Abstract
Filopodia are fine actin-based cellular projections used for both environmental sensing and cell motility, and they are essential organelles for metazoan cells. In this study, we reconstruct the origin of metazoan filopodia and microvilli. We first report on the evolutionary assembly of the filopodial molecular toolkit and show that homologs of many metazoan filopodial components, including fascin and myosin X, were already present in the unicellular or colonial progenitors of metazoans. Furthermore, we find that the actin crosslinking protein fascin localizes to filopodia-like structures and microvilli in the choanoflagellate Salpingoeca rosetta. In addition, homologs of filopodial genes in the holozoan Capsaspora owczarzaki are upregulated in filopodia-bearing cells relative to those that lack them. Therefore, our findings suggest that proteins essential for metazoan filopodia and microvilli are functionally conserved in unicellular and colonial holozoans and that the last common ancestor of metazoans bore a complex and specific filopodial machinery.
Keywords: fascin, choanoflagellate, Capsaspora, Cdc42, formin evolution, gelsolin evolution, filopodia, pseudopodia
Introduction
A dynamic cytoskeletal and membrane system is a hallmark of the eukaryotic cell. It allows cells to change cell shape to carry out motility, phagocytosis, and other key functions (Fletcher and Mullins 2010). Cell motility, in particular, is a common feature among eukaryotes that often requires specialized organelles. There are two main classes of cellular structures responsible for cell motility in eukaryotes: tubulin-based cilia and flagella, conspicuous in eukaryotes as diverse as choanoflagellates, ciliates and dinoflagellates, and actin-based filopodia and lamellipodia, which allow cells to crawl along surfaces through amoeboid movement (Soldati and Meissner 2004).
Filopodia are finger-like structures based upon 10–30 parallel bundled actin filaments whose growing/barbed ends orient toward the filopodial tip. Here, many proteins accumulate and form the so-called tip complex, which controls actin monomer addition to filament ends (Small et al. 2002; Bohil et al. 2006; Faix and Rottner 2006; Gupton and Gertler 2007; Mattila and Lappalainen 2008; Lundquist 2009; Mellor 2010; Nambiar et al. 2010). In metazoans, filopodia function as sensory and exploratory organelles and typically display active protrusive and retractile motility (Yang and Svitkina 2011). Filopodia also contribute to cell adhesion (Schäfer et al. 2010) and mediate many essential, metazoan-specific phenomena, including growth cone guidance, wound-healing, embryonic development, and angiogenesis and they serve as precursors for dendritic spines in neurons (Magie et al. 2007; Mattila and Lappalainen 2008; Mellor 2010). In contrast, the other major type of actin-based cell protrusion (Small et al. 2002; Faix and Rottner 2006), the lamellipodium, is a flat sheet-like structure based upon a branched network of actin filaments (Mattila and Lappalainen 2008; Small et al. 2008; Vallotton and Small 2009). Filopodia often emerge from lamellipodial sheets and both structures share some molecular components, although a characteristic filopodial molecular architecture has been described (Mattila and Lappalainen 2008).
Filopodia-like structures (often also called pseudopodia) are known in cells from diverse other eukaryotic lineages. These cellular protrusions have historically been defined as “filopodia” based primarily on morphological characteristics (i.e., being long slender cellular protrusions) and the presence of actin filaments (Yang and Svitkina 2011). Although a hallmark of metazoan filopodia is their dynamic nature (i.e., protrusion and retraction), little is known about the dynamic properties of filopodia-like structures in non-metazoans (Adl et al. 2012; Cavalier-Smith 2013). Among bikonts, filopodia-like structures are found in excavates (e.g., Naegleria gruberi; Preston and King 2005), stramenopiles (Pawlowski 2008), and rhizarians (Cavalier-Smith 2003; Pawlowski 2008; Ota et al. 2011), in which filopodia represent one of the defining morphological characteristics of the group (Pawlowski 2008; Brown et al. 2012). In contrast, no filopodia-like structures have been so far described in other bikont clades, such as plants or alveolates. Filopodia are most abundant and diversified in the other major eukaryotic supergroup Amorphea (also known as “unikonts” or “podiates”) (Adl et al. 2012; Derelle and Lang 2012; Cavalier-Smith 2013), in which filopodia-like structures have been reported in amoebozoans, apusozoans (Cavalier-Smith and Chao 2010), and several independent opisthokont lineages, including nucleariids (the sister group of fungi) (Mikrjukov and Mylnikov 2001; Zettler et al. 2001), filastereans (Cavalier-Smith 2003), choanoflagellates (Leadbeater and Morton 1974; Dayel et al. 2011), and metazoans (see fig. 1 for their phylogenetic relationships). In contrast, no filopodia-like structures have been described in fungi.
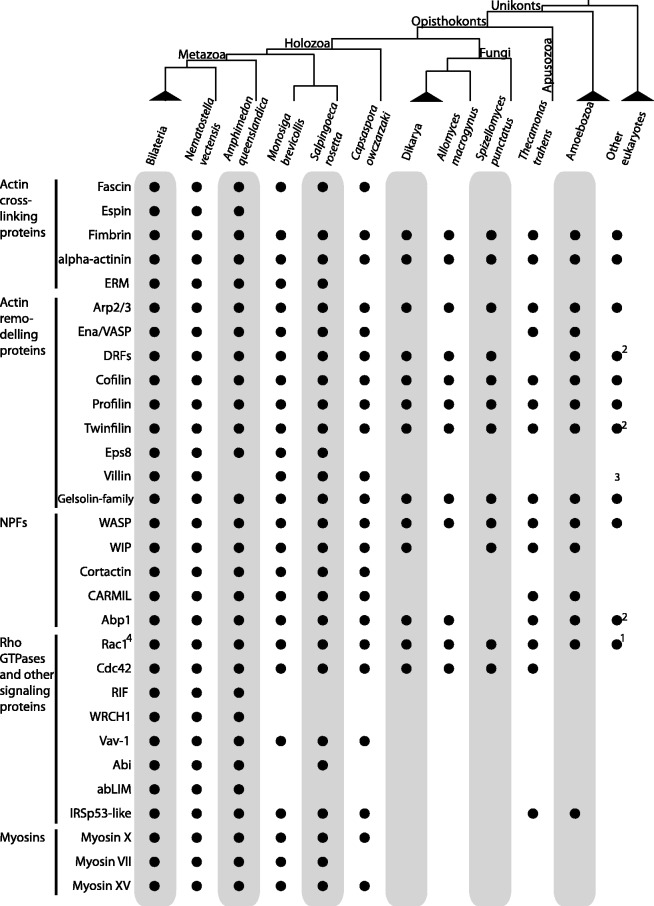
Fig. 1.
