Abstract
In corals, biocalcification is a major function that may be drastically affected by ocean acidification (OA). Scleractinian corals grow by building up aragonitic exoskeletons that provide support and protection for soft tissues. Although this process has been extensively studied, the molecular basis of biocalcification is poorly understood. Notably lacking is a comprehensive catalog of the skeleton-occluded proteins—the skeletal organic matrix proteins (SOMPs) that are thought to regulate the mineral deposition. Using a combination of proteomics and transcriptomics, we report the first survey of such proteins in the staghorn coral Acropora millepora. The organic matrix (OM) extracted from the coral skeleton was analyzed by mass spectrometry and bioinformatics, enabling the identification of 36 SOMPs. These results provide novel insights into the molecular basis of coral calcification and the macroevolution of metazoan calcifying systems, whereas establishing a platform for studying the impact of OA at molecular level. Besides secreted proteins, extracellular regions of transmembrane proteins are also present, suggesting a close control of aragonite deposition by the calicoblastic epithelium. In addition to the expected SOMPs (Asp/Glu-rich, galaxins), the skeletal repertoire included several proteins containing known extracellular matrix domains. From an evolutionary perspective, the number of coral-specific proteins is low, many SOMPs having counterparts in the noncalcifying cnidarians. Extending the comparison with the skeletal OM proteomes of other metazoans allowed the identification of a pool of functional domains shared between phyla. These data suggest that co-option and domain shuffling may be general mechanisms by which the trait of calcification has evolved.
Keywords: biomineralization, calcium carbonate skeleton, scleractinian, proteomics, evolution
Introduction
It is generally accepted that anthropogenic CO2 emissions cause deleterious effects, not only on the atmosphere (i.e., global warming) but also on seawater chemistry. Since pre-industrial times, the average pH of seawater has fallen from 8.2 to 8.1, and this ocean acidification (OA) is thought to have significantly affected marine organisms in a variety of ways (Caldeira and Wickett 2003; Kline et al. 2012), including impacts on important physiological functions such as calcification (Orr et al. 2005; Cohen et al. 2009; Kroeker et al. 2013). This process is essential for a wide range of marine metazoans and fulfils a diverse array of important physiological and ecological roles (Cusack and Freer 2008). In the neritic domain of tropical and subtropical regions, the most prominent calcium carbonate producers are cnidarians, in particular scleractinians (stony corals), where coral reefs have an annual net production ranging from 5 to 126 mol CaCO3 m−2 year−1 (Milliman and Droxler 1996; Gattuso et al. 1998). Recent studies have shown that OA and increasing sea surface temperatures decrease coral calcification rates (Cohen and Mcconnaughey 2003; Langdon and Atkinson 2005; Orr et al. 2005; De’ath et al. 2009; Kleypas and Yates 2009; Pandolfi et al. 2011; Chan and Connolly 2013). However, most of the data supporting these negative effects are obtained at the organism/colony level and only few studies have considered the underlying molecular and cellular mechanisms. Of particular significance is the fact that the molecular machinery of coral biomineralization (the “calcification toolkit”) is, as yet, poorly characterized. Thus, a better understanding of the process is required, in particular at the level of the mineralizing space, where CaCO3 crystals are formed and shaped (Allemand et al. 2011). A number of candidate genes for roles in coral calcification have been identified for being expressed in the calicoblastic ectoderm (Zoccola et al. 2004, 2009; Moya et al. 2008). This number was recently increased with high-throughput analyses (Meyer et al. 2011; Shinzato et al. 2011; Moya et al. 2012), whereas microarray studies led to the identification of the SCRiPs, a family of small cysteine-rich proteins showing molecular features suggestive of an involvement in calcification processes (Sunagawa et al. 2009). Amongst the most obvious candidates for roles in coral calcification are the genes that encode skeletal organic matrix proteins (SOMPs), that is, proteins secreted by the calcifying tissues—the calicoblastic ectoderm in corals—which are occluded in the skeleton during its formation (Tambutté et al. 2008). These proteins are the main components of the skeletal organic matrix (OM), which in corals contains also polysaccharides (Goldberg 2001), glycoproteins (Dauphin and Cuif 1997), and lipids (Farre et al. 2010). The SOMPs have been characterized biochemically “in bulk” (i.e., without fractionation) in different coral species (Cuif, Dauphin, Gautret 1999; Cuif, Dauphin, Freiwald, et al. 1999; Ingalls et al. 2003; Puverel et al. 2005), the compositional analyses have shown that it is enriched in aspartic acid and, in lesser extent, in glutamic acid and glycine. This feature, together with the presence of saccharidic moieties, makes the OM unusually acidic. Moreover, its interaction with calcium carbonate was observed experimentally in the scleractinians Balanophyllia europaea (Goffredo et al. 2011), Acropora digitifera, Lophelia pertusa, Montipora caliculata (Falini et al. 2013) as well as by OM produced by coral cells of Stylophora pistillata (Helman et al. 2008), suggesting an important role of the OM in coral calcification.
Although several OM proteins have been extensively studied in other calcifying metazoans, such as molluscs (Marin et al. 2012) and sea urchins (Veis 2011), their characterization is still in its infancy for corals. Only recently an approach combining proteomics with genomics was applied unveiling a set of partial and complete sequences of proteins in the skeleton of the hexacoral S. pistillata (Drake et al. 2013). Prior to this study, only the full sequence of two SOMPs, galaxin from the scleractinian Galaxea fascicularis (Fukuda et al. 2003), and scleritin from the octocoral Corallium rubrum (Debreuil et al. 2012) were published and a number of other partial sequences from octocorals (Rahman and Isa 2005; Rahman et al. 2005; Rahman, Isa, Takemura, et al. 2006; Rahman, Isa, Uehara 2006) and scleractinians (Puverel et al. 2005).
One prerequisite for understanding the process of coral calcification is a comprehensive survey of the skeletal matrix proteins. In this article, we address this requirement, by describing 36 extracellular proteins that constitute the SOMP repertoire, or “skeletome,” of the scleractinian coral Acropora millepora. This is the first large-scale survey of the proteins present in the skeleton of a member of the acroporid family and the second in cnidarians. These proteins give new insights on the molecular tools required for controlling the deposition of the calcium carbonate and provide a platform for investigating the impact of environmental factors on calcification at the molecular level. In addition, they open novel perspectives on the mechanisms of evolution of SOMPs within the Cnidaria and, more broadly, within other calcifying metazoans.
Results and Discussion
Analysis of the Matrix on Gel
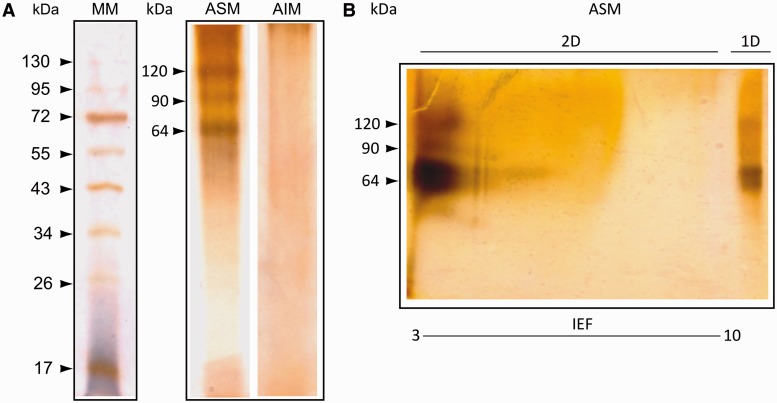
Coral skeleton was treated with bleach twice to remove contaminating tissue, symbionts, and bacteria, that can be entrapped in the highly porous skeleton (supplementary fig. S1, Supplementary Material online), the second treatment being conducted after the skeleton had been reduced to fine powder (granulometry < 200 µm). The residual powder was then extracted with 10% acetic acid, yielding acid-soluble matrix (ASM) and acid-insoluble matrix (AIM) fractions, assumed to be contaminant-free and closely associated with the aragonite skeleton. This extraction process yielded approximately 0.034 ± 0.01% of ASM and 0.23 ± 0.03% (wt/wt) of AIM relative to the skeleton weight. 1D-gel electrophoresis of the ASM extract (fig. 1A) showed three main discrete bands at around 120, 90, and 64 kDa embedded in a diffuse background of more dispersed “smearing” macromolecules, whereas the AIM was characterized by a smear lacking discrete bands. On the 2D gel (fig. 1B), the 90 and 64 kDa fractions, and, to a lesser extent, the 120 kDa fraction, in the ASM are characterized by large spots circa pI 3, suggesting that these macromolecules are very acidic.
Fig. 1.
Electrophoretic analysis of the ASM and AIM after AgNO3 staining on (A) 12% poly-acrylamide SDS-polyacrylamide gel electrophoresis (PAGE) gel; (B) 4–10% precast poly-acrylamide gel using an IPG (3–10) strip in the first dimension, under denaturing conditions.
Identification and Characterization of SOMPs
Raw data generated by LC-MS/MS on the ASM and AIM were analyzed with the software MASCOT to search against a pooled database consisting of transcripts and predicted proteins of A. millepora and the genus Symbiodinium.
