Abstract
Little is currently known about the genetic complexity of quantitative behavioral variation, the types of genes involved, or their effects on intermediate phenotypes. Here, we conduct a genome-wide association study of Drosophila melanogaster courtship song variation using 168 sequenced inbred lines, and fail to find highly significant associations. However, by combining these data with results from a well-powered Evolve and Resequence (E&R) study on the same trait, we provide statistical evidence that some power to associate genotype and phenotype is available. Genes that are significant in both analyses are enriched for expression in the nervous system, and affect neural development and synaptic growth when perturbed. Quantitative complementation at one of these loci, Syntrophin-like 1, supports a hypothesis that variation at this locus affects variation in the inter-pulse interval of courtship song. These results suggest that experimental evolution may provide an approach for genome-scale replication in Drosophila.
Keywords: association studies, population genetics, behavior, artificial selection, courtship, Drosophila
Introduction
Biologists have long been amazed at the amount of individual variation within populations. Though much of this individuality can be attributed to nonheritable sources, an impressive fraction of it results from genetic variability (Lewontin 1974; Kendler and Greenspan 2006). In a modest but growing list of case studies, the link between a particular genetic variant and variation in a trait has been made, and these cases have proven very illuminating (Stern and Orgogozo 2008; Chan et al. 2009; Wittkopp et al. 2009; Baxter et al. 2010; Martin and Orgogozo 2013). It is difficult, however, to discover general principles among these data due to case-by-case differences in ascertainment bias, methodology, and lack of consistency with respect to traits or model systems (Stern and Orgogozo 2008; Kopp 2009; Martin and Orgogozo 2013). Recent work in human genetics illustrates that genome-wide association studies (GWAS) not only provide a comprehensive and consistent methodology to compare variation among different traits but also show that large sample sizes are required to obtain even modest statistical power (Allen et al. 2010; Yang et al. 2010; Makowsky et al. 2011). A new resource that will facilitate GWAS is now available in the Drosophila melanogaster model system: a collection of 168 completely sequenced inbred lines (Mackay et al. 2012). Though this sample is quite small relative to successful human GWAS, the ability to quantify replicate individuals of each genotype in a common environment may increase power considerably.
Here, we use these lines to investigate the interpulse interval (IPI) of male courtship song. Males in many Drosophila species produce a series of auditory pulses (a “pulse song”) when courting females, and the pause between pulses (IPI) varies greatly among species (Cowling and Burnet 1981; Ewing and Miyan 1986; Gleason and Ritchie 1998). Previous work supports a hypothesis that the courtship song IPI is under sexual selection within species and selection for species recognition between species (Bennet-Clark and Ewing 1969; Ritchie et al. 1999; Talyn and Dowse 2004; Immonen and Ritchie 2012). This evolutionary relevance, combined with the ability to efficiently quantify IPI, makes this trait a good model behavior for genotype–phenotype studies.
Results
Genome-Wide Association Study
To conduct a GWAS of IPI, we recorded 3,956 individual males singing to a standardized female genotype. Of the RAL lines available from the Bloomington Drosophila Stock Center, we were able to characterize 191, and draft genome sequences were available for 168 of these in “Freeze 1” of the Drosophila Genomic Research Panel (DGRP) (supplementary table S1, Supplementary Material online). An average of 16 replicate males were recorded per inbred line in these lines, allowing us to estimate that a significant proportion of IPI variation arises from genetic variation (broad-sense heritability, as estimated from the among inbred-line component of variance = 0.46). To investigate the genetic basis of this variation, we quantified the association between IPI and each of 2.46 million single nucleotide polymorphisms (SNPs). This set of variants includes all SNPs present in a minimum of four lines that met a set of quality filtering standards described in Mackay et al. (2012). Similar to reported results for starvation resistance, startle response, and chill-coma recovery using these lines (Mackay et al. 2012), 142 SNPs are significant at a nominal P < 10−5 threshold, and 31 are significant at a nominal P < 10−6. However, by comparing the observed distribution of P values with 1,000 permuted data sets, the estimated false discovery rate (FDR) at each of these thresholds is found to be considerable: FDR = 0.75 for P < 10−5, FDR = 0.59 for P < 10−6. Indeed, based on these permutations, enrichment of true associations is comparable at P < 10−3 and P < 10−4 thresholds (FDR = 0.79 and 0.69, respectively). As permutations can only partially control for confounds such as population stratification or variable haplotype length across the genome, the true FDR might be even higher than this analysis suggests. Additional information on the GWAS is provided in supplementary data, Supplementary Material online.