Phylogenetic distribution of diverse proteins associated with filopodia and related actin-based cellular protrusions. A black dot indicates the presence of clear homologs, whereas absence of a dot indicates that a homolog was not detected in that taxon. 1Based on Boureux et al. (2007). 2Only present in the excavates Naegleria gruberi and Trichomonas vaginalis. 3Plants have a villin-like protein that, despite having the same domain architecture as metazoans, is not phylogenetically related with holozoan villin (supplementary fig. S5, Supplementary Material online). 4Rac1 RhoGTPases are key filopodia-inducers in amoebozoans, but not in metazoans (Dumontier et al. 2000).
Moreover, microvilli, which are another type of fine actin-based cell protrusion, are restricted to holozoan lineages; good examples are the apical collar of microvilli in choanoflagellates and sponge choanocytes (Karpov and Leadbeater 1998; Gonobobleva and Maldonado 2009) and the apical microvilli of epithelial cells (DeRosier and Tilney 2000).
Two possible scenarios may account for the patchy distribution of filopodia in the eukaryotic tree. One possibility is that filopodial structures and their specific molecular components evolved independently several times during eukaryotic evolution (Pawlowski 2008). Alternatively, filopodia-like structures may have been present in ancestral eukaryotes and secondarily lost in multiple lineages. In this latter case, all filopodiated eukaryotes may share the same molecular toolkit for the formation of actin-based cellular protrusions. To differentiate between these two options, it is critical to determine the evolutionary history of proteins required for metazoan filopodia formation. The molecular composition of filopodia in metazoans is well described (Gupton and Gertler 2007; Mattila and Lappalainen 2008; Mellor 2010) and includes actin-crosslinking proteins, actin-remodeling proteins, nucleation promoting factors (NPFs), Rho GTPases and other signaling proteins, and motor proteins. Filopodia from the amoebozoan Dictyostelium discoideum contain many, but not all, of the molecular components of metazoan filopodia (Faix and Rottner 2006), whereas the composition of filopodia from other non-metazoans remains largely unknown. Furthermore, some proteins characteristic of metazoan filopodia have also been detected in microvilli and lamellipodia (DeRosier and Tilney 2000; Small et al. 2002; Tilney et al. 2004; Gupton and Gertler 2007; Mattila and Lappalainen 2008).
By deciphering the evolutionary history of metazoan filopodial genes, as well as by experimentally analyzing the expression and subcellular localization of metazoan filopodial components in non-metazoans, we aim to investigate the ancestry of the molecular toolkit for filopodia formation in metazoans (Gupton and Gertler 2007; Mattila and Lappalainen 2008).
We analyzed the genomes of diverse unicellular and colonial relatives of Metazoa, including the filasterean Capsaspora owczarzaki, the choanoflagellates Salpingoeca rosetta and Monosiga brevicollis, the apusozoan Thecamonas trahens, and the early branching fungi Spizellomyces punctatus and Allomyces macrogynus (Ruiz-Trillo et al. 2007, 2008; King et al. 2008; Fairclough et al. 2013) for metazoan filopodial proteins. We find that while some components of metazoan filopodia evolved relatively recently and are only detected in metazoans, choanoflagellates, and C. owczarzaki, others are ancient and evolved before the divergence of holozoans from other eukaryotes. Moreover, we show that unicellular holozoans produce filopodia-like structures and that the filopodia marker protein fascin localizes to filopodia-like structures and the microvillar collar in the choanoflagellate S. rosetta. Finally, gene transcription analyses of filopodial toolkit genes suggest subfunctionalization of some of the components in different S. rosetta life history stages and reveal that the transcription of filopodial genes is correlated with the presence of filopodia-like structures in C. owczarzaki. Taken together, these data show that the origin of several key components of filopodia formation predates the origin of metazoans and suggest that at least some of these proteins perform similar functions in unicellular and colonial relatives of metazoans.
Results and Discussion
Origin of the Filopodial Genetic Toolkit
To investigate the origin and evolutionary history of proteins required for metazoan filopodia formation, we performed a taxon-rich genomic survey of the metazoan filopodial toolkit (Mattila and Lappalainen 2008). To classify as many proteins as possible, we performed similarity searches and, when possible, phylogenetic analyses.
Actin-Crosslinking Proteins
Mechanical cohesion of filopodia is achieved by actin cross-linking proteins, including fascin, espin, fimbrin (also known as plastin), alpha-actinin, and in some cases ERM (Ezrin–Radixin–Moesin) proteins. Our data show that fimbrin and alpha-actinin, which are present not only in filopodia, but also in other actin-based structures (Mellor 2010), are present in all eukaryotes examined in this study. ERM proteins, which link the actin cytoskeleton to the membrane (Bretscher et al. 2002; Hoeflich and Ikura 2004; Niggli and Rossy 2008; McClatchey 2012), and fascin, a critical filament-bundling protein in metazoan filopodia, are both restricted to holozoans (fig. 1; supplementary fig. S2B, Supplementary Material online) (Ruiz-Trillo et al. 2008). Espin, on the other hand, is restricted more narrowly to metazoans, where it is expressed in a limited number of cell types and specialized filopodial structures, including sterocilia (Mellor 2010) (fig. 1). The restriction of both microvilli and ERM proteins to choanoflagellates and metazoans raises the possibility that the evolution of ERM proteins contributed to the origin of microvilli (Nambiar et al. 2010) in their last common ancestor (supplementary figs. S1 and S2A, Supplementary Material online). Moreover, the finding of fascin and other metazoan filopodial proteins in non-metazoans raises the possibility that they functioned in filopodia in the Urmetazoa.
Actin-Remodeling Proteins
Most actin-remodeling proteins are widespread among eukaryotes (fig. 1). This is the case with the Arp2/3 complex, a major actin-remodeling factor (Pollard 2007), which convergently elongates actin filaments from the cortical actin meshwork and is regulated, in metazoans, by WASP and cortactin (Weaver et al. 2003). The seven subunits of this complex are present in almost all eukaryotes, suggesting this is an ancient protein network.
Formins are also involved in actin filament formation, but instead of bundling an existing actin meshwork, formins nucleate actin filaments de novo. Our phylogenetic analyses show that Diaphanous-related formins (DRFs) (Chalkia et al. 2008), those with the domain structure GBD-FH3-FH2-DAD, are present in all Amorphea investigated as well as in Excavata (supplementary figs. S3 and S4, Supplementary Material online). Interestingly, Ena/VASP, a multifaceted actin-regulatory protein with essential roles in filopodia formation and elongation that is essential for DRF-based de novo actin nucleation (Schirenbeck et al. 2006), is exclusive to Amorphea and appears to have been secondarily lost in Fungi (fig. 1). Therefore, although DRF-like formins are present in some Excavata, it is unclear whether actin nucleation based on formins is truly an ancestral mechanism in eukaryotes, as they lack Ena/VASP.