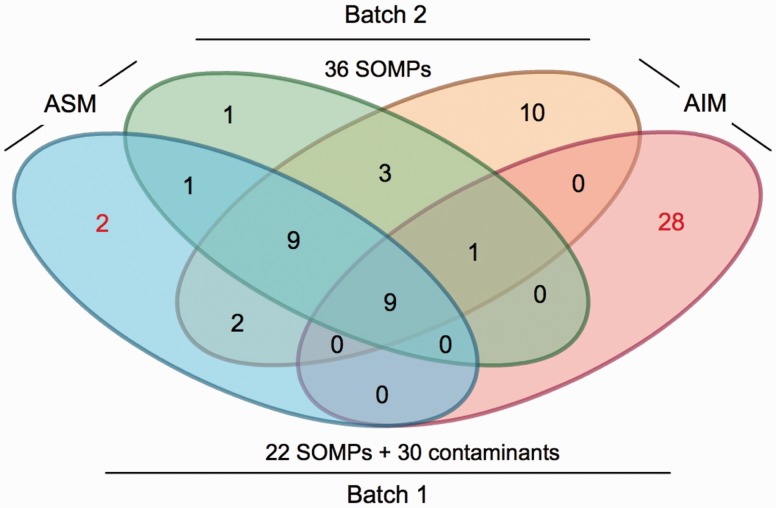
This procedure was initially made on the AIM and ASM prepared after only a single (whole skeleton) bleach treatment (“first batch”), but was then repeated on OM fractions (“second batch”) after the second bleaching step (i.e., on skeleton reduced to fine powder) outlined earlier. This enabled to analyze the effect of extended bleaching on removal of contaminants (fig. 2). Although the proteomic analyses of the “first batch” revealed intracellular coral proteins and proteins of symbiont origin (Ramos-Silva et al. 2013), the second batch was free of such contaminants, enabling the detection of 43 unique A. millepora proteins likely to constitute the SOMP repertoire, that is, proteins that are strongly associated to the skeleton. Of these 43 proteins, 36 assignments could be made with high confidence (i.e., with more than one unique peptide matching the sequence, supplementary table S1, Supplementary Material online), whereas seven sequences each with only a single peptide match were dropped from the list (supplementary table S2, Supplementary Material online) despite their properties being generally consistent with those of the high-confidence data set. Figure 3 provides a schematic overview of the domain structure and general properties of the Acropora SOMPs that were classified based on the in silico sequence analyses. From the 36 SOMPs identified, 22 proteins were common to both ASM and AIM (supplementary table S1, Supplementary Material online). Similar ASM and AIM protein repertoires have also been observed in the case of mollusc shell matrices (Marie et al. 2009). On the other hand, 12 SOMPs were exclusively associated with the insoluble organic fraction of the skeleton, and only 2 with the soluble matrix (supplementary table S1, Supplementary Material online). Possible causes for this apparent bias (fig. 2) between matrices include the over/under representation of certain peptides that is inherent to the proteomic approach, and the fact that some proteins from the acid-soluble fraction may bind acid-insoluble components.
Fig. 2.
Comparison of the proteins identified by proteomics on the ASM and AIM in two different conditions, batch 1 and batch 2. Batch 1 consisted of treating the skeletal fragments with sodium hypochlorite once, whereas batch 2 consisted of batch 1 followed by a subsequent similar treatment on the skeletal-sieved powder (<200 µm). Extracts from batch 1 showed evidence of contamination with the identification of specific intracellular proteins from Acropora millepora (tubulins, histones, ATP-synthase) and proteins from zooxanthellae: 2 contaminants of 23 identifications in the ASM and 28 contaminants of 38 identifications in the AIM. In contrast, no contaminants were identified in batch 2, indicating that a second bleach treatment on powder is effective in removing potential sources of contamination and is required for obtaining exclusively SOMPs. The 22 SOMPs identified in batch 1 were also present in batch 2.
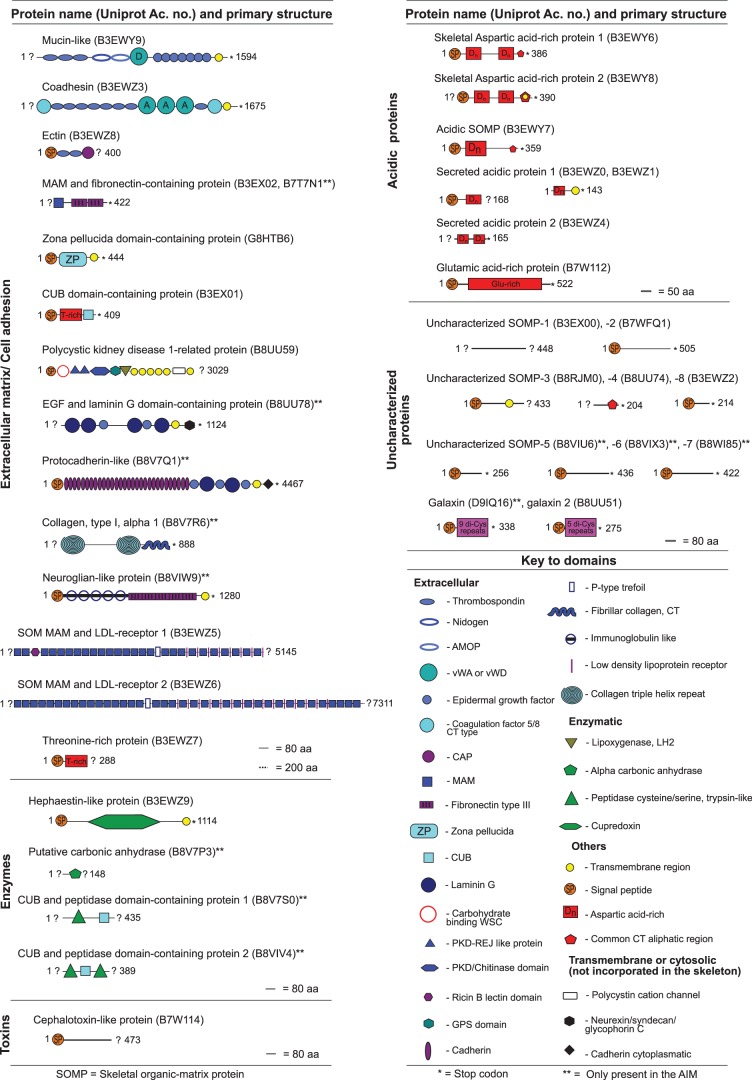
Fig. 3.
List of SOMPs identified by proteomics in the acid soluble and insoluble matrices extracted from Acropora millepora skeleton. Proteins were named according to the characterization of their primary structure.
Acidic Proteins
In both ASM and AIM, the most abundant proteins (high emPAI values; Ishihama et al. 2005; supplementary table S1, Supplementary Material online) were acidic (fig. 1B), that is, with pI < 4.5 (Marin and Luquet 2007) and rich in negatively charged residues (Asp, Glu). Asp and Glu-rich proteins are supposed to interact directly with calcium carbonate crystals promoting crystal nucleation, determining the growth axes and inhibiting the crystal growth (Wheeler et al. 1981; Addadi et al. 1987). Because of their polyanionic character, Asp and Glu-rich proteins usually have high-capacity, but low-affinity, calcium-binding properties (Maurer et al. 1996). Among the A. millepora SOMPs, six acidic proteins were identified (fig. 3), including two similar (48.8% identity; supplementary fig. S2, Supplementary Material online) proteins each containing approximately 20% of Asp residues and named as SAARP1 (B3EWY6) and 2 (B3EWY8) (secreted acidic Asp-rich protein). These proteins are likely to belong to a family of highly acidic proteins conserved across scleractinians. Indeed two Asp-rich proteins, named coral acid-rich proteins (CARPs), were recently identified in S. pistillata skeleton (Drake et al. 2013). Similarly to CARPs, SAARPs contain two Asp-rich regions intercalated by two nonacidic regions (fig. 3). In particular, CARP 4 (M1SN56) exhibits the highest identity percentage with the two SAARPs, 60.6% and 45.2%, for SAARP 1 and 2, respectively. However, the predicted protein sequences of CARP4 and CARP5 (M1RYM2) are shorter than SAARPs and lack a putative peptide signal required for targeting these proteins to the secretory pathway. Interestingly, the four acidic proteins also show significant identity, in the nonacidic regions, with the protein fragment P27 (M1TA76) identified in the skeleton of S. pistillata (Drake et al. 2013) (supplementary fig. S3, Supplementary Material online); however, this similarity is not understood from a functional viewpoint.
The two SAARPs together with a third protein, acidic SOMP (B3EWY7), corresponded to the identifications with the highest emPAI scores (supplementary table S1, Supplementary Material online). The acidic SOMP has lower Asp content (9.9%) and its sequence does not contain two Asp-rich regions as in SAARPs (fig. 3). Still, it shows significant similarities in the nonacidic region with both SAARPs (supplementary fig. S4, Supplementary Material online).
The three other skeletal proteins included the secreted acidic proteins Amil-SAP1 (B3EWZ0, B3EWZ1), Amil-SAP2 (B3EWZ4) (19% and 21.6% of Asp and Glu, respectively), and one Glu-rich protein (B7W112). Amil-SAP1 and Amil-SAP2 are the counterparts of Adi-SAP1 (supplementary fig. S5, Supplementary Material online) and Adi-SAP2, respectively, which have been hypothesized to have roles in calcification along with seven other proteins, Adi-SAP3 to -SAP9, in Acropora digitifera, on the basis of their high Asp plus Glu content (Shinzato et al. 2011). Of these acidic proteins, only SAP1 and two exhibit a predicted signal peptide, and are confirmed here to be true SOMPs. BLASTP searches with the six acidic SOMPs retrieved significant matches (E value < 10−4) with proteins in the UniprotKB database. Interestingly, among the best matches for Amil-SAP1 and the SAARPs were shell matrix proteins (aspeins) from Pinctada fucata, P. maxima, and Isognomon perna. However, in these cases, similarity scores might be largely misleading due to the relatively low complexity of the sequences, dominated by acidic residues; note also that none of these SOMPs had significant similarity to annotated domains from the InterPro database. Moreover, these data confirm the direct involvement of acidic proteins in skeletal formation in A. millepora.
