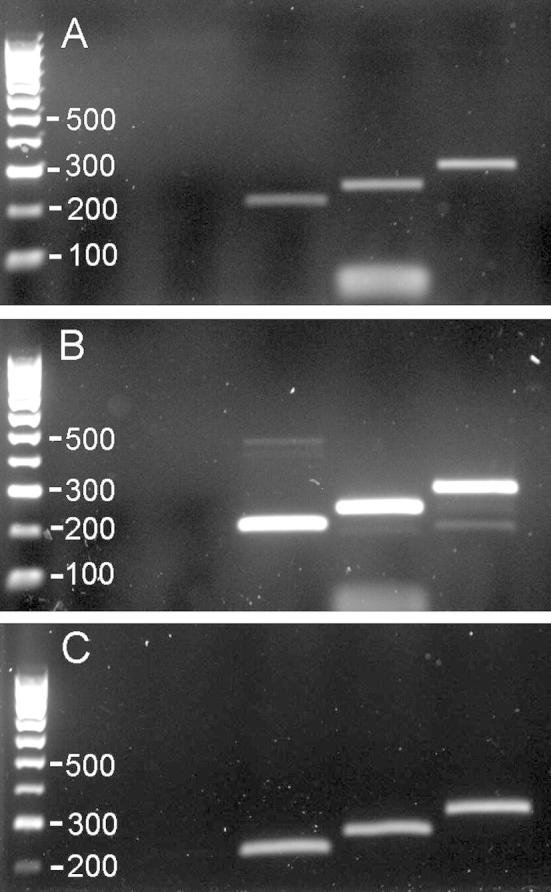
Organ of Corti RT-PCR for Pitpnm genes. RT-PCR results showing bands for Pitpnm1 (left), Pitpnm2 (centre) and Pitpnm3 (right) from cDNA from the organ of Corti of three separate mice (A–C). Ladder band size is shown on each figure; the Pitpnm bands are between 200 and 350 bp in size.
Discussion
The expression pattern of Pitpnm1 in the inner ear is both time- and cell-specific. The most intense expression at any stage is always seen in the inner hair cell, starting at E18.5 and remaining present there in adult ears. It is interesting that constant expression of Pitpnm1 at prenatal stages and in adults was seen in both inner hair cells and vestibular hair cells, while expression in the outer hair cells was transient, appearing to spread from the apex towards the base between P0 and P3, then retract from the base and become limited to the apex by P5. By 9 weeks old there was no expression detected in the outer hair cells. Both the inner and vestibular hair cells are responsible for transducing the sensory stimuli to afferent neural activity, while the outer hair cells, a feature specific to mammals, modulate the sensitivity of the basilar membrane. The difference in expression of Pitpnm1 may reflect these differences in function, specifically, the need for motility in outer hair cells. Pitpnm1 is known to inhibit RhoA, a GTPase important for the regulation of outer hair cell motility (Kalinec et al., 2000), and thus the absence of Pitpnm1 in mature outer hair cells may be a necessary feature specific to the outer hair cells to enable them to function. However, the role of Pitpnm1 in the vestibular and inner hair cells, a role which must either be unnecessary in outer hair cells or performed by another protein, remains unknown.
It is intriguing that despite the very specific expression in hair cells Pitpnm1 is not required for normal hearing. Although several of the older Pitpnm1−/− mice have raised thresholds, so do some of the wildtypes (Fig. 3F, I). The C57BL/6N strain is known to suffer from progressive deafness, and it is likely that this is causing the raised thresholds in some of the older animals tested. The absence of an auditory phenotype in mice lacking Pitpnm1 implies that the reduced Pitpnm1 expression seen in the Mir96 mutant does not contribute to its phenotype (Lewis et al., 2009).
The homozygous null mice showed very little difference to their wildtype siblings across a wide range of tests (http://www.sanger.ac.uk/mouseportal/phenotyping/MCTJ). Where they do differ, the differences are sex-specific, and in the case of the decreased circulating cholesterol levels of the male homozygotes, the readings cover a wide range of values, from higher than the wildtypes to much lower, and the difference may not be biologically meaningful. However, the potential link to hypocalcaemia is interesting because Pitpnm1 is a calcium-binding protein (Lev et al., 1999).
The lack of detection of Pitpnm1 by immunohistochemistry in either eye or ear in homozygous mutants (Fig. 2) demonstrates that the knockout is efficient. However, there are three Pitpnm genes, all of which are homologous to the Drosophila retinal degeneration B gene, and all of which are expressed in the organ of Corti (Fig. 4), so it is possible that one or both of the others are compensating for the lack of Pitpnm1. A knockout of Pitpnm2 has been studied and was found to have no phenotype despite specific expression in the retina (Lu et al., 2001), and it is notable that Pitpnm1 is also expressed in the retina (Fig. 2B) but no eye or vision abnormalities were observed in the Pitpnm1 knockout. No Pitpnm3 knockouts have been reported. It would be interesting to study a double or triple knockout, and determine if the lack of a phenotype in Pitpnm1 and Pitpnm2 null mice is due to compensation by the other Pitpnm family members.
Author contributions
The expression study was carried out by F.C. and ABR was done by S.P. M.L. did the staining of the mutant mouse ear and eye, and the RT-PCR on the three Pitpnm genes. The study was designed by K.P.S. and M.L., and the paper was written by F.C., M.L. and K.P.S.