Other proteins involved in actin remodeling include cofilin, which has depolymerizing activity, profilin, which sequesters actin monomers (Revenu et al. 2004), and several proteins, such as Eps8, Twinfilin, Villin, and other Gelsolin family proteins (supplementary fig. S5, Supplementary Material online), which are involved in capping (Mattila and Lappalainen 2008) (i.e., they stabilize filament ends by binding them and inhibiting actin monomer association or dissociation). Most of these other actin regulators are widespread among eukaryotes or Amorphea (fig. 1), except for the capping protein Eps8, which is specific to metazoans and choanoflagellates. Eps8 is a direct binding partner of ERM proteins (discussed earlier) and, together, they stimulate formation of microvilli (Zwaenepoel et al. 2012). Villin seems to be restricted to holozoans as well, although plants have a villin-like protein with similar domain architecture but with uncertain affinity to other known gelsolin-domain proteins (supplementary fig. S5, Supplementary Material online). Villin is a multi-faceted actin-remodeling protein that, in metazoans, is usually associated with microvillar formation, particularly in intestinal cells (Silacci et al. 2004; Khurana and George 2008; Nambiar et al. 2010).
In sum, we find that some actin-remodeling proteins have an ancient eukaryotic origin whereas others evolved later in evolution, either at the stem of Holozoa (villin and Eps8) or at the stem of Amorphea (Ena/VASP).
Nucleation Promoting Factors
NPFs activate the Arp2/3 complex (Goley and Welch 2006). Some important NPFs in metazoan filopodia are WASP (and the associated WASP-interacting protein [WIP]) (Veltman and Insall 2010; Kollmar et al. 2012) and Cortactin (Weaver et al. 2001; Goley and Welch 2006). In metazoans, WASP is a direct target of Cdc42 RhoGTPase, mediating the activation of the Arp2/3 complex (Takenawa and Miki 2001; Antón et al. 2007; Faix et al. 2009; Mellor 2010), which is also activated by Cortactin (Kinley et al. 2003; Ren et al. 2009), while WIP is responsible for inactivating WASP (Antón et al. 2007).
Our data show that WASP is widespread among eukaryotes (fig. 1; supplementary fig. S6, Supplementary Material online), whereas WIP is restricted to Amorphea and Cortactin to holozoans (fig. 1). It is worth mentioning that a major regulator of Cortactin, c-Src tyrosine kinase (Weaver et al. 2001, 2003), is a component of the metazoan integrin adhesome that also originated in the holozoan lineage (Sebé-Pedrós et al. 2010; Suga et al. 2012).
Rho GTPases and Other Signaling Proteins
RhoGTPases are key regulators of actin dynamics and play important roles as major switches in filopodia formation (Ridley 2006; Ladwein and Rottner 2008; Faix et al. 2009). They act through two types of actin nucleators: WASP and DRFs (Ridley 2006). Cdc42 RhoGTPase, thought to be exclusive to opisthokonts (Boureux et al. 2007), appears to be the primary filopodia-inducing RhoGTPase in metazoans (Ridley 2006). In Dictyostelium, which has filopodia but lacks Cdc42, Rac1 GTPases induce filopodia (Vlahou and Rivero 2006), whereas in plants, the Rac1-related Rop GTPases can also stimulate actin polymerization (Boureux et al. 2007). We find that Rac1-type RhoGTPases are ancestral within Amorphea, and, interestingly, that Cdc42 is not specific to opisthokonts (Boureux et al. 2007), as it is also present in the apusozoan T. trahens, sister group of opisthokonts (fig. 1; supplementary fig. S7, Supplementary Material online). RIF and WRCH1, which induce filopodia in some metazoan cell types (Faix et al. 2009), are specific to Metazoa (fig. 1).
Beside RhoGTPases, the signaling proteins abLIM, Abi, Vav-1, and IRSp53-like also regulate filopdia formation. These proteins evolved during different episodes in eukaryotic evolutionary history. IRSp53-like proteins are ancestral Amorphea proteins (supplementary fig. S9, Supplementary Material online), Vav-1 is specific to holozoans (fig. 1), Abi specific to metazoans and choanoflagellates (fig. 1) and abLIM specific to metazoans (supplementary fig. S8, Supplementary Material online).
Motor Proteins: Myosins
Myosins are a large protein family present in all eukaryotes and are essential for cell trafficking along actin filaments (Richards and Cavalier-Smith 2005; Odronitz and Kollmar 2007). The MyTH4-FERM domain myosins (named according to their protein domain composition) have special relevance for filopodia function and formation. For example, the metazoan myosin X (Tuxworth et al. 2001; Berg and Cheney 2002; Nagy et al. 2008; Nambiar et al. 2010) is essential for filopodia formation (Bohil et al. 2006) and for the transport of proteins such as integrins (Breshears et al. 2010) or Ena/VASP protein to the filopodial tip (Zhang et al. 2004; Sousa and Cheney 2005). Two other MyTH4-FERM myosins, myosin VII and myosin XV, are also important for filopodia function (Breshears et al. 2010). Our analysis shows that these three myosins (X, VII, and XV) emerged at the origin of Holozoa (fig. 1).