Unique Uncharacterized Proteins
High-throughput approaches such as proteomics and transcriptomics have often revealed completely novel proteins that lack conserved domains or significant database matches that would allow to classify them into families and to hypothesize about their functions (Ramos-Silva et al. 2012). Galaxin exemplifies the case of such “orphans” because its function remains unknown (Reyes-Bermudez et al. 2009). In A. millepora skeleton, we identified two distinct galaxins along with further eight uncharacterized “orphan” SOMPs (fig. 3) referred here as USOMPs. These proteins do not constitute a single group and, with the exception of three proteins, exhibit no significant similarities. Two of these exceptions are galaxin proteins, and the third is provided by USOMP-4 (B8UU74), with a region near the C-terminus that has similarity to three of the acidic proteins discussed earlier (supplementary fig. S4, Supplementary Material online). The presence of multiple galaxin-related genes has previously been reported (Reyes-Bermudez et al. 2009; Shinzato et al. 2011); one of the galaxins identified here corresponds to GenBank ADI50283.1, a previously known gene from A. millepora (Reyes-Bermudez et al. 2009). The second sequence (galaxin-2, B8UU51) is orthologous with galaxin 2 from A. digitifera (Shinzato et al. 2011). The two A. millepora galaxins have 31.5% identity overall, the di-Cys repeat region being most similar. Note that the di-Cys pattern might indicate a role in the assembly and support of the macromolecular framework (Fukuda et al. 2003). Indeed a multiple alignment with five galaxins and similar sequences from Nematostella vectensis (GIs: 156374951 and 156377965) and Hydra magnipapillata (GI: 221103149) shows the conservation of at least nine double Cys motifs (supplementary fig. S6, Supplementary Material online). Similarly in S. pistillata skeleton, three proteins were identified, P12 (M1GFJ4), P16 (M1SN43), and P22 (M1T243) (Drake et al. 2013), which can fit in our description of uncharacterized proteins. From these, P16 and P22 are completely unique, whereas P12 shows similarities with USOMP8 (supplementary table S6, Supplementary Material online).
Extracellular Matrix-Like Proteins
Other SOMPs, although in some cases lacking clear overall matches, contain conserved regions or domains. The largest group comprises those with one or more domains usually associated with extracellular matrix (ECM) or cell-adhesion proteins from other metazoans. Amongst the 15 ECM-related proteins identified in the skeletal OM of A. millepora most contained multiple domains (fig. 3). Several of these ECM-related SOMPs resembled human cell-adhesion proteins such as mucin-4 (mucin-like SOMP, B3EWY9) and hemicentin-1 (coadhesin, B3EWZ3) (supplementary table S1, Supplementary Material online), which are heavily glycosylated and rich in cysteine residues, thereby forming disulfide bridges involved in modulating their adhesive properties. SOMPs that exhibit mucin-like or co-adhesin signatures have previously been reported in the mineralizing matrices of molluscs (Marin et al. 2000; Suzuki et al. 2009; Joubert et al. 2010), such as thrombospondin type 1 repeats, von Willebrand factor type A and epidermal growth factor-like domains, which are typical of ECM proteins involved in cell–cell and cell–substrate adhesion and in the binding of other macromolecules (Engel 1991; Mosher and Adams 2012). Moreover, many ECM domains present in Acropora’s skeletal proteome have been detected in previous high-throughput proteomic studies on the OM of molluscs, sea urchin, chicken eggshell, and coral (supplementary table S5, Supplementary Material online) but only the polycystic kidney disease 1 (PKD1)-related protein (B8UU59) contains domains (lipoxygenase, egg-jelly receptor, and PKD/chitinase) not previously found in proteomic analyses of other skeletal organic matrices. On the other hand some of the data sets used in these comparisons consist of broader lists of proteins, including some of intracellular origin that may be derived from cell debris and are most likely not involved in biomineralization (Mann et al. 2010; Zhang et al. 2012). Thus, comparisons were interpreted carefully and taking into account other evidences such as the expression in skeleton secreting-tissues (Jonchère et al. 2010; Marie et al. 2012; Zhang et al. 2012), the direct interaction with CaCO3 or the detection in the calcifying mediums (Hincke et al. 2000) (supplementary table S5, Supplementary Material online).
From the 15 ECM-related SOMPs, 8 have homology domains with proteins from the S. pistillata skeletal proteome (supplementary tables S5 and S6, Supplementary Material online): mucin-like, coadhesin, ectin (B3EWZ8), epidermal growth factor (EGF) and laminin G domain-containing protein (B8UU78), Meprin, A-5 protein, and receptor protein-tyrosine phosphatase Mu (MAM) and LDL-receptor domain-containing protein 1 (B3EWZ5) and 2 (B3EWZ6), zona pellucida domain-containing protein (G8HTB6), and protocaherin-like (B8V7Q1). Still, apart from protocadherin, we could not confirm candidate orthologous for these SOMPs in S. pistillata because most sequences correspond to fragments. Four ECM-related SOMPs were exclusively detected in the AIM, two of them having homologs usually found in the central nervous system in metazoans (Bieber et al. 1989; Frank and Kemler 2002)—protocadherin-like and neuroglian-like protein (B8VIW9), also the EGF and laminin G domain-containing protein and finally collagen (B8V7R6). The latter is homologous to the human alpha-2 type I collagen (P08123), which is found in bone (Glimcher 1959; Veis and Sabsay 1987). Fibrils of collagen are generally in close contact with the mineral phase and remain as an insoluble component of the organic fraction throughout the extraction process. Although collagen has been detected in the spicules of gorgonian (Goldberg 1974; Kingsley et al. 1990) and scleractinian corals (Drake et al. 2013), this is the first report of the occurrence of collagen type I in the OM of a coral skeleton.
Enzymes
Four of the SOMPs identified correspond to three types of enzymes (fig. 3). The first enzyme corresponds to a 148 amino acid (aa) fragment (B8V7P3) of an alpha-type carbonic anhydrase (CA), a zinc metalloenzyme that catalyzes the reversible hydration of carbon dioxide. CA activity has been detected in the skeletal OM of both zooxanthellate (Acropora hebes; Isa and Yamazato 1984) and azooxanthellate (Tubastrea aurea; Tambutté et al. 2006) scleractinians.
In the case of the CA identified here, its best BLAST hit in UniprotKB is the α-CA from S. pistillata, STPCA-2 (C0IX24) (supplementary table S1, Supplementary Material online). This enzyme was previously shown to be highly active compared with other CAs and was localized in the oral endoderm and aboral tissue (Bertucci et al. 2011). Despite its high similarity with the cytosolic human carbonic anhydrase II, STPCA-2 has unusual features such as a putative signal peptide and an insert of 30 amino acids between positions 212 and 243. Moreover, the same protein was also identified in the skeleton of S. pistillata by proteomics (Drake et al. 2013). These results, together with the data presented here for Acropora, directly link one specific CA to skeleton formation. Similarly, to a mechanism proposed for some molluscs (Miyamoto et al. 1996), the CA found here may display its enzymatic function in the extracellular calcifying medium, and be subsequently occluded in the growing skeleton.
The second SOMP (B3EWZ9), hephaestin-like, is a secreted member of the copper oxidase family with 1,114 aa length, containing a copper-binding site and a cupredoxin domain. Hephaestin proteins function as copper-dependent ferroxidases, mainly involved in iron and copper metabolism at membranes (Vulpe et al. 1999; Lang et al. 2012). Although to date there is no experimental evidence of activity of this type in mineralizing matrices, it is reasonable to propose that the hephaestin-like protein catalyzes the oxidation of iron (from Fe2+ to Fe3+) in the compartment where aragonite precipitation occurs. It was previously suggested that corals may entrap iron in the skeleton as a detoxification mechanism when high concentrations of the metal are present in the reef (Brown et al. 1991). Alternatively, the incorporation of iron in the skeleton of A. millepora may be a normal process, regulated by hephaestin. Interestingly, cupredoxin domains have also been identified in the shell of the mollusc Crassostrea gigas (in the L-ascorbate oxidase, K1PLZ9; Laccase-15, K1QQA2 and Laccase-18, K1PMS4) and in the tooth and spicules of the echinoderm S. purpuratus (in a protein similar to hephaestin, H3JMP5) (supplementary table S5, Supplementary Material online). Finally, the third enzymatic function assigned is observed in two SOMPs (B8V7S0 and B8VIV4), exclusive to the AIM. It corresponds to proteases containing peptidase S1/S6, chymotrypsin/Hap and C1r/C1s, Uegf, Bone morphogenetic protein 1 (CUB) domains.
Even though they are present at relatively lower abundance (supplementary table S1, Supplementary Material online), the enzymes detected here may play crucial roles in the supply of bicarbonate ions at the site of calcification (CA), in the regulation of iron and copper metabolism (hephaestin), and in the assembly/cleavage of the OM (proteases).
Toxin-Like
One SOMP (B7W114) corresponds to a secreted protein that has high similarity (50%) with the N-terminus of a toxin (of 1,052 aa long) from the cephalopod Sepia esculenta (SE-cephalotoxin) (B2DCR8; BLASTP E value = 8−43), whereas a number of toxins have been characterized from cubozoans (box jellyfish) (Brinkman et al. 2012) and anthozoans (sea anemones) (Frazão et al. 2012). However, very little is known about toxins in scleractinian corals, though studies have demonstrated antibacterial activity of the mucus (Gochfeld and Aeby 2008; Geffen et al. 2009).
The level of identity between B7W114 and the cephalotoxin (supplementary fig. S7, Supplementary Material online) is suggestive of conserved function. If the toxic character of the toxin-like SOMP is later confirmed, we may hypothesize that this protein acts in the coral skeleton in a similar way to the lysozyme in the chicken eggshell (Hincke et al. 2000), which due to its well-known anti-microbial properties was suggested to have a protective function in the eggshell in addition to its ability to interact with calcium carbonate.