Mackay et al. (2012) report that a multiple regression containing only the 6–10 most significant SNPs can explain 60–90% of variation in all traits examined. Similarly, we estimated the variance in IPI explained by the five most significant SNPs in the real data and compared it with the variance explained by the five most significance SNPs in a small number of permuted data sets. Though the multiple regression on the true data explained 46% of the variation in IPI (consistent with Mackay et al.), the five most significant SNPs in the permuted data can “explain” considerable variation as well (26% to 42% across five permutations), suggesting that this method produces spurious results.
Evolve and Resequence: Combined Analysis
By combining GWAS results with previous data, we compared evidence for two possible models: one in which quantitative variation in this trait is due mainly to a few major effect variants and another where variation is due to a larger number of modest-effect causal loci. To weigh evidence for these competing hypotheses, we compared the GWAS results reported here with results from our previous “Evolve and Resequence” (E&R) study (Turner and Miller 2012). In this previous work, we performed 14 generations of divergent selection on IPI in four replicate populations that were created by combining the sequenced inbred lines used here for the GWAS. By resequencing these experimental populations, we associated a large number of SNPs with IPI with very high confidence (13,343 SNPs at 0.005 FDR) (Turner and Miller 2012). Though powerful, this approach also has its weaknesses: because a population-based sequencing approach is used, it is difficult to estimate the fraction of significant variants that are directly affected by selection versus those affected by linked selection. In combination with the E&R approach, however, the sequenced inbred lines of the DGRP have utility, as linkage disequilibrium (LD) patterns are known across the genome. Because LD decays to r2 = 0.2 in less than 50 bp (Mackay et al. 2012), these lines are an ideal way to determine if a large fraction of the 13,343 significant SNPs identified using E&R independently affect IPI, or if differentiation of so many variants was caused by linkage to a few large effect variants. Because selection was performed on populations created by mixing the sequenced inbred lines used in the GWAS, this comparison is straightforward and appropriate.
To test the “few large effects” model, we attempted to replicate the SNPs with P < 10−5 in the GWAS using the E&R study. After 14 generations, the average IPI values of divergently selected populations in the E&R study were different by more than two starting standard deviations (Turner and Miller 2012). Thus, if most initial variation in IPI was due to a few large effect variants, these variants should be highly differentiated. This model is firmly rejected: of the 13,343 most differentiated variants in the E&R study, zero of them also had P < 10−5 in the GWAS study.
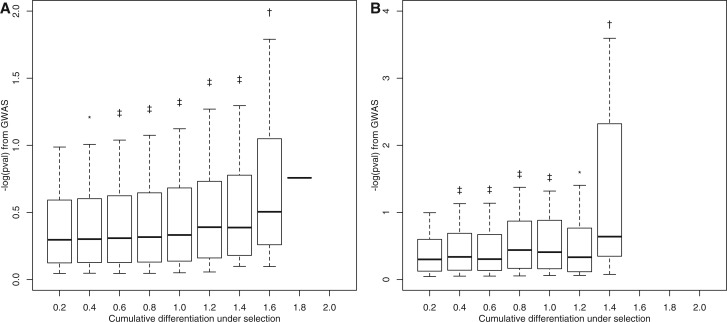
To investigate the alternative model implicated by our E&R study, in which a large number of causal alleles are present in the population, we attempted to validate the E&R results using the GWAS data at a genome-wide level. Figure 1 shows the joint distribution of GWAS P values and degree of differentiation under selection for variants from two frequency slices in the GWAS: the most common SNPs (present in 45–50% of the inbred lines) and the least common SNPs for which association was attempted (present in 2.5–5.0% of the inbred lines). For SNPs in both categories, those that were more differentiated under artificial selection have a significant enrichment of low P values compared with those that differentiated little. This pattern is not due to a few highly significant SNPs, as statistical significance is unchanged by excluding SNPs that had P < 10−5 in the GWAS study. If the low LD among sequenced lines truly indicates that SNPs are statistically independent from all but their close neighbors, the genome-wide correspondence between the amount of evolution under selection and GWAS P value would support a highly polygenic model (but see the discussion presented later).
Fig. 1.