Filopodia-like Structures in Unicellular and Colonial Relatives of Metazoans
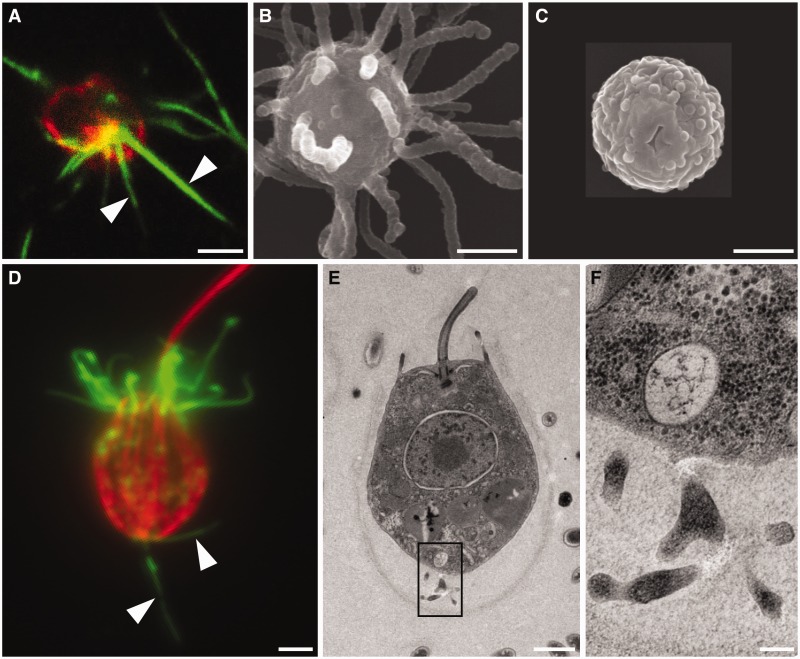
The richness of the filopodial toolkit in unicellular and colonial holozoans prompted us to investigate the presence, abundance, and distribution of filopodia-like structures in these close relatives of metazoans. We stained for polymerized actin (using phalloidin) and tubulin (using anti-tubulin antibodies) in C. owczarzaki and S. rosetta. In C. owczarzaki, multiple 1–20 µm long bundles of actin microfilaments can be found in filopodiated stage cells (fig. 2A). Scanning electron microscopy (SEM) confirms the presence of multiple long filopodia-like structures in this cell stage of C. owczarzaki (fig. 2B), in contrast with the naked cystic cell stage (fig. 2C). In the choanoflagellate S. rosetta actin microfilaments were detected in two distinct sites: in the apical collar of actin-filled microvilli and in basally positioned 1–10 µm long cellular protrusions that resemble filopodia (fig. 2D) (Leadbeater 1979; Dayel et al. 2011). Supporting the inference that the basal actin microfilaments are associated with filopodia, transmission electron microscopy (TEM) of thin sections through S. rosetta cells shows the presence of multiple basally positioned cellular processes (fig. 2E and F, black rectangle). Thus, our data show the presence of multiple long, actin-filled cellular projections that resemble filopodia in two close relatives of metazoans.
Fig. 2.
Filopodia-like structures in close relatives of metazoans. (A) Capsaspora owczarzaki filopodiated cells bear multiple long bundles of actin microfilaments, as revealed by staining with phalloidin (green). The cell periphery is revealed by staining with antibodies against beta-tubulin (red). SEM shows the presence of multiple long filopodia-like structures in C. owczarzaki filopodiated cells (B), but not in nonfilopodiated, cystic cells (C). (D) In Salpingoeca rosetta, attached cells bear actin microfilaments in the apical collar of microvilli and in basally positioned long cellular protrusions that resemble filopodia. Salpingoeca rosetta cells were stained with phalloidin (green) and antibodies against beta-tubulin (red). (E) TEM of thin sections through a choanoflagellate shows the presence of basally positioned cellular processes (indicated with black rectangle), shown in higher magnification in (F). Scale bars (A–E: 1 µM, F: 200 nm).
Fascin Localizes to Filopodia-Like Structures and Actin-Filled Microvilli in S. rosetta
In metazoans, fascin functions as a filament-bundling protein that localizes to filopodia, and in some cell types also to microvilli (DeRosier and Tilney 2000; Kureishy et al. 2002; Tilney et al. 2004). Given that we identified clear fascin homologs in S. rosetta, M. brevicollis, and C. owczarzaki (fig. 1; supplementary fig. S2B, Supplementary Material online) (see above), we next investigated whether fascin homologs in choanoflagellates might function in filopodia-like structures and microvilli as they do in the metazoans. Western blot analysis showed that a commercially available fascin antibody, which was originally raised against the human protein fascin, recognizes a single band of the expected size when used to probe S. rosetta lysate (fig. 3D). The S. rosetta genome encodes two fascin paralogs with predicted molecular weights of 54.3 and 54.6 kDa. Thus, we performed immunolocalization studies of fascin in S. rosetta. Interestingly, fascin localizes to the basal filopodia-like structures and to the actin-filled collar of S. rosetta (fig. 3A and B). These data suggest a functional conservation of the critical filopodial protein fascin between choanoflagellates and Metazoa.
Fig. 3.
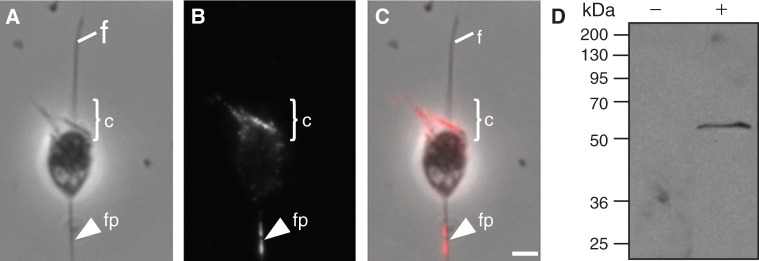
Subcellular localization of Fascin in Salpingoeca rosetta. (A) Phase contrast microscopy shows the morphology of a fixed S. rosetta cell. (B, C) Immunolocalization studies reveal that Fascin localizes to a basal filopodia-like structure (fp) and to the apical actin filled collar (c). (D) Western blot analysis shows that S. rosetta cell lysate probed with Fascin antibodies detect a single band of approximately 55 kDa (+). No signal was detected when primary Fascin antibody was omitted (−). f, flagellum; c, microvilli collar; fp, filopodia. Scale bar: 1 µM.
Expression of Filopodial Genes in C. owczarzaki and S. rosetta
To further investigate the putative functional homology of filopodial genes between metazoans and their unicellular relatives, we analyzed the expression levels of diverse filopodial genes between different life history stages of C. owczarzaki and S. rosetta.
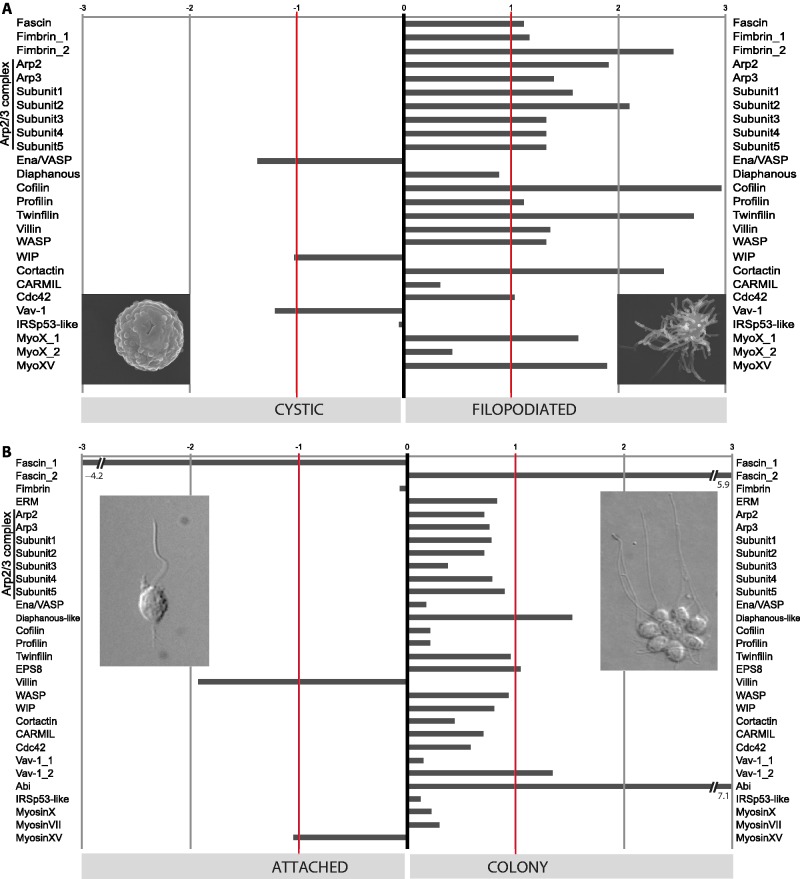
Capsaspora owczarzaki can differentiate into at least two different cell types, an attached cell type that has filopodia-like structures (fig. 2B) and a naked, nonfilopodial, cystic form that is not attached to the substrate (fig. 2C). We investigated the expression of filopodial gene homologs in these two cell types. Homologs of most of the genes involved in metazoan filopodia, such as fascin, myosin X, and Cortactin, are upregulated in the adherent filopodial form of C. owczarzaki (fig. 4A). The differential expression of homologs of filopodial genes in filopodial and nonfilopodial life stages of C. owczarzaki is consistent with the hypothesis that there is functional homology between C. owczarzaki and metazoan filopodia.