Proteins with Transmembrane Domains
The occurrence of transmembrane (TM) proteins associated to calcium carbonate biominerals has been explained as contamination by soft tissues (Zhang et al. 2012). However, with the double-bleaching procedure used in the present case, this hypothesis is very unlikely and we suggest another explanation. Eleven SOMPs identified among the different groups described above contain putative TM domains. Ten of these SOMPs have a single TM domain located at their C-terminus (fig. 3), the exception being the PKD1-related protein, which is predicted to contain 11 TM helices, 4 of which are part of the polycystin cation channel domain. SOMPs with TM domains represent about one half of the ECM group, one protein (hephaestin-like) in the enzyme group, one protein (uncharacterized SOMP-3) of the “orphan” group and two in the acidic group (SAARP 2 and SAP-1). We propose that the TM proteins detected here are cleaved in their extracellular region, becoming subsequently occluded in the forming biomineral. Three lines of evidence support this hypothesis. First, without exception, the 75 peptides detected by LC-MS/MS that correspond to the 11 TM domain-containing SOMPs belong to the extracellular region located at the N-terminal side of the proteins (supplementary table S1, Supplementary Material online). Second two proteases were detected amongst the SOMPs having chymotrypsin/Hap domains that may constitute the required tool for cleaving the extracellular region of the TM domain-containing SOMPs. Third, the 11 TM-containing SOMPs possess predicted chymotrypsin cleavage sites in their extracellular regions (supplementary table S1, Supplementary Material online). Specific cleavage sites for chymotrypsin and TM domains are illustrated in figure 4 for two SOMPs: SAARP2 and zona pellucida domain-containing protein. These lines of evidence are congruent with the recent data on the spicule matrix proteins of the sea urchin S. purpuratus (Mann et al. 2010), so the mechanism proposed here may apply more generally.
Fig. 4.
Primary structures of TM SAARP2 and zona pellucida domain-containing protein, including: putative peptide signals (underlined), codon stop (*), TM domains (□), peptides identified by MSMS (red) and chymotrypsin high specificity cleavage sites (residues [FYW] not before P highlighted in green) with more than 80% of cleavage probability.
If this hypothesis is later confirmed by other approaches, this may call for a complete change of paradigm in the biomineralization field, whatever the biological model studied. Until now, the “molecular tools” controlling the mineralization process were all supposed to be secreted outside the calcifying epithelial cells to interact with the inorganic precursors of mineralization. The data presented here suggest that membrane-bound proteins may also contribute to the process via their extracellular domains, which are subsequently incorporated in the biomineral after being cleaved by peptidases. Furthermore, it suggests that there is a true connection between the epithelium and the mineral front, and that the mineral deposition is accomplished under the direct guidance of the cell surfaces, rather than remotely, as “classical” views tend to show, in particular in mollusc shell models (Wilbur and Saleuddin 1983).
SOMPs in Early Stages of Calcification Affected by High CO2
The availability of transcriptome data derived from A. millepora primary polyps (Moya et al. 2012) permitted investigation of the impact of elevated CO2 on expression of the SOMPs identified here. Comparisons with the data sets submitted with the NCBI Gene Expression Omnibus database (GEO) under the accession number GSE33016 confirmed that genes corresponding to all of the SOMPs are expressed during the early stages of calcification and at least 26 of these were up regulated, whereas 4 were down regulated in polyps exposed to higher concentrations of CO2 (supplementary table S4, Supplementary Material online). SOMPs are therefore appropriate molecular markers for the investigation of the impacts of elevated CO2 on Acropora calcification.
Homology Comparison between Acropora, N. vectensis, and H. magnipapillata
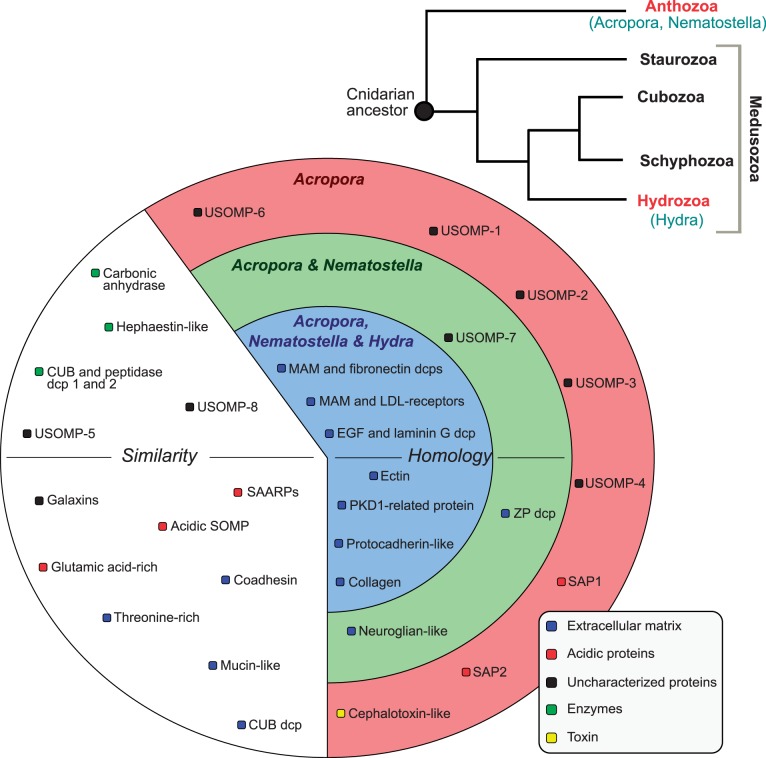
In an attempt to unravel the origins of the A. millepora “skeletome”, homologs of the 36 biomineralization proteins were searched amongst the predicted proteins (using BLASTP) and corresponding transcripts (BLASTN and TBLASTX) from the three cnidarians for which whole genome sequence data are presently available—A. digitifera, a scleractinian coral which diverged from A. millepora in the Mio-Pliocene (Van Oppen et al. 2001), the sea anemone N. vectensis, and H. magnipapillata. Sea anemones are classified as a distinct Order (Actinaria) from corals (Scleractinia) but in the same subclass (Zoantharia = Hexacorallia) within Class Anthozoa. Although sea anemones are considered to be “close,” but skeleton-less, relatives of corals, Hydra is a phylogenetically remote cnidarian belonging to Class Hydrozoa (Collins 2002; Ball et al. 2004; Collins et al. 2006). This approach enabled proteins that are involved in Acropora biomineralization but have ancient cnidarian origins (i.e., are present in Hydra as well as Acropora) to be resolved from those that may be anthozoan-specific but not restricted to calcifying anthozoans (i.e., present in Nematostella as well as Acropora) and those that are so far unique to stony corals. Clear orthologs of each of the 36 A. millepora SOMPs could be identified in A. digitifera (supplementary tables S1 and S3, Supplementary Material online), confirming that all the sequences assigned to A. millepora are of coral origin and are not derived from zooxanthellae.
BLAST comparisons against the predicted proteins of N. vectensis and H. magnipapillata, indicate that eight of the SOMPs do not have counterparts in the two noncalcifying cnidarians: five “orphan” proteins (USOMPs), SAP-1, SAP-2, and the SOMP similar to cephalotoxin (outer circle, fig. 5). Indeed, SAPs were previously suggested to be restricted to members of the genus Acropora (Shinzato et al. 2011).
Fig. 5.
Resume of the results of similarity searches with BLAST and homology detection (by global alignment, domain architecture comparison and the NC method) using the 36 SOMPs (and corresponding transcripts) from Acropora millepora and the genomes of A. digitifera, Nematostella vectensis, and Hydra magnipapillata. Proteins in the outer circle (red) do not have similarities (and homologs) in the predicted proteins of Nematostella or Hydra. Proteins in the middle circle (green) have homologs in Acropora and Nematostella. Proteins from the inner circle (blue) have homologs in Acropora, Nematostella, and Hydra. SOMPs in the white region of the circle show considerable similarity with proteins from Nematostella and Hydra but their homology is not certain. The phylogenetic tree on the upright side represents the relationships previously purposed between Cnidaria (Collins et al. 2006); dcp, domain containing protein.
To date, a BLAST search against the cnidarian sequence data on NCBI with these proteins only retrieved orthologous sequences in other Acropora species (supplementary table S3, Supplementary Material online) and low sequence similarities (E value < 10−5) in Porites astreoides (USOMP-2), Montastrea faveolata (USOMP-2, USOMP-4), Aiptasia pallida (USOMP-1, USOMP-3), Anemonia viridis (USOMP-3), and Clytia hemisphaerica (similar to cephalotoxin). In particular, the apparent absence of homologs of the cephalotoxin-like sequence in other cnidarians is surprising given the similarity between the Acropora and cuttlefish (Sepia) proteins. Note that the SE-cephalotoxin (B2DCR8) identified in the salivary glands of the cuttlefish Sepia esculenta (Cephalopoda, Mollusca) does not exhibit any significant similarity to known proteins (Ueda et al. 2008). In turn, the striking similarity (50%) between the Acropora and Sepia sequences (supplementary fig. S6, Supplementary Material online) suggests that the ancestor of these proteins predates the divergence of molluscs and corals (i.e., 550–600 Ma or more) but could have been lost in most of the other eumetazoan lineages. Such a genome restructuring, that is, massive loss, has been recently documented via large-scale comparisons of distantly related genomes: for example, the gastropod Lottia and Nematostella share exclusively 89 gene families that are not retrieved in other phyla investigated (Ptitsyn and Moroz 2012).
Amongst the other SOMPs, we have distinguished similar (white circle area, fig. 5) from homologous (green and blue circles, fig. 5) proteins taking into account the statistical significance of BLAST searches, the percentage of identity (cutoff 30%) in the pairwise global alignment, the common domain architecture and the fact that many of the SOMPs are mosaic proteins, that is, proteins with multiple domains (Bork 1992; Doolittle 1992) (supplementary table S3, Supplementary Material online). In particular for those proteins from the ECM-like group and enzymes, it is difficult to infer homology merely on the basis of BLAST searches (generally highly significant within a certain domain) together with identity percentages and alignment coverage (generally low). To overcome this difficulty, the Neighborhood Correlation (NC) method was applied to the 3 genomes. This method has been developed and used to accurately identify homologs in complex multidomain families (Song et al. 2008; Joseph and Durand 2009), enabling the prediction of similar proteins (due to domain insertion) from homologous proteins (due to common ancestor).