Correlation between evolution under selection and GWAS P value. The x axis is the cumulative differentiation for variants under artificial selection: there were two pairs of divergently selected populations, so a maximum value of 2 is achieved if a variant reciprocally fixes twice. The y axis is the P value distribution for each variant bin from the GWAS, on a −log10 scale. Box plots show median (thick line), outer quartiles (box) and outer dectiles (whiskers). Symbols indicate significantly lower P values vs. the initial, least differentiated bin in a Kolmogorov–Smirnov test: *P < 0.05, †P < 0.005, ‡P < 10−4. (A) The most common SNPs (present in 45–50% of inbred lines); n for each bin = 49,787; 37,727; 23,562; 11,874; 4,962; 1,684; 448; 43; 1. (B) The least common SNPs present in at least 4 lines (present in 2.5–5.0% of inbred lines); n = 480,795; 71,813; 19,286; 4,919; 1,054; 259; 25 for each bin.
To investigate some potential causes of this pattern, we performed the same analysis with several permuted GWAS data sets. As in previous permutations, the vector of phenotypes in the GWAS was randomized, but other data structure was maintained. As shown in the supplementary data, Supplementary Material online, the pattern disappears for the common variants when phenotypes are permuted. Four of 35 possible comparisons in the permuted data sets have P < 0.05, but this is expected due to multiple tests. The single lowest P value across all permuted data sets was 1.4E−3, whereas the true data ranged from 5.4E−4 to 1.0E−15. The permuted data were less clear for the rarest variants in the genome. Though none of the permuted data sets were nearly as significant as the true data, more low P values were present than expected by chance in the permuted data. One potential explanation for this is that some of these variants are in fact common, but appear rare due to missing data or genotyping error. Under this scenario, some “rare” variants are likely to have both lower P values by chance in the GWAS, and higher differentiation due to drift under selection (if they are in fact common).
Candidate Genes
Among the most significant variants in the E&R study (FDR < 0.005), 101 also have P < 0.01 in the GWAS (supplementary table S2, Supplementary Material online). These SNPs are in the exons, introns, or within 1 kb of 42 genes in the D. melanogaster genome annotation (supplementary table S3, Supplementary Material online). Although little is known about genes that might affect IPI (Gleason 2005), we investigated whether these 42 genes were a nonrandom set using data from FlyAtlas (Chintapalli et al. 2007) (available via FlyMine [Lyne et al. 2007] and FlyBase [McQuilton et al. 2012]). FlyAtlas is a tissue-specific gene expression atlas, allowing comparisons of the expression level of genes in 26 specific tissues to the whole organism (table 1). The tissue with the largest proportion of expression enrichment for the genes putatively associated with IPI was the brain, followed by the head, eye, larval central nervous system (CNS), and thoracicoabdominal ganglion. This pattern is characteristic of genes with strong expression in the nervous system: the 37 most brain-biased genes in the genome show strong enrichment in these same 5 tissue fractions (Lyne et al. 2007; Thomas et al. 2012). Though it can be difficult to compare these data with a null expectation that accounts for all possible sources of bias, the genome as a whole does not show enrichment in these tissues, and the enrichment of brain expression in IPI-associated genes is significant (Fisher’s exact test P = 0.016). Consistent with these results, the single most enriched Biological Process (Huang et al. 2009) annotation among these genes is axon guidance (8.7-fold, not significant after correction for multiple tests; supplementary table S4, Supplementary Material online). Though this enrichment is due to only three genes with this annotation (Dscam, Lim3, and beat-Ib), several other IPI-associated genes are annotated with related functions (beat-IIIc, Dif, and CG12484) and contribute to the enrichment of brain gene expression. In addition, two associated genes (SNAP25 and Syn1) are known to regulate synaptic growth at the neuromuscular junction.
Table 1.
Expression Levels of Candidates Genes in Select Tissues, Relative to Expression in the Whole Organism.