Fig. 4.
Expression of filopodial and related genes in unicellular holozoans. (A) Log2-fold expression (see Materials and Methods) of Capsaspora owczarzaki filopodial genes between filopodiated and cystic stages. (B) Log2-fold expression of Salpingoeca rosetta filopodial genes between attached and colonial stages. Red lines highlight 2-fold expression differences. For clarity, negative values indicate overexpression in one stage compared with the other, and vice versa.
Salpingoeca rosetta can differentiate into at least five distinct cell types, including three solitary cell types (slow swimmers, fast swimmers, and substrate attached cells) and two colonial forms (rosettes and chains) (Dayel et al. 2011). Both attached cells and colonial cells have been previously reported to produce filopodia-like structures (Leadbeater 1979; Dayel et al. 2011). In attached cells, filopodia-like structures may mediate the attachment to environmental substrates both by searching the environment for suitable attachment sites and by contributing to the construction of a goblet-shaped attachment structure called a theca. In colonies, filopodia-like structures extend from the basal pole of cells in most, but not all, rosette colonies and may contribute to colony formation or stabilization. When we compared the expression of homologs of filopodial genes between attached cells and colonies (chains and rosettes; fig. 4B), most were not differentially expressed, consistent with the hypothesis that cells in both life history stages form filopodia-like structures. Surprisingly, however, some of the filopodial gene homologs were differentially expressed, suggesting that the molecular composition of filopodia-like structures in different life stages might be specialized. For example, one S. rosetta fascin homolog, Fascin1, is upregulated in attached cells, whereas Fascin2 is upregulated in colonies, suggesting subfunctionalization. Other genes upregulated in colonies are Diaphanous-like, Vav-1 and Abi, whereas Villin and Myosin XV are upregulated in attached cells. This raises the possibility that the different patterns of expression in different types of filopodiated cells in S. rosetta may contribute to cell differentiation.
Evolutionary Assembly of the Metazoan Filopodial Toolkit
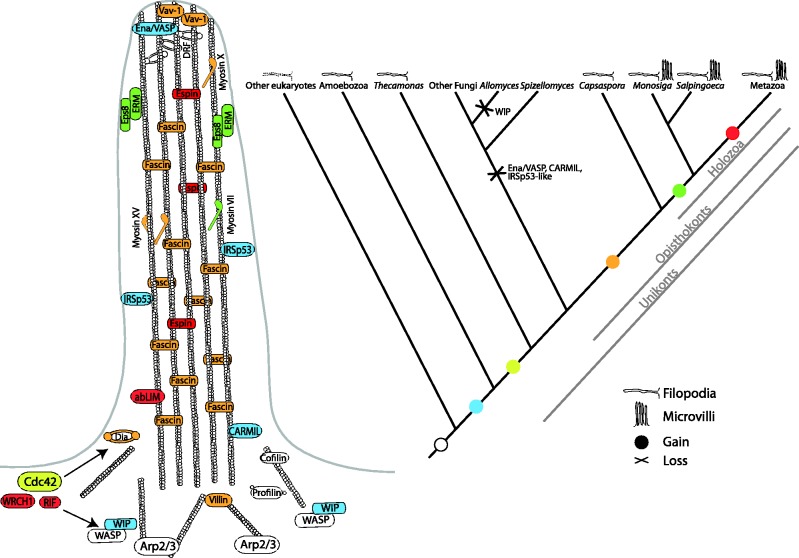
Our evolutionary reconstruction suggests a gradual assembly of the metazoan filopodial toolkit (fig. 5). Many actin remodeling and crosslinking proteins, such as fimbrin, alpha-actinin, profilin, cofilin, and twinfilin, are ancient. It is likely that Arp2/3-WASP-DRF-based filopodia formation (with DRF as the anti-capping agent instead of VASP), coupled with RhoGTPase regulation, was the ancestral eukaryotic mechanism, rather than the formin-based mechanism, because demonstration of DRF-based filopodia formation (independent of Arp2/3 and without the presence of VASP protein, known to be essential for formin-based filopodia formation [Schirenbeck et al. 2006], discussed earlier) awaits demonstration in non-amorphean taxa. Later, the Ena/VASP protein evolved in the amorphean clade, where two independent mechanisms of filopodia formation, Arp2/3-WASP-VASP, and DRFs-VASP have been demonstrated in amoebozoans and in metazoans.
Fig. 5.
Evolution of the metazoan filopodial toolkit. Schematic representation of the metazoan filopodial toolkit (left), with colors indicating the inferred evolutionary origin of each gene (white, eukaryotes; blue, Amorphea; yellow, opisthokonts + apusozoans; orange, holozoans; green, choanoflagellates + metazoans; red, metazoans). The cladogram (right) represents gains and losses and the presence of filopodia and microvilli in different groups. Dashed filopodia in other eukaryotes represent the presence of filopodia only in some groups (see main text).
It was only later, in the stem of the holozoan lineage, that the metazoan filopodia-specific toolkit was established. This includes Cdc42 signalling as an initiator of filopodia formation and also the control of filopodia formation through Tyrosine kinase signaling (involving Src and Abl cytoplasmic TyrK). The metazoan filopodia toolkit also includes fascin as the main actin-bundling protein, specific motor proteins myosin X, VII, and XV and other proteins such as cortactin, Vav-1 and Abi. Our expression data suggest that this complex complement is, indeed, functionally conserved in unicellular holozoans, as most filopodial genes are overexpressed in C. owczarzaki filopodial cells. Moreover the main actin-bundling protein fascin localizes to filopodia and microvilli in S. rosetta and in many metazoan cell types, suggesting that fascin functioned as an actin-crosslinking protein in the filopodia and microvilli of the Urmetazoa.