This combined analysis implied that at least three SOMPs have homologs in the anthozoan but not in H. magnipapillata (middle circle, fig. 5): USOMP-7, neuroglian-like and zona pellucida domain-containing protein, whereas other 9 SOMPs, all from the ECM-like group, have homologs in the three genomes (inner circle, fig. 5). Several of these conserved ECM-like SOMPs are also found in vertebrates, such as the protocadherin-like, collagen, and PKD1-related proteins (supplementary table S1, Supplementary Material online). Whether these proteins function in similar ways in the skeletal matrix and in the ECM remains to be clarified. However, these results strongly corroborate the idea of a modern skeletal matrix derived from the recruitment of noncalcifying ECM-components.
Conversely, homologs for 16 of the SOMPS could not be confidently identified in the noncalcifying cnidarians due to specific factors: incompleteness of at least one of the sequences in study (putative carbonic-anhydrase, peptidase, Glu-rich, and USOMP-5), low identity percentages (cutoff 30%) (hephaestin-like, galaxins, SAARPs, acidic SOMP, and USOMP-8), different combination of domain architectures and neighborhood coefficients above the threshold (mucin-like, coadhesin, threonine-rich, and CUB domain-containing protein, peptidase) (supplementary table S3, Supplementary Material online). In consequence, these SOMPs were considered as “similar to” and not confirmed as homologous or nonhomologous (white circle area, fig. 5). Interestingly, homologs of hephaestin are present in both chordates and the symbiotic sea anemone Anemonia viridis whereas only low similarity matches were identified in N. vectensis and H. magnipapillata (supplementary fig. S8, Supplementary Material online). Also the SOMP mucin-like has similar domain architecture in the C-terminus side (extracellular nidogen, adhesion associated domain in mucin-4 and other proteins, von Willebrand factor D, and EGF) to the human mucin-4 found in chordates; however, the same domain combination was not identified in the genomes of Nematostella and Hydra (supplementary table S3, Supplementary Material online).
Conclusions
The work described here represents the first comprehensive survey of the skeletal molecular toolkit of the scleractinian coral A. millepora. In total, 36 proteins were identified from the acid soluble and insoluble skeletal organic matrices. The proteins of these two fractions overlapped significantly, only 2 proteins being exclusively associated with the soluble fraction and 12 with the insoluble fraction. It is unclear how complete this survey is; trypsin-resistant skeletal proteins, in particular those that do not possess suitable trypsin cleavage sites, may not have been detected. It is also possible that the calicoblastic cells secrete proteins that guide the calcification process but are not occluded in the skeleton and would therefore not be detectable by the approach used here.
One tenet of our analyses is that the classical view of a coral skeletal matrix composed primarily or exclusively of acidic proteins (Mitterer 1978; Constantz and Weiner 1988) is eclipsed by the idea of a more complex matrix consisting of a diverse range of proteins with a larger spectrum of different potential functions. Whilst acidic proteins remain quantitatively key players in calcification (as aptly illustrated by the 2D gel), other SOMPs, some present in lower amount, are also likely to fulfill important roles in the deposition of the skeleton. The second tenet of our analysis is that the SOMPs are not exclusively secreted. Some SOMPs are membrane-bound proteins, the extracellular domains of which are subsequently cleaved in the mineralizing space and occluded in the growing biomineral.
What kind of information on the macroevolution of calcifying matrices can be inferred from this analysis? The evolutionary picture that emerges from our analysis is multifaceted. Whilst all of the SOMPs identified in A. millepora have homologs encoded in the genome of Acropora digitifera, surprising few of these are actually unique to the scleractinians, that is, present in the two acroporids but absent from noncalcifying cnidarians (N. vectensis and H. magnipapillata). These coral-restricted proteins may be “novelties” in the calcification repertoire. The timing of this innovation might have occurred deep in the Paleozoic when scleractinian corals emerged (∼450 Ma) (Stolarski et al. 2011). In contrast, a considerable proportion of the ECM-like SOMPs identified here have homologous proteins in N. vectensis and H. magnipapillata, suggesting that their origins predate the divergence of the classes Anthozoa and Hydrozoa (Ball et al. 2004), which occurred in the Cambrian or earlier. Each of these proteins, and a number of others subsequently lost from N. vectensis and H. magnipapillata (typified by the proteins similar to cephalotoxin, hephaestin-like, and mucin-like), was presumably co-opted to function in calcification when this trait arose in the scleractinian lineage. Broadening the comparison with include other OM proteomes from metazoans revealed that most of the domains represented in the A. millepora skeletal proteome also occur in other OM, pleading for the concept of a pool of “shared domains” potentially involved in biomineralization. However, only few of the proteins containing these common domains are predicted to be homologous, suggesting that they have been independently recruited to roles in calcification at different times across the various taxa with this trait. Beyond macroevolutionary considerations, the data set presented here constitutes an important tool for understanding the molecular bases of calcification in stony corals, and for quantifying the impact of environmental changes (such as OA), on this process.
Materials and Methods
Skeletal Collection and SEM Observations
Acropora millepora colonies were collected at the Great Barrier Reef in Australia (Pioneer Bay, Orpheus Island) in November 2010, prior to the annual spawning event. Mother colonies that died after spawning were used to collect the skeletal material; animal tissue, symbionts, and other micro-organisms were removed by immersion in NaOCl (5%, vol/vol) for 72 h. The skeletal material was then rinsed with purified water, dried and mechanically fragmented. The skeleton microstructure was observed with a tabletop Scanning Electron Microscope (SEM, Hitachi TM-1000) under an acceleration voltage of 15 keV. Mirror polished transverse and longitudinal sections were observed after etching in EDTA (1 % wt/vol, 3 min), rinsed with water and dried. The skeletal fragments were then split in two batches. The first one was not submitted to further treatment before mineral dissolution for matrix extraction, whereas the second batch was treated further (discussed later).
OM Extraction
Skeletal fragments of the second batch were reduced to powder (Fritsch Pulverisette 14), which was subsequently sieved (particle size below 200 µm). The powder was bleached in NaOCl solution (10 times dilution, 0.26% active chlorine) for 5 h and washed with milli-Q water several times until no trace of NaOCl was left. This treatment allows removing organic exogenous or endogenous contaminants that can be entrapped in the highly porous skeleton (supplementary fig. S1, Supplementary Material online), while keeping intact the most tightly bound skeletal matrix components (Gaffey and Bronnimann 1993). The extraction was performed according to a published procedure (Marin et al. 2001). In brief, the dried powder put in suspension in cold water was decalcified overnight in acetic acid (10% vol/vol) at 4 °C with an electronic burette (Titronic Universal, Schott, Mainz, Germany). The solution was centrifuged: the organic pellet (AIM) was rinsed several times with Milli-Q water and freeze-dried. The supernatant (ASM) was treated on an ultrafiltration cell (Amicon 400 mL, 10 kDa cutoff membrane) for volume reduction, then once concentrated, dialyzed several days against water at 4 °C, and freeze-dried. The extraction was performed in duplicate (2 × 30 g of skeletal powder under the same NaOCl treatment) to check the reproducibility of the results.
ASM/AIM Analysis on 1D and 2D Gel Electrophoresis
The skeletal matrix—both ASM and AIM fractions—was analyzed on 1D electrophoresis and stained with silver nitrate (Morrissey 1981). The ASM was directly denatured with Laemmli buffer (Laemmli 1970) according to standard conditions while the AIM was only partly solubilized by Laemmli buffer. The solubilized fraction is defined as the Laemmli-soluble/acetic acid insoluble fraction (LS/AIM). Electrophoresis was performed on discontinuous 12% acrylamide mini-gels at 100 V for 15 min and 150 V for 1 h. For the 2D electrophoresis, immobilized pH gradient (IPG) strips (ReadyStrip, BioRad) were loaded with 180 μg of ASM dissolved in 180 μL of rehydratation buffer (6 M urea, 2 M thiourea, 4% [wt/vol] Chaps, 20 mM dithiothreitol, 0.1% ampholytes, 0.001% bromophenol blue) and rehydrated overnight at 50 V (25 °C) in a PROTEAN IEF cell (BioRad). Focusing was carried out at 250 V for 15 min, followed by 4,000 V for 2 h and 4,000 V until 10,000 Vh. Subsequently the IPG strips were equilibrated by transfer for 10 min into 1 mL of equilibration buffer I (6 M urea, 2% sodium dodecyl sulphate [SDS], 375 mM Tris/HCl pH 8.8, 20% glycerol) with 2% (wt/vol) dithiothreitol, followed by 10 min in equilibration buffer II, with 2.5% (wt/vol) iodoacetamide instead. Strips were rinsed in 25 mM Tris, 192 mM glycine and 0.1% SDS (TGS) and placed on the top of precast NuPAGE(R) BisTris Novex SDS-polyacrylamide gels (4–10%) (Invitrogen, Carlsbad, CA) with an overlay solution of 0.5% agarose/TGS (wt/vol). Electrophoresis was performed at 200 V for 35 min. Both 1D and 2D gels were stained with silver nitrate (Morrissey 1981).
Proteomic Analysis
The AIM and ASM were prepared for proteomic analysis according to a routine procedure including reduction, alkylation, trypsin digestion, drying, and solubilization in TFA (Marie et al. 2009, 2010). MS analyses were performed in duplicate on a LTQ Velos (ThermoScientific, France) instrument in the positive ion mode. The ion source was equipped with a picoTip emitter as nanospray needle (FS360-75-30-CE-5-C10.5, NewObjective, USA) operating at 1.5 kV. The acquisition was done with the Excalibur 2.1 software (ThermoScientific). Typically, two scan events were used: at first, m/z 400–1,600 survey scan MS with enhanced resolution; second, data-dependent scans MS/MS on the 20 most intense ions from the previous event. The spectra were recorded using dynamic exclusion of previously analyzed ions for 0.6 min. The MS/MS normalized collision energy was set to 35 eV.