Acknowledgements
We thank the Sanger Institute Mouse Genetics Project for generating the mutant mice, Rosalind Lacey for animal care and assistance with ABR, Neil Ingham for help with the ABR graphs, and Vinit Mahajan, Mary-Ann Mahajan, Stephen Tsang and Valerie Vancollie for unpublished data on the eye, as reported in the heatmap at http://www.sanger.ac.uk/mouseportal/. This work was supported by the Wellcome Trust (Grant No. 098051).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
F.A. Carlisle, Email: francesca.carlisle@gmail.com.
S. Pearson, Email: sp9@sanger.ac.uk.
K.P. Steel, Email: karen.steel@kcl.ac.uk.
M.A. Lewis, Email: morag.lewis@kcl.ac.uk.
References
- Aikawa Y., Hara H., Watanabe T. Molecular cloning and characterization of mammalian homologues of the Drosophila retinal degeneration B gene. Biochem Biophys Res Commun. 1997;236:559–564. doi: 10.1006/bbrc.1997.7009. [DOI] [PubMed] [Google Scholar]
- Aikawa Y., Kuraoka A., Kondo H., Kawabuchi M., Watanabe T. Involvement of PITPnm, a mammalian homologue of Drosophila rdgB, in phosphoinositide synthesis on Golgi membranes. J Biol Chem. 1999;274:20569–20577. doi: 10.1074/jbc.274.29.20569. [DOI] [PubMed] [Google Scholar]
- Baron C.L., Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- Davis A. Whurr; London: 1995. Hearing in adults. [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Harris W.A., Stark W.S. Hereditary retinal degeneration in Drosophila melanogaster. A mutant defect associated with the phototransduction process. J Gen Physiol. 1977;69:261–291. doi: 10.1085/jgp.69.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham N.J., Pearson S., Steel K.P. Using the auditory brainstem response (ABR) to determine sensitivity of hearing in mutant mice. Curr Protoc Mouse Biol. 2011;1:279–287. doi: 10.1002/9780470942390.mo110059. [DOI] [PubMed] [Google Scholar]
- Jamora C., Yamanouye N., Van Lint J., Laudenslager J., Vandenheede J.R., Faulkner D.J., Malhotra V. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell. 1999;98:59–68. doi: 10.1016/S0092-8674(00)80606-6. [DOI] [PubMed] [Google Scholar]
- Kalinec F., Zhang M., Urrutia R., Kalinec G. Rho GTPases mediate the regulation of cochlear outer hair cell motility by acetylcholine. J Biol Chem. 2000;275:28000–28005. doi: 10.1074/jbc.M004917200. [DOI] [PubMed] [Google Scholar]
- Kuhn S., Johnson S.L., Furness D.N., Chen J., Ingham N., Hilton J.M., Steffes G., Lewis M.A., Zampini V., Hackney C.M., Masetto S., Holley M.C., Steel K.P., Marcotti W. MiR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc Natl Acad Sci USA. 2011;108:2355–2360. doi: 10.1073/pnas.1016646108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Hernandez J., Martinez R., Chen A., Plowman G., Schlessinger J. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol Cell Biol. 1999;19:2278–2288. doi: 10.1128/mcb.19.3.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.A., Quint E., Glazier A.M., Fuchs H., De Angelis M.H., Langford C., van Dongen S., Abreu-Goodger C., Piipari M., Redshaw N., Dalmay T., Moreno-Pelayo M.A., Enright A.J., Steel K.P. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V., Argov R., Dahan N., Ramachandran S., Amarilio R., Shainskaya A., Lev S. Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Mol Cell. 2004;14:319–330. doi: 10.1016/s1097-2765(04)00214-x. [DOI] [PubMed] [Google Scholar]
- Litvak V., Dahan N., Ramachandran S., Sabanay H., Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- Litvak V., Shaul Y.D., Shulewitz M., Amarilio R., Carmon S., Lev S. Targeting of Nir2 to lipid droplets is regulated by a specific threonine residue within its PI-transfer domain. Curr Biol. 2002;12:1513–1518. doi: 10.1016/s0960-9822(02)01107-7. [DOI] [PubMed] [Google Scholar]
- Litvak V., Tian D., Carmon S., Lev S. Nir2, a human homolog of Drosophila melanogaster retinal degeneration B protein, is essential for cytokinesis. Mol Cell Biol. 2002;22:5064–5075. doi: 10.1128/MCB.22.14.5064-5075.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Peng Y.W., Shang J., Pawlyk B.S., Yu F., Li T. The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 2001;107:35–41. doi: 10.1016/s0306-4522(01)00337-2. [DOI] [PubMed] [Google Scholar]
- Lu C., Vihtelic T.S., Hyde D.R., Li T. A neuronal-specific mammalian homolog of the Drosophila retinal degeneration B gene with expression restricted to the retina and dentate gyrus. J Neurosci. 1999;19:7317–7325. doi: 10.1523/JNEUROSCI.19-17-07317.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P., Flors C., Olson M.F. Constitutively active RhoA inhibits proliferation by retarding G(1) to S phase cell cycle progression and impairing cytokinesis. Eur J Cell Biol. 2009;88:495–507. doi: 10.1016/j.ejcb.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servotte S., Zhang Z., Lambert C.A., Ho T.T., Chometon G., Eckes B., Krieg T., Lapiere C.M., Nusgens B.V., Aumailley M. Establishment of stable human fibroblast cell lines constitutively expressing active Rho-GTPases. Protoplasma. 2006;229:215–220. doi: 10.1007/s00709-006-0204-0. [DOI] [PubMed] [Google Scholar]
- Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., de Jong P.J., Stewart A.F., Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Litvak V., Toledo-Rodriguez M., Carmon S., Lev S. Nir2, a novel regulator of cell morphogenesis. Mol Cell Biol. 2002;22:2650–2662. doi: 10.1128/MCB.22.8.2650-2662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]