In the common ancestor of choanoflagellates and metazoans, the complexity of the filopodial apparatus was further expanded as filopodial specialization in the form of the microvillar collar evolved. Our findings support the hypothesis that microvilli and filopodia are related, with microvilli reusing part of the filopodial toolkit, while also depending on the function of proteins like ERM and Eps8, which are restricted to choanoflagellates and metazoans. Finally, in metazoans the toolkit further expanded, particularly with the evolution of new RhoGTPases that are known to act instead of Cdc42 in specific cell-types.
Our analyses suggest that other non-metazoan eukaryotic lineages evolved their specific filopodial toolkits based on an ancient molecular machinery that included core actin linking proteins (profilin, twinfilin, fimbrin, cofilin, and others) and an ancestral filopodia formation mechanism (Arp2/3-WASP-DRFs). We hypothesize this mechanism was deployed under the control of different signaling triggers (e.g., Cdc42 in metazoans and Rac1 in amoebozoans) together with different specific co-factors. The recently reported NET superfamily (Deeks et al. 2012), a plant specific membrane-actin cytoskeleton adaptor protein, exemplifies this idea of convergence (in this case, to mediate the interaction between the membrane and the actin cytoskeleton). Therefore, we infer that metazoan-type filopodia originated at the stem of Holozoa, built upon many ancient proteins and acting with some more recently evolved (i.e., holozoan-specific) molecular components.
In any case, the study of the cell biology and genome content of filopodiated chlorarachniophytes (Rhizaria) (Ota and Vaulot 2012), labyrinthulomycetes, other filopodiated stramenopiles (Tsui et al. 2009; Gómez et al. 2011), and filopodiated Excavata, such as N. gruberi (Preston and King 2005), will be crucial to gain new insights into the question of whether there is a common, functionally homologous, molecular toolkit underlying all eukaryotic filopodia.
Conclusions
Our study reconstructs in detail the evolutionary assembly of the metazoan filopodial and microvillar molecular toolkit. We find that many components of the metazoan filopodial toolkit are paneukaryotic, whereas other elements evolved in stem Amorphea. Finally, a number of metazoan filopodial and microvillar components evolved in stem holozoans. Moreover, some of the components of metazoan microvilli appeared concomitantly with the evolution of the feeding collar at the stem of choanoflagellates and metazoans and likely played a crucial role in the feeding mode of the Urmetazoan, as it does in choanoflagellates.
We further demonstrate that fascin is expressed both in filopodia-like structures and microvilli in the choanoflagellate S. rosetta and that filopodial genes are differentially upregulated in C. owczarzaki’s filopodial cell-stage. This suggests functional conservation of the metazoan filopodial toolkit in both choanoflagellates and C. owczarzaki. Given the predicted homology of filopodia between metazoans and their unicellular relatives, we hypothesize that the existence of a complex filopodial toolkit in the ancestors of metazoans may have contributed to the origin of metazoans, by being co-opted to function in cell–cell and cell-matrix adhesion functions within a multicellular context.
Materials and Methods
Gene Searches and Phylogenetic Analysis
A primary search was performed using the basic local alignment sequence tool (BLAST: BlastP and TBlastN) using Homo sapiens and C. owczarzaki proteins as queries against Protein and Genome databases with the default BLAST parameters and an e-value threshold of e−5 at the National Center for Biotechnology Information (NCBI), the Joint Genome Institute (JGI), the Broad Institute (for S. rosetta and S. punctatus), as well as the Amphimedon queenslandica genome database (www.metazome.net; last accessed June 2013). For some proteins, we also performed Hmmer searches using HMMER3.0b2 (Eddy 1998) to confirm that we were retrieving all orthologs.
We performed searches using the following taxon sampling: seven metazoans (H. sapiens, Drosophila melanogaster, Daphnia pulex, Capitella teleta, Lottia gigantea, Nematostella vectensis, and A. queenslandica), two choanoflagellates (M. brevicollis and S. rosetta), one filasterean (C. owczarzaki), four fungi (Laccaria bicolor, Saccharomyces cerevisiae, A. macrogynus, and S. punctatus), one apusozoan (T. trahens), two amoebozoans (Acanthamoeba castellanii and D. discoideum), three viridiplantae (Arabidopsis thaliana, Ostreococcus taurii, and Chlamydomonas reinhardtii), two excavates (Trichomonas vaginalis and N. gruberi), and three chromalveolates (Thalassiosira pseudonana, Tetrahymena thermophila, and Toxoplasma gondii).
Alignments were constructed using the MAFFT v.6 online server (Katoh et al. 2002) and then manually inspected and edited using Geneious software. Only those species and those positions that were unambiguously aligned were included in the final analyses. Maximum likelihood (ML) phylogenetic trees were estimated by RaxML (Stamatakis 2006) using the PROTGAMMAWAG + Γ + I model, which uses the WAG amino acid exchangeabilities and accounts for among-site rate variation with a four category discrete gamma approximation and a proportion of invariable sites. Statistical support for bipartitions was estimated by performing 100-bootstrap replicates using RaxML with the same model. Bayesian analyses were performed with MrBayes3.2 (Huelsenbeck and Ronquist 2001), using the LG + Γ + I model of evolution, with four chains, a subsampling frequency of 100 and two parallel runs. Runs were stopped when the average standard deviation of split frequencies of the two parallel runs was <0.01, usually at around 18,000,000 generations. The two LnL graphs were checked and an appropriate burn-in length established. Bayesian posterior probabilities were used to assess the confidence values of each bipartition.
Cell Culture and Microscopy
Salpingoeca rosetta cultures enriched for attached cells were maintained in artificial sea water and split 1:5 every 3 days. For immunofluorescence, the cells were grown to a density of 106 cells/mL and carefully scraped off from the surface of the culture flasks. Cells were then pelleted by spinning for 10 min at 4,000 × g and resuspended in a small volume of artificial seawater. Approximately 0.4 mL of the cells were applied to poly-l-lysine coated coverslips, left to attach for 30 min.
Capsaspora owczarzaki cells were grown on coverslides in ATCC medium 1034 (modified PYNFH medium) for two days and directly fixed.