LC was performed on an Ultimate 3000 nano-LC system (Dionex, Voisins Le Bretonneux, France). Chromatographic separation of peptides was obtained with a C18 PepMap micro-precolumn (5 μm; 0.3 mm × 5 mm) for a 3 min desalting and a C18 PepMap nano-column (3 μm; 100 Å; 75 μm × 150 mm) with a gradient elution at a flow rate of 300 nL/min. Eluent A was a mixture of 95% H2O, 5% CH3CN, and 0.1% formic acid (vol/vol). Eluent B was a mixture of 20% H2O, 80% CH3CN, and 0.1% formic acid (vol/vol). The gradient program was from 0% B to 50% B over 120 min and 100% B for 10 min.
In Silico Analysis of the SOMPs
The data obtained with the LTQ-velos system were used for protein identification with the MASCOT search engine (version 2.1, Matrix Science, London, UK). MS/MS raw data were searched with carbamidomethylation as fixed modification, and methionine oxidation, asparagine, and glutamine de-amidation as variable modifications. The tolerance of the precursor and fragment masses was set to 0.4 Da. Proteins identified with at least two distinct peptides were considered valid assignments. The corresponding peptide sequences were validated by manually inspection of the MS/MS spectra. The subject of MASCOT searches was a pooled protein database comprising the 6-frame translated nucleotide sequences from different publicly available sources in April 2012: A. millepora sequences were downloaded from NCBI including 101,380 plus 15,389 sequences from the nucleotide and EST databases, respectively. EST (7,964) and nucleotide (5,112) data from the genus Symbiodinium were also included together with the assembled EST of two Symbiodinium strains (Mf104b – clade B [76,284 EST] and KB8 – clade A [72,152 EST]) (Bayer et al. 2012) from http://medinalab.org/zoox (last accessed October 3, 2012). The proteins identified through MASCOT were used in similarity searches against the UniprotKB/Swissprot database. Subsequently, protein sequences were analyzed for the presence of signal peptides with Signal IP 4.0 (http://www.cbs.dtu.dk/services/SignalP/, last accessed February 15, 2013), TM domains with TMHMM v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/, last accessed February 15, 2013) and further characterized for homologous domains, and regions using the InterproScan platform (http://www.ebi.ac.uk/Tools/pfa/iprscan/, last accessed February 15, 2013). In addition, the proteins with TM domains were also analyzed for potential cleavage sites cleaved by proteases with PeptideCutter (http://web.expasy.org/peptide_cutter/, last accessed December 11, 2012).
Homology Analysis and Protein Comparisons at the Domain Level
Homology analysis were performed using the 36 A. millepora sequences involved in biomineralization (transcripts and proteins) against the predicted coding genes of Acropora digitifera (http://marinegenomics.oist.jp/genomes/gallery, last accessed November 2, 2011), N. vectensis (http://www.uniprot.org/, last accessed February 14, 2013), and H. magnipapillata (ftp://ftp.jgi-psf.org/pub/JGI_data/Hydra_magnipapillata/, last accessed February 14, 2013). First, searches with a local BLAST (version 2.2.25+) (Altschul et al. 1990) were performed using the transcripts (and protein sequences) of the 36 SOMPs from A. millepora against: 1) the coding genes of A. digitifera (TBLASTN), with default parameters; 2) the coding genes from N. vectensis and H. magnipapillata (TBLASTX), with default parameters; 3) the predicted proteins of the three cnidarian genomes (BLASTP), with and without SEG (i.e., low complexity filter on query sequence) (Wootton and Federhen 1993). The best hit for each SOMP (E value threshold <10−4) was selected and the sequences globally aligned using Needleman–Wunsch algorithm for pairwise aligment (cutoff: 30% identity, expect for mosaic proteins) (supplementary table S3, Supplementary Material online). The best matches were also manually compared for their domain architecture. Second, the NC method (http://www.neighborhoodcorrelation.org/, last accessed December 12, 2012) (Song et al. 2008) was implemented for homology identification between the three genomes (A. digitifera vs. N. vectensis and A. digitifera vs. H. magnipapillata). Multidomain proteins with high confidence NC coefficients (>0.8) were considered strictly homologs (Joseph and Durand 2009). Orthologs of SOMPs in A. digitifera genome were selected and subsequently analyzed for the presence of the homologs in N. vectensis and H. magnipapillata. The SOMPs and corresponding best matches in the three genomes that were not complete sequences but showed significant blast scores (E value < 10−4), were not assessed for their homology and just considered similar.
High-throughput proteomic data sets obtained from OMs occluded in CaCO3 structures were collected from the literature and their primary sequences analyzed with InterProScan: Strongylocentrotus purpuratus (tooth: Mann et al. 2008a; spicules: Mann et al. 2010; test and spine: Mann et al. 2008b), Gallus gallus (eggshell: Mann et al. 2006; Miksík et al. 2010), Lottia gigantea (shell: Marie et al. 2013), Crassostrea gigas (shell: Zhang et al. 2012), and Pinctada (shell: Marie et al. 2012). The complete set of signatures obtained in these proteomes (InterPro entries - domains and repeats) was compared with those present in the SOMPs.
Supplementary Material
Supplementary tables S1–S6 and figures S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
P.R.S. carried out the protein extractions, proteomic, and sequence analysis, and drafted the manuscript. J.K. participated in the design of the study and data analysis. L.H. collected and prepared the coral samples prior to extraction and contributed with scientific input for the manuscript. N.G. performed new coral decalcifications to check the reproducibility of the extraction procedure and contributed with technical input. I.Z.C. performed the mass spectrometry experiments. B.M. and D.J.M. contributed to the interpretation of the data and writing of the manuscript. F.M. participated in the design of the study, data analysis and drafting of the manuscript. This work was supported by the EU FP7 Marie Curie Initial Training Network BIOMINTEC (PITN-GA-2008-215507, www.biomintec.de, coordinator H.C. Schröder) and by the EU FP7 Knowledge Based Bio-Economy project BioPreDyn grant 289434 (www.biopredyn.eu) to P.R.S. and by the ANR project (ACCRO-EARTH, ref. BLAN06-2_159971, Gilles Ramstein, LSCE; during the period 2007–2011) and the COST project TD0903 (“Biomineralix”, www.biomineralix.eu, 2009–2013) to P.R.-S., B.M., N.G., and F.M. The authors thank Dr J.M. Joseph for the helpful feedback about the Neighborhood Correlation method.
References
- Addadi L, Moradian J, Shay E, Maroudas NG, Weiner S. A chemical model for the cooperation of sulfates and carboxylates in calcite crystal nucleation: relevance to biomineralization. Proc Natl Acad Sci U S A. 1987;84:2732–2736. doi: 10.1073/pnas.84.9.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand D, Tambutté É, Zoccola D, Tambutté S. Coral calcification, cells to reefs. In: Dubinsky Z, Stambler N, editors. Coral reefs: an ecosystem in transition. Dordrecht (The Netherlands): Springer; 2011. pp. 119–150. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ball EE, Hayward DC, Saint R, Miller DJ. A simple plan—cnidarians and the origins of developmental mechanisms. Nat Rev Genet. 2004;5:567–577. doi: 10.1038/nrg1402. [DOI] [PubMed] [Google Scholar]
- Bayer T, Aranda M, Sunagawa S, Yum LK, Desalvo MK, Lindquist E, Coffroth MA, Voolstra CR, Medina M. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PloS One. 2012;7:e35269. doi: 10.1371/journal.pone.0035269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci A, Tambutté S, Supuran CT, Allemand D, Zoccola D. A new coral carbonic anhydrase in Stylophora pistillata. Mar Biotechnol. 2011;13:992–1002. doi: 10.1007/s10126-011-9363-x. [DOI] [PubMed] [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Bork P. Mobile modules and motifs. Curr Opin Struct Biol. 1992;2:413–421. [Google Scholar]
- Brinkman DL, Mulvenna J, Konstantakopoulos N, Hodgson WC, Isbister GK, Seymour JE, Burnell JN. 106 Molecular diversity of box jellyfish toxins. Toxicon. 2012;60:148–149. [Google Scholar]
- Brown BE, Tudhope AW, Le Tissier MDA, Scoffin TP. A novel mechanism for iron incorporation into coral skeletons. Coral Reefs. 1991;10:211–215. [Google Scholar]
- Caldeira K, Wickett ME. Anthropogenic carbon and ocean pH. Nature. 2003;425:365. doi: 10.1038/425365a. [DOI] [PubMed] [Google Scholar]
- Chan NCS, Connolly SR. Sensitivity of coral calcification to ocean acidification: a meta-analysis. Glob Change Biol. 2013;19:282–290. doi: 10.1111/gcb.12011. [DOI] [PubMed] [Google Scholar]
- Cohen AL, McConnaughey TA. Geochemical perspectives on coral mineralization. Rev Mineral Geochem. 2003;54:151–188. [Google Scholar]
- Cohen AL, McCorkle DC, De Putron S, Gaetani GA, Rose KA. Morphological and compositional changes in the skeletons of new coral recruits reared in acidified seawater: insights into the biomineralization response to ocean acidification. Geochem Geophys Geosyst. 2009;10 Q07005, doi:10.1029/2009GC002411. [Google Scholar]
- Collins AG. Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J Evolution Biol. 2002;15:418–432. [Google Scholar]
- Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol. 2006;55:97–115. doi: 10.1080/10635150500433615. [DOI] [PubMed] [Google Scholar]