| Genes Significant in E&R and GWAS |
Whole Genome | |||
|---|---|---|---|---|
| Upregulated | Downregulated | Up/Down | Up/Down | |
| Brain | 19 | 12 | 1.58 | 0.64 |
| Head | 15 | 12 | 1.25 | 0.64 |
| Eye | 17 | 15 | 1.13 | 0.53 |
| Larval CNS | 15 | 14 | 1.07 | 0.88 |
| Thoracicoabdominal ganglion | 15 | 14 | 1.07 | 0.63 |
| Crop | 12 | 13 | 0.92 | 0.55 |
| S2 cells | 13 | 15 | 0.87 | 0.69 |
| Testes | 13 | 16 | 0.81 | 0.49 |
| Salivary gland | 10 | 14 | 0.71 | 0.59 |
| Midgut | 11 | 16 | 0.69 | 0.45 |
| Carcass | 10 | 15 | 0.67 | 0.53 |
| Heart | 10 | 16 | 0.63 | 0.40 |
| Larval carcass | 11 | 18 | 0.61 | 0.46 |
| Male accessory gland | 7 | 12 | 0.58 | 0.49 |
| Hindgut | 8 | 15 | 0.53 | 0.56 |
| Larval hindgut | 8 | 16 | 0.50 | 0.54 |
| Larval fat body | 6 | 12 | 0.50 | 0.43 |
| Larval trachea | 10 | 20 | 0.50 | 0.56 |
| Larval midgut | 8 | 18 | 0.44 | 0.39 |
| Larval tubule | 8 | 20 | 0.40 | 0.47 |
| Tubule | 10 | 28 | 0.36 | 1.08 |
| Larval salivary gland | 7 | 20 | 0.35 | 0.52 |
| Ovary | 7 | 20 | 0.35 | 0.92 |
| Fat body | 6 | 21 | 0.29 | 0.44 |
| Virgin spermatheca | 6 | 21 | 0.29 | 0.42 |
| Mated spermatheca | 5 | 18 | 0.28 | 0.44 |
Note.—Brain and Eye categories are both significant at P < 0.05.
Validation of Candidate Genes
An ideal way to validate these putative associations would be to replace the allele in a reference strain with each of the two natural alleles, and then measure the IPI of these otherwise genetically identical strains. However, due to the potentially high rate of false discovery in these data, this approach may prove inefficient. Instead, we used a quantitative complementation approach (Pasyukova et al. 2000; Service 2004). This method compares the trait values of four F1 genotypes: a strain with each natural allele is crossed to a strain with a mutated copy of the candidate gene and a control strain. When the natural strains are crossed to the mutant strain, the natural alleles at that locus are functionally hemizygous, but when crossed to the control strain, any difference between the natural alleles will be reduced by the presence of an additional reference allele. If the natural alleles affect the phenotype, this should manifest as a significant interaction term in a specific direction. Genotypes with the putative long-IPI allele should have a longer IPI when combined with the mutant strain versus the control strain and/or genotypes with the putative short-IPI allele should have a shorter IPI when combined with the mutant strain vs. the control strain.
We attempted validation at two genes: Syntrophin-like 1 (Syn1) and Down syndrome cell adhesion molecule (Dscam). For Syn1, we crossed strains with each natural allele to a strain with a modified Minos transposable element (Mi{ET1}) inserted into a coding exon of Syn1 (Metaxakis et al. 2005); a strain with the same genotype, except for the insertion, was available to use as the control. For Dscam, we used a strain with an engineered deletion of 300 kb that removed approximately 45 genes including the target locus. This mutation is homozygous lethal and is therefore maintained in a heterozygous state over a nonrecombining balancer chromosome. We used hybrid offspring with the balancer chromosome as controls, and compared them with hybrid offspring with the deletion.
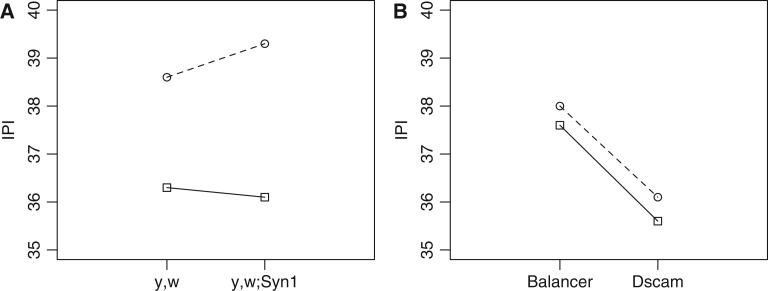
Quantitative complementation at the Syn1 gene supports a hypothesis that this gene harbors natural variation affecting IPI (fig. 2). The main effect of wild type line (RAL-365 vs. RAL-555) was highly significant (P < 2e−16), but the main effect of genomic background (control strain vs. insertion) was not (P = 0.08). Indeed, the marginal means of individuals with the control strain was very similar to the flies with the insertion in Syn1 (37.6 ms vs. 37.8 ms), indicating that the genomic background is well controlled. The IPI of the strain with the putative long-IPI allele become longer when crossed to the insertion strain, and the putative short-IPI strain became shorter, as expected if variation at Syn1 affects IPI. The interaction is slight compared with the main effects, however, and a very large data set was required to obtain statistical significance (P = 0.04, N = 898 individuals).