For both S. rosetta and C. owczarzaki, cells were fixed for 5 min with 6% acetone and for 15 min with 4% formaldehyde. The coverslips were washed gently four times with 100 mM Pipes at pH 6.9, 1 mM EGTA, and 0.1 mM MgSO4 (PEM), incubated for 30 min in blocking solution (PEM+: 1% BSA, 0.3% Triton X-100), 1 h in primary antibodies solution (in PEM+), and after further washes (PEM+), 1 h in the dark with fluorescent secondary antibodies (1:100 in PEM+, Alexa Fluor 488 goat anti-mouse, and Alexa Fluor 568 goat anti-rabbit; Invitrogen) and washed again four times (PEM). To visualize F-actin coverslips were incubated for 15 min in the dark with rhodamine phallidin (6 U/ml in PEM; Molecular Probes). After 3 washes (PEM), coverslips were mounted onto slides with Fluorescent Mounting Media (4 μL; Prolong Gold Antifade, Invitrogen). The following primary antibodies have been used: mouse monoclonal antibody against β-tubulin (E7, 1:400; Developmental Studies Hybridoma Bank); mouse monoclonal antibody against Fascin (ab78487, 1:100; Abcam). Images were taken with a 100× oil immersion objective on an inverted Leica microscope.
For SEM, C. owczarzaki cells were fixed for 1 h with 2.5% glutaraldehyde and 1 h with 1% osmium tetroxide, followed by sequential dehydration with ethanol. Next, drying critical point was performed and samples were coated with carbon. Samples were observed in a Hitachi S-4100 microscope.
For TEM, choanoflagellate cells were concentrated by gentle centrifugation, loaded into 100-µm deep specimen carriers and high pressure frozen in a Bal-Tec HPM 010 high pressure freezer (Bal-Tec AG, Liechtenstein). Freeze-substitution was performed over 2 h by the SQFS method of McDonald and Webb (2011), then infiltrated with Eponate 12 resin and polymerized in a Pelco Biowave research microwave oven (Ted Pella, Inc., Redding, CA) over a period of 2 h. Sections were cut at 70-nm thickness, poststained with uranyl acetate and lead citrate, and viewed in a Tecnai 12 transmission EM (FEI Inc., Hillsboro, OR) operating at 120 kV. Images were recorded on a Gatan Ultrascan 1000 CCD camera.
Gene Expression Analyses
Total RNA from C. owczarzaki’s described life stages was extracted using Trizol reagent. Libraries were sequenced with 76 base pair reads on an Illumina HiSeq instrument (Illumina). mRNA was isolated from S. rosetta cultures enriched for colonial and attached cells using the RNAeasy (Qiagen) and Oligotex (Qiagen) kits. Libraries were sequenced with 68 base paired-end reads on an Illumina GAII instrument (Illumina) following manufacturer’s recommendations. In both cases, fragments per kilobase per million reads mapped per CDS was calculated and colonial and attached values averaged and log2 transformed (resulting in negative or positive values corresponding to overexpression in one particular cell stage or the other, this relationship is arbitrary and only for visualization purposes).
Supplementary Material
Supplementary figures S1–S9 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Matt Welch for providing critical feedback on the manuscript. They also thank the Joint Genome Institute and Broad Institute for making data publicly available. N.K. is a Senior Fellow in the Integrated Microbial Biodiversity Program of the Canadian Institute for Advanced Research. This work was supported by an Institució Catalana de Recerca i Estudis Avançats contract, a European Research Council Starting Grant (ERC-2007-StG- 206883), a grant (BFU2011-23434) from Ministerio de Economía y Competitividad (MINECO) to I.R.-T., funding from NIH NIGMS GM089977 to N.K., pregraduate Formacion Profesorado Universitario grant from MICINN to A.S-P., a Deutsche Forschungsgemeinschaft (DFG) postdoctoral fellowship to P.B., and the Canadian Research Chair program grant to B.F.L.
References
- Adl SM, Simpson AGB, Lane CE, et al. (25 co-authors) The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–514. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón IM, Jones GE, Wandosell F, Geha R, Ramesh N. WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 2007;17:555–562. doi: 10.1016/j.tcb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci U S A. 2006;103:12411. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breshears LM, Wessels D, Soll DR, Titus MA. An unconventional myosin required for cell polarization and chemotaxis. Proc Natl Acad Sci U S A. 2010;107:6918–6923. doi: 10.1073/pnas.0909796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Brown MW, Kolisko M, Silberman JD, Roger AJ. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr Biol. 2012;22:1–5. doi: 10.1016/j.cub.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Phylogeny of Choanozoa, Apusozoa, and other Protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur J Protistol. 2013;49:115–178. doi: 10.1016/j.ejop.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny and evolution of Apusomonadida (Protozoa: Apusozoa): new genera and species. Protist. 2010;161:549–576. doi: 10.1016/j.protis.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M. Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol. 2008;25:2717–2733. doi: 10.1093/molbev/msn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Calcutt JR, Ingle EKS, et al. (12 co-authors) A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr Biol. 2012;22:1–6. doi: 10.1016/j.cub.2012.06.041. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lang BF. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 2012;29:1277–1289. doi: 10.1093/molbev/msr295. [DOI] [PubMed] [Google Scholar]
- DeRosier DJ, Tilney LG. F-actin bundles are derivatives of microvilli: what does this tell us about how bundles might form? J Cell Biol. 2000;148:1–6. [PMC free article] [PubMed] [Google Scholar]
- Dumontier M, Höcht P, Mintert U, Faix J. Rac1 GTPases control filopodia formation, cell motility, endocytosis, cytokinesis and development in Dictyostelium. J Cell Sci. 2000;113:2253–2265. doi: 10.1242/jcs.113.12.2253. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, et al. (15 co-authors) Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Gómez F, Moreira D, Benzerara K, López-García P. Solenicola setigera is the first characterized member of the abundant and cosmopolitan uncultured marine stramenopile group MAST-3. Environ Microbiol. 2011;13:193–202. doi: 10.1111/j.1462-2920.2010.02320.x. [DOI] [PubMed] [Google Scholar]
- Gonobobleva E, Maldonado M. Choanocyte ultrastructure in Halisarca dujardini (Demospongiae, Halisarcida) J Morphol. 2009;270:615–627. doi: 10.1002/jmor.10709. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Gertler FB. Filopodia: the fingers that do the walking. Sci Signal. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Ikura M. Radixin: cytoskeletal adopter and signaling protein. Int J Biochem Cell Biol. 2004;36:2131–2136. doi: 10.1016/j.biocel.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JPP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Karpov S, Leadbeater BSC. Cytoskeleton structure and composition in choanoflagellates. J Eukaryot Microbiol. 1998;45:361–367. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, George SP. Regulation of cell structure and function by actin-binding proteins: villin’s perspective. FEBS Lett. 2008;582:2128–2139. doi: 10.1016/j.febslet.2008.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook M, Young S, Kuo A. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;21:4300–4305. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinley AW, Weed SA, Weaver AM, Karginov AV, Bissonette E, Cooper JA, Parsons JT. Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr Biol. 2003;13:384–393. doi: 10.1016/s0960-9822(03)00107-6. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Lbik D, Enge S. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res Notes. 2012;5:1–23. doi: 10.1186/1756-0500-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureishy N, Sapountzi V, Prag S, Anilkumar N, Adams JC. Fascins, and their roles in cell structure and function. Bioessays. 2002;24:350–361. doi: 10.1002/bies.10070. [DOI] [PubMed] [Google Scholar]