- Constantz B, Weiner S. Acidic macromolecules associated with the mineral phase of scleractinian coral skeletons. J Exp Zool. 1988;248:253–258. [Google Scholar]
- Cuif J-P, Dauphin Y, Gautret P. Compositional diversity of soluble mineralizing matrices in some recent coral skeletons compared to fine-scale growth structures of fibres: discussion of consequences for biomineralization and diagenesis. Int J Earth Sci. 1999;88:582–592. [Google Scholar]
- Cuif J-P, Dauphin Y, Freiwald A, Gautret P, Zibrowius H. Biochemical markers of zooxanthellae symbiosis in soluble matrices of skeleton of 24 Scleractinia species. Comp Biochem Physiol Part A Mol Integr Physiol. 1999;123:269–278. [Google Scholar]
- Cusack M, Freer A. Biomineralization: elemental and organic influence in carbonate systems. Chem Rev. 2008;108:4433–4454. doi: 10.1021/cr078270o. [DOI] [PubMed] [Google Scholar]
- Dauphin Y, Cuif JP. Isoelectric properties of the soluble matrices in relation to the chemical composition of some Scleractinian skeletons. Electrophoresis. 1997;18:1180–1183. doi: 10.1002/elps.1150180726. [DOI] [PubMed] [Google Scholar]
- De’ath G, Lough JM, Fabricius KE. Declining coral calcification on the Great Barrier Reef. Science. 2009;323:116–119. doi: 10.1126/science.1165283. [DOI] [PubMed] [Google Scholar]
- Debreuil J, Tambutté E, Zoccola D, Deleury E, Guigonis J-M, Samson M, Allemand D, Tambutté S. Molecular cloning and characterization of first organic matrix protein from sclerites of red coral, Corallium rubrum. J Biol Chem. 2012;287:19367–19376. doi: 10.1074/jbc.M112.352005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF. Reconstructing history with amino acid sequences. Proteome Sci. 1992;1:191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JL, Mass T, Haramaty L, Zelzion E, Bhattacharya D, Falkowski PG. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci U S A. 2013;110:3788–3793. doi: 10.1073/pnas.1301419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. Common structural motifs in proteins of the extracellular matrix. Curr Opin Cell Biol. 1991;3:779–785. doi: 10.1016/0955-0674(91)90050-9. [DOI] [PubMed] [Google Scholar]
- Falini G, Reggi M, Fermani S, Sparla F, Goffredo S, Dubinsky Z, Levi O, Dauphin Y, Cuif J-P. Control of aragonite deposition in colonial corals by intra-skeletal macromolecules. J Struct Biol. 2013;pii:S1047–8477. doi: 10.1016/j.jsb.2013.05.001. (13)00115-9. [DOI] [PubMed] [Google Scholar]
- Farre B, Cuif J-P, Dauphin Y. Occurrence and diversity of lipids in modern coral skeletons. Zoology. 2010;113:250–257. doi: 10.1016/j.zool.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- Frazão B, Vasconcelos V, Antunes A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an overview. Mar Drugs. 2012;10:1812–1851. doi: 10.3390/md10081812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda I, Ooki S, Fujita T, Murayama E, Nagasawa H, Isa Y, Watanabe T. Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem Biophys Res Commun. 2003;304:11–17. doi: 10.1016/s0006-291x(03)00527-8. [DOI] [PubMed] [Google Scholar]
- Gaffey SJ, Bronnimann CE. Effects of bleaching on organic and mineral phases in biogenic carbonates. J Sediment Res. 1993;63:752–754. [Google Scholar]
- Gattuso J-P, Frankignoulle M, Wollast R. Carbon and carbonate metabolism in coastal aquatic ecosystems. Annu Rev Ecol Syst. 1998;29:405–434. [Google Scholar]
- Geffen Y, Ron EZ, Rosenberg E. Regulation of release of antibacterials from stressed scleractinian corals. FEMS Microbiol Lett. 2009;295:103–109. doi: 10.1111/j.1574-6968.2009.01590.x. [DOI] [PubMed] [Google Scholar]
- Glimcher M. Molecular biology of mineralized tissues with particular reference to bone. Rev Mod Phys. 1959;31:359–393. [Google Scholar]
- Gochfeld D, Aeby G. Antibacterial chemical defenses in Hawaiian corals provide possible protection from disease. Mar Eco Prog Ser. 2008;362:119–128. [Google Scholar]
- Goffredo S, Vergni P, Reggi M, Caroselli E, Sparla F, Levy O, Dubinsky Z, Falini G. The skeletal organic matrix from Mediterranean coral Balanophyllia europaea influences calcium carbonate precipitation. PLoS One. 2011;6:e22338. doi: 10.1371/journal.pone.0022338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg WM. Evidence of a sclerotized collagen from the skeleton of a gorgonian coral. Comp Biochem Physiol B Comp Biochem. 1974;49:525–529. doi: 10.1016/0305-0491(74)90188-6. [DOI] [PubMed] [Google Scholar]
- Goldberg WM. Acid polysaccharides in the skeletal matrix and calicoblastic epithelium of the stony coral Mycetophyllia reesi. Tissue Cell. 2001;33:376–387. doi: 10.1054/tice.2001.0191. [DOI] [PubMed] [Google Scholar]
- Helman Y, Natale F, Sherrell RM, Starovoytov V, Gorbunov MY, Falkowski PG. Extracellular matrix production and calcium carbonate precipitation by coral cells in vitro. Proc Natl Acad Sci U S A. 2008;105:54–58. doi: 10.1073/pnas.0710604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincke MT, Gautron J, Panheleux M, Garcia-ruiz J, Mckee MD. Identification and localization of lysozyme as a component of eggshell membranes and eggshell matrix. Matrix Biol. 2000;19:443–453. doi: 10.1016/s0945-053x(00)00095-0. [DOI] [PubMed] [Google Scholar]
- Ingalls AE, Lee C, Druffel ER. Preservation of organic matter in mound-forming coral skeletons. Geochim Cosmochim Acta. 2003;67:2827–2841. [Google Scholar]
- Isa Y, Yamazato K. The distribution of carbonic anhydrase in a staghorn coral, Acropora hebes (Dana) Galaxea. 1984;3:25–36. [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Jonchère V, Réhault-Godbert S, Hennequet-Antier C, Cabau C, Sibut V, Cogburn LA, Nys Y, Gautron J. Gene expression profiling to identify eggshell proteins involved in physical defense of the chicken egg. BMC Genomics. 2010;11:57. doi: 10.1186/1471-2164-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JM, Durand D. Family classification without domain chaining. Bioinformatics. 2009;25:i45–i53. doi: 10.1093/bioinformatics/btp207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert C, Piquemal D, Marie B, Manchon L, Pierrat F, Zanella-Cléon I, Cochennec-Laureau N, Gueguen Y, Montagnani C. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics. 2010;11:613. doi: 10.1186/1471-2164-11-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RJ, Tsuzaki M, Watabe N, Mechanic GL. Collagen in the spicule organic matrix of the gorgonian Leptogorgia virgulata. Biol Bull. 1990;179:207–213. doi: 10.2307/1541771. [DOI] [PubMed] [Google Scholar]
- Kleypas JA, Yates KK. Coral reefs and ocean acidification. Oceanography. 2009;22:108–117. [Google Scholar]
- Kline DI, Teneva L, Schneider K, et al. (24 co-authors) A short-term in situ CO2 enrichment experiment on Heron Island (GBR) Sci Rep. 2012;2:1–9. doi: 10.1038/srep00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker KJ, Kordas RL, Crim R, Hendriks IE, Ramajo L, Singh GS, Duarte CM, Gattuso J-P. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob Change Biol. 2013;19:1884–1896. doi: 10.1111/gcb.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lang M, Braun CL, Kanost MR, Gorman MJ. Multicopper oxidase-1 is a ferroxidase essential for iron homeostasis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2012;109:13337–13342. doi: 10.1073/pnas.1208703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon C, Atkinson MJ. Effect of elevated pCO2 on photosynthesis and calcification of corals and interactions with seasonal change in temperature/irradiance and nutrient enrichment. J Geophys Res. 2005;110:1–16. [Google Scholar]
- Mann K, Macek B, Olsen JV. Proteomic analysis of the acid-soluble organic matrix of the chicken calcified eggshell layer. Proteomics. 2006;6:3801–3810. doi: 10.1002/pmic.200600120. [DOI] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M. In-depth, high-accuracy proteomics of sea urchin tooth organic matrix. Prot Sci. 2008a;6:33. doi: 10.1186/1477-5956-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Prot Sci. 2008b;10:1–10. doi: 10.1186/1477-5956-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Wilt FH, Poustka AJ. Proteomic analysis of sea urchin (Strongylocentrotus purpuratus) spicule matrix. Prot Sci. 2010;8:1–12. doi: 10.1186/1477-5956-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Jackson DJ, Ramos-Silva P, Zanella-Cléon I, Guichard N, Marin F. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage specific novelties. FEBS J. 2013;280:214–232. doi: 10.1111/febs.12062. [DOI] [PubMed] [Google Scholar]
- Marie B, Joubert C, Tayalé A, Zanella-Cléon I, Belliard C, Piquemal D, Cochennec-Laureau N, Marin F, Gueguen Y, Montagnani C. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci U S A. 2012;109:20986–20991. doi: 10.1073/pnas.1210552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Marin F, Marie A, Bédouet L, Dubost L, Alcaraz G, Milet C, Luquet G. Evolution of nacre: biochemistry and proteomics of the shell organic matrix of the cephalopod Nautilus macromphalus. ChemBioChem. 2009;10:1495–1506. doi: 10.1002/cbic.200900009. [DOI] [PubMed] [Google Scholar]
- Marie B, Zanella-Cléon I, Le Roy N, Becchi M, Luquet G, Marin F. Proteomic analysis of the acid-soluble nacre matrix of the bivalve Unio pictorum: detection of novel carbonic anhydrase and putative protease inhibitor proteins. ChemBioChem. 2010;11:2138–2147. doi: 10.1002/cbic.201000276. [DOI] [PubMed] [Google Scholar]