Fig. 2.
Quantitative complementation tests. Each point is the median IPI of an F1 genotype; dashed lines and circles are F1s with RAL-555 (with putative long-IPI alleles at both loci) and solid lines and squares are F1s with RAL-365 (with putative short-IPI alleles). (A) Complementation test of Syn1 and (B) complementation test of Dscam.
If all alleles are additive, the difference between insertion F1s and control F1s (0.9 ms) should estimate the difference in effect between natural alleles. This difference is similar in magnitude to the effect size estimated from the GWAS. The A/T polymorphism at position 21,749,958 on the left arm of the third chromosome falls within an intron of Syn1 and was the SNP that was among the most significant in the combined GWAS + E & R data set. The IPI of inbred lines with the T allele averaged 1.4 ms shorter than those with the A allele. This SNP was at high frequency (0.44) in the DGRP population, and had a P value of 0.0003 in the GWAS (uncorrected for multiple tests). It was also among the most divergent variants in the E&R study: the T allele was at frequencies of 0.91 and 1.00 in the short-IPI populations, but only 0.32 and 0.28 in the long-IPI populations.
Significant results in a quantitative complementation assay are subject to alternative interpretations (Service 2004). For example, if the mutated allele interacts epistatically with alleles at other loci that differ between the wild-type strains, this could produce a significant interaction term. This is likely to be a much greater problem when using deficiency stocks, which delete a large number of loci, and comparing the F1s to balancer chromosomes. Moreover, the similar magnitude of the interaction term, in the predicted direction, supports the hypothesis that this interaction detected here is due to allelic differences at Syn1 rather than elsewhere in the genome.
In contrast to results at Syn1, complementation at the Dscam gene provided no evidence for natural variation (fig. 2). In the RAL collection, an intron of this gene contained a T/C polymorphism with similar allele frequency (0.44) and effect size estimate (1.0 ms) as the variant at Syn1. It was less significant in the GWAS (P = 0.0089), but highly differentiated in the E&R study in the expected direction (0.10 and 0.46 in the short-IPI populations, 0.81 and 0.95 in the long-IPI populations). In the complementation test, however, there was no evidence for a significant interaction (P = 0.52). The marginal mean of flies with the balancer chromosome is much lower than flies with the deficiency (38.2 vs. 36.8), indicating a less-controlled genomic background; this is not surprising because the balancer chromosome is expected to have a different genotype from the deletion chromosome at many sites. Despite this difference between control and deletion F1s, the two natural strains changed in a parallel way, contrary to the expectation if natural variation in IPI was due in part to variation at Dscam.
Discussion
The results presented here suggest that GWAS on 168 sequenced Drosophila lines has insufficient power to determine the genetic basis of variation in IPI. Although permutations indicate enrichment of true associations in these data, this slight enrichment is insufficient to treat SNPs exceeding P < 10−5 or P < 10−6 thresholds as highly enriched data sets (FDR = 0.75 for P < 10−5, FDR = 0.59 for P < 10−6). As the distributions of low P values reported here for IPI are similar to those previously reported for chill coma recovery, starvation resistance, and startle response (Mackay et al. 2012), these are potentially general conclusions. This seems paradoxical at first, as Mackay et al. report that a multiple regression containing only the 6–10 most significant SNPs can explain 60–90% of variation in all traits they examined. However, this result may be spurious, as multiple regression models built with IPI data are able to “explain” variation even when permuted. This is likely due to the circular nature of building and testing models with the same data (see also Ober et al. 2012).
Despite the lack of power in the GWAS, the sequenced lines may have utility when combined with other sources of data. When combined with an E&R study, which had different strengths and weaknesses than the GWAS, these lines appear to support a highly polygenic model for IPI variation. The overall enrichment of low GWAS P values among SNPs that became differentiated under selection is intriguing. This pattern does not appear to be a trivial consequence of variable power, as it remains when starting SNP frequency is controlled (fig. 1). The pattern is somewhat suspect for rare variants, as qualitatively (if not quantitatively) similar patterns were found in some permuted data sets (supplementary data, Supplementary Material online). For common variants, however, observed data were very distinct from permuted data. Because of the very low LD among sequenced lines, this pattern seems to support an extremely polygenic model. It should be noted, however, that there may be nonindependence among SNPs in the sequenced lines that is not captured in the LD measurement. Indeed, the recent analysis by Ober et al. (2012) suggests that there is limited haplotype diversity in this population, and this might indicate that the results in figure 1 could be created by a more modest number of causal variants.