- Ladwein M, Rottner K. On the Rho’d: the regulation of membrane protrusions by Rho-GTPases. FEBS Lett. 2008;582:2066–2074. doi: 10.1016/j.febslet.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Leadbeater BSC. Developmental and ultrastructural observations on two stalked marine Choanoflagellates, Acanthoecopsis spiculifera Norris and Acanthoeca spectabilis Ellis. Proc R Soc Lond B Biol Sci. 1979;204:57–66. doi: 10.1098/rspb.1979.0012. [DOI] [PubMed] [Google Scholar]
- Leadbeater BSC, Morton C. A microscopical study of a marine species of Codosiga James-Clark (Choanoflagellata) with special reference to the ingestion of bacteria. Biol J Linn Soc Lond. 1974;6:337–347. [Google Scholar]
- Lundquist EA. The finer points of filopodia. PLoS Biol. 2009;7:e1000142. doi: 10.1371/journal.pbio.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev Biol. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- McClatchey AI. ERM proteins. Curr Biol. 2012;22:R784–R785. doi: 10.1016/j.cub.2012.07.057. [DOI] [PubMed] [Google Scholar]
- McDonald KL, Webb RI. Freeze substitution in 3 hours or less. J Microsc. 2011;243:227–233. doi: 10.1111/j.1365-2818.2011.03526.x. [DOI] [PubMed] [Google Scholar]
- Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803:191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Mikrjukov KA, Mylnikov AP. A study of the fine structure and the mitosis of a lamellicristate amoeba, Micronuclearia podoventralis gen. et sp. nov. (Nucleariidae, Rotosphaerida) Eur J Protistol. 2001;37:15–24. [Google Scholar]
- Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci U S A. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, Tyska MJ. Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell Mol Life Sci. 2010;67:1239–1254. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol. 2008;40:344–349. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S, Eikrem W, Edvardsen B. Ultrastructure and molecular phylogeny of Thaumatomonads (Cercozoa) with emphasis on Thaumatomastix salina from Oslofjorden, Norway. Protist. 2011;163:560–573. doi: 10.1016/j.protis.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Ota S, Vaulot D. Lotharella reticulosa sp. nov.: a highly reticulated network forming chlorarachniophyte from the Mediterranean Sea. Protist. 2012;163:91–104. doi: 10.1016/j.protis.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Pawlowski J. The twilight of Sarcodina: a molecular perspective on the polyphyletic origin of amoeboid protists. Protistology. 2008;5:281–302. [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Preston T, King C. Locomotion and phenotypic transformation of the Amoeboflagellate Naegleria gruberi at the water/air interface. J Eukaryot Microbiol. 2005;50:245–251. doi: 10.1111/j.1550-7408.2003.tb00128.x. [DOI] [PubMed] [Google Scholar]
- Ren G, Crampton MS, Yap AS. Cortactin: coordinating adhesion and the actin cytoskeleton at cellular protrusions. Cell Motil Cytoskeleton. 2009;66:865–873. doi: 10.1002/cm.20380. [DOI] [PubMed] [Google Scholar]
- Revenu C, Athman R, Robine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol. 2004;5:635–646. doi: 10.1038/nrm1437. [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PWH, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MWMW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Schäfer C, Born S, Möhl C, Houben S, Kirchgeßner N, Merkel R. Dependence of adhesion, actin bundles, force generation and transmission on filopodia. Cell Adh Migr. 2010;4:2:215–225. doi: 10.4161/cam.4.2.10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TEB, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci U S A. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Roger A, Lang BF, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- Small J, Auinger S, Nemethova M, Koestler S, Goldie K, Hoenger A, Resch G. Unravelling the structure of the lamellipodium. J Microsc. 2008;231:479–485. doi: 10.1111/j.1365-2818.2008.02060.x. [DOI] [PubMed] [Google Scholar]
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- Soldati D, Meissner M. Toxoplasma as a novel system for motility. Curr Opin Cell Biol. 2004;16:32–40. doi: 10.1016/j.ceb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Sousa AD, Cheney RE. Myosin-X: a molecular motor at the cell’s fingertips. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Suga H, Dacre M, De Mendoza A, Shalchian-Tabrizi K, Manning G, Ruiz-Trillo I. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci Signal. 2012;5:ra35–ra35. doi: 10.1126/scisignal.2002733. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Guild GM. Microvilli appear to represent the first step in actin bundle formation in Drosophila bristles. J Cell Sci. 2004;117:3531–3538. doi: 10.1242/jcs.01215. [DOI] [PubMed] [Google Scholar]
- Tsui CKM, Marshall W, Yokoyama R, Honda D, Lippmeier JC, Craven KD, Peterson PD, Berbee ML. Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol Phylogenet Evol. 2009;50:129–140. doi: 10.1016/j.ympev.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Tuxworth RI, Weber I, Wessels D, Addicks GC, Soll DR, Gerisch G, Titus MA. A role for myosin VII in dynamic cell adhesion. Curr Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- Vallotton P, Small JV. Shifting views on the leading role of the lamellipodium in cell migration: speckle tracking revisited. J Biosci. 2009;122:1955–1958. doi: 10.1242/jcs.042036. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880. doi: 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahou G, Rivero F. Rho GTPase signaling in Dictyostelium discoideum: insights from the genome. Eur J Cell Biol. 2006;85:947–959. doi: 10.1016/j.ejcb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Young ME, Lee W-L, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Yang C, Svitkina T. Filopodia initiation—focus on the Arp2/43 complex and formins. Cell Adh Migr. 2011;5:402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettler LA, Nerad TA, O’Kelly CJ, Sogin ML. The nucleariid amoebae: more protists at the animal-fungal boundary. J Eukaryot Microbiol. 2001;48:293–297. doi: 10.1111/j.1550-7408.2001.tb00317.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, Bhaskar A, Cheney RE, Stromblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel I, Naba A, Da Cunha MML, Del Maestro L, Formstecher E, Louvard D, Arpin M. Ezrin regulates microvillus morphogenesis by promoting distinct activities of Eps8 proteins. Mol Biol Cell. 2012;23:1080–1094. doi: 10.1091/mbc.E11-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.