- Marin F, Corstjens P, De Gaulejac B, De Vrind-De Jong E, Westbroek P. Mucins and molluscan calcification. Molecular characterization of mucoperlin, a novel mucin-like protein from the nacreous shell layer of the fan mussel Pinna nobilis (Bivalvia, pteriomorphia) J Biol Chem. 2000;275:20667–20675. doi: 10.1074/jbc.M003006200. [DOI] [PubMed] [Google Scholar]
- Marin F, Le Roy N, Marie B. The formation and mineralization of mollusk shell. Front Biosci (Schol Ed). 2012;4:1099–1125. doi: 10.2741/s321. [DOI] [PubMed] [Google Scholar]
- Marin F, Luquet G. Unusually acidic proteins in biomineralization. In: Bauerlein E, editor. Handbook of biomineralization—biological aspects and structure formation. Weinheim (Germany): Wiley-VCH; 2007. pp. 273–90. [Google Scholar]
- Marin F, Pereira L, Westbroek P. Large-scale fractionation of molluscan shell matrix. Protein Expr Purif. 2001;23:175–179. doi: 10.1006/prep.2001.1487. [DOI] [PubMed] [Google Scholar]
- Maurer P, Hohenester E, Engel J. Extracellular calcium-binding proteins. Curr Opin Cell Biol. 1996;8:609–617. doi: 10.1016/s0955-0674(96)80101-3. [DOI] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol. 2011;20:3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x. [DOI] [PubMed] [Google Scholar]
- Miksík I, Sedláková P, Lacinová K, Pataridis S, Eckhardt A. Determination of insoluble avian eggshell matrix proteins. Anal Bioanal Chem. 2010;397:205–214. doi: 10.1007/s00216-009-3326-3. [DOI] [PubMed] [Google Scholar]
- Milliman JD, Droxler AW. Neritic and pelagic carbonate sedimentation in the marine environment: ignorance is not bliss. Geologische Rundschau. 1996;85:496–504. [Google Scholar]
- Mitterer RM. Amino acid composition and metal binding capability of the skeletal protein of corals. Bull Mar Sci. 1978;28:173–180. [Google Scholar]
- Miyamoto H, Miyashita T, Okushima M, Nakano S, Morita T, Matsushiro A. A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci U S A. 1996;93:9657–9660. doi: 10.1073/pnas.93.18.9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JH. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Mosher DF, Adams JC. Adhesion-modulating/matricellular ECM protein families: a structural, functional and evolutionary appraisal. Matrix Biol. 2012;31:1–7. doi: 10.1016/j.matbio.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Moya A, Huisman L, Ball EE, Hayward DC, Grasso LC, Chua CM, Woo HN, Gattuso J-P, Forêt S, Miller DJ. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO(2)-driven acidification during the initiation of calcification. Mol Ecol. 2012;21:2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x. [DOI] [PubMed] [Google Scholar]
- Moya A, Tambutté S, Bertucci A, Tambutté E, Lotto S, Vullo D, Supuran CT, Allemand D, Zoccola D. Carbonic anhydrase in the scleractinian coral Stylophora pistillata: characterization, localization, and role in biomineralization. J Biol Chem. 2008;283:25475–25484. doi: 10.1074/jbc.M804726200. [DOI] [PubMed] [Google Scholar]
- Orr JC, Fabry VJ, Aumont O, et al. (28 co-authors) Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature. 2005;437:681–686. doi: 10.1038/nature04095. [DOI] [PubMed] [Google Scholar]
- Pandolfi JM, Connolly SR, Marshall DJ, Cohen AL. Projecting coral reef futures under global warming and ocean acidification. Science. 2011;333:418–422. doi: 10.1126/science.1204794. [DOI] [PubMed] [Google Scholar]
- Ptitsyn A, Moroz LL. Computational workflow for analysis of gain and loss of genes in distantly related genomes. BMC Bioinformatics. 2012;13(1 Suppl):S5. doi: 10.1186/1471-2105-13-S15-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puverel S, Tambutté E, Pereira-Mouriès L, Zoccola D, Allemand D, Tambutté S. Soluble organic matrix of two Scleractinian corals: partial and comparative analysis. Comp Biochem Physiol B Biochem Mol Biol. 2005;141:480–487. doi: 10.1016/j.cbpc.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Isa Y. Characterization of proteins from the matrix of spicules from the alcyonarian, Lobophytum crassum. J Exp Mar Biol Ecol. 2005;321:71–82. [Google Scholar]
- Rahman MA, Isa Y, Takemura A, Uehara T. Analysis of proteinaceous components of the organic matrix of endoskeletal sclerites from the alcyonarian Lobophytum crassum. Calcif Tissue Int. 2006;78:178–185. doi: 10.1007/s00223-005-0253-y. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Isa Y, Uehara T. Proteins of calcified endoskeleton: II partial amino acid sequences of endoskeletal proteins and the characterization of proteinaceous organic matrix of spicules from the alcyonarian, Synularia polydactyla. Proteomics. 2005;5:885–893. doi: 10.1002/pmic.200401130. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Isa Y, Uehara T. Studies on two closely related species of octocorallians: biochemical and molecular characteristics of the organic matrices of endoskeletal sclerites. Mar Biotechnol (NY). 2006;8:415–424. doi: 10.1007/s10126-005-6150-6. [DOI] [PubMed] [Google Scholar]
- Ramos-Silva P, Benhamada S, Le Roy N, et al. (11 co-authors) Novel molluskan biomineralization proteins retrieved from proteomics: a case study with Upsalin. ChemBioChem. 2012;13:1067–1078. doi: 10.1002/cbic.201100708. [DOI] [PubMed] [Google Scholar]
- Ramos-Silva P, Marin F, Kaandorp J, Marie B. Biomineralization toolkit: The importance of sample cleaning prior to the characterization of biomineral proteomes. Proc Natl Acad Sci U S A. 2013;110:E2144–E2146. doi: 10.1073/pnas.1303657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Bermudez A, Lin Z, Hayward DC, Miller DJ, Ball EE. Differential expression of three galaxin-related genes during settlement and metamorphosis in the scleractinian coral Acropora millepora. BMC Evol Biol. 2009;9:178. doi: 10.1186/1471-2148-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, et al. (13 co-authors) Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Song N, Joseph JM, Davis GB, Durand D. Sequence similarity network reveals common ancestry of multidomain proteins. PLoS Comput Biol. 2008;4:e1000063. doi: 10.1371/journal.pcbi.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarski J, Kitahara MV, Miller DJ, Cairns SD, Mazur M, Meibom A. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol Biol. 2011;11:316. doi: 10.1186/1471-2148-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, DeSalvo MK, Voolstra CR, Reyes-Bermudez A, Medina M. Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS One. 2009;4:e4865. doi: 10.1371/journal.pone.0004865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Saruwatari K, Kogure T, Yamamoto Y, Nishimura T, Kato T, Nagasawa H. An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science. 2009;325:1388–1390. doi: 10.1126/science.1173793. [DOI] [PubMed] [Google Scholar]
- Tambutté S, Tambutté E, Zoccola D, Allemand D. Organic matrix and biomineralization of scleractinian corals. In: Bauerlein E, editor. Handbook of biomineralization—biological aspects and structure formation. Weinheim (Germany): Wiley-VCH; 2008. pp. 243–259. [Google Scholar]
- Tambutté S, Tambutté E, Zoccola D, Caminiti N, Lotto S, Moya A, Allemand D, Adkins J. Characterization and role of carbonic anhydrase in the calcification process of the azooxanthellate coral Tubastrea aurea. Mar Biol. 2006;151:71–83. [Google Scholar]
- Ueda A, Nagai H, Ishida M, Nagashima Y, Shiomi K. Purification and molecular cloning of SE-cephalotoxin, a novel proteinaceous toxin from the posterior salivary gland of cuttlefish Sepia esculenta. Toxicon. 2008;52:574–581. doi: 10.1016/j.toxicon.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Van Oppen MJ, McDonald BJ, Willis B, Miller DJ. The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting, or morphological convergence? Mol Biol Evol. 2001;18:1315–1329. doi: 10.1093/oxfordjournals.molbev.a003916. [DOI] [PubMed] [Google Scholar]
- Veis A. Organic matrix-related mineralization of sea urchin spicules, spines, test and teeth. Front Biosci. 2011;16:2540. doi: 10.2741/3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veis A, Sabsay B. The collagen of mineralized matrices. In: Peck WA, editor. Bone and mineral research. Vol. 5. New York (NY): Elsevier Science; 1987. p. 15. [Google Scholar]
- Vulpe CD, Kuo YM, Murphy TL, Cowley L, Askwith C, Libina N, Gitschier J, Anderson GJ. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat Genet. 1999;21:195–199. doi: 10.1038/5979. [DOI] [PubMed] [Google Scholar]
- Wheeler AP, George JW, Evans CA. Control of calcium carbonate nucleation and crystal growth by soluble matrx of oyster shell. Science. 1981;212:1397–1398. doi: 10.1126/science.212.4501.1397. [DOI] [PubMed] [Google Scholar]
- Wilbur K, Saleuddin AS. Shell formation. In: Wilbur K, Saleuddin AS, editors. The Mollusca. Vol. 4. Physiology, Part 1. New York: Academic Press; 1983. pp. 235–287. [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Computers Chem. 1993;17:149–163. [Google Scholar]
- Zhang G, Fang X, Guo X, et al. (85 co-authors) The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- Zoccola D, Moya A, Béranger GE, Tambutté E, Allemand D, Carle GF, Tambutté S. Specific expression of BMP2/4 ortholog in biomineralizing tissues of corals and action on mouse BMP receptor. Mar Biotechnol. 2009;11:260–269. doi: 10.1007/s10126-008-9141-6. [DOI] [PubMed] [Google Scholar]
- Zoccola D, Tambutté E, Kulhanek E, Puverel S, Scimeca J-C, Allemand D, Tambutté S. Molecular cloning and localization of a PMCA P-type calcium ATPase from the coral Stylophora pistillata. Biochim Biophys Acta. 2004;1663:117–126. doi: 10.1016/j.bbamem.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.