Our combined analysis is similar to a recent analysis Huang et al. (2012). These authors also mixed inbred lines from this collection, imposed artificial selection, resequenced the populations and compared results with a GWAS. Huang et al. found that zero of the SNPs with P < 10−5 in the GWAS were significant in the second experiment for chill coma recovery, starvation resistance, and startle response. This failure to replicate is in agreement with our results, and is to be expected if the FDRs in the GWAS is very high. However, rather than concluding that SNPs with P < 10−5 in the GWAS may be false discoveries, these authors propose that the lack of correspondence is due to epistasis. The evidence supporting this hypothesis was the nonrandom association of gene function with SNPs in both analyses. We consider this is be a low standard of evidence, as it is difficult to form a null hypothesis for the expected associations with gene function that include all possible sources of bias. It therefore seems more parsimonious to conclude that the failure to replicate is due to a high FDR. The genome-wide correspondence between experiments that we found was not reported by Huang et al. It remains to be determined whether this is due to differences in the design or execution of the selection experiment or differences in the architecture of the genotype–phenotype relationships for different traits.
Genes with replicated associations between experiments are found to be a nonrandom subset of the genome. This may indicate that, despite the apparent complexity of IPI variation, the combined data sets have sufficient power to locate some of the genes affecting this behavior. As this conclusion rests entirely upon the nonrandom gene annotations of overlapping genes, the standard of evidence is modest at best. It is likely that gene length, heterozygosity, the allele-frequency spectrum, and other parameters are also nonrandomly associated with gene function, so it is possible that this result is due to unmodeled biases. We therefore attempted to validate two of the most intriguing candidates using quantitative complementation. Though we find no evidence that Dscam harbors natural variation affecting IPI, complementation at Syn1 was successful. This increases the standard of evidence supporting a causal association between alleles of Syn1 and IPI, though nonallelic complimentation provides an alternative interpretation of our results (Service 2004). The Syn1 gene is one of two syntrophins in the D. melanogaster genome, which are homologous to a larger syntrophin gene family in humans (Nagai et al. 2010). When the Drosophila syntrophins are knocked down, flies have locomotory defects and synaptic overgrowth at the neuromuscular junction (Nagai et al. 2010). Consistent with the high expression of Syn1 in the nervous system, knock-down of this gene (together with Syn2) in the nervous system recapitulates these defects, but expression in muscle does not (Chintapalli et al. 2007; Nagai et al. 2010). Drosophila produce their courtship song by extending and vibrating one wing, so differential enervation of the indirect flight muscles is a possible intermediate phenotype to the effects on IPI (Bennet-Clark and Ewing 1969). The overall overrepresentation of genes expressed in the nervous system may indicate that differences in the morphology or physiology of the nervous system could be a mechanism connecting other genetic variation to courtship song variation as well. It will be interesting to investigate the generality of these results by determining the genetic basis of IPI variation in other species, the divergence in IPI between species, and the genetic basis of other behaviors in D. melanogaster.
Materials and Methods
Drosophila Stocks and Culture
The RAL collection of inbred lines, created in the lab of Trudy Mackay (Ayroles et al. 2009; Mackay et al. 2012), were obtained from the Bloomington Drosophila Stock Center. These lines were collected by the Mackay lab in Raleigh, NC, USA, and each line underwent full-sibling inbreeding for 20 generations to eliminate most genetic variation (Ayroles et al. 2009). Flies were maintained in 25 × 95 mm vials on molasses medium in standard Drosophila incubators at 25 °C under a 12-h light/dark cycle. The outbred mixture of these lines used for E&R was created by stochastically mass-mating 173 of these lines and allowing them to mix to the F5 generation, as previously described (Turner and Miller 2012).
To quantitatively compliment the association at Syn1 (discussed later), we used Bloomington stock 23336: y1 w67c23; MiET1Syn1MB01557. This stock has an engineered Minos transposable element inserted into a coding exon of Syn1 (Bellen et al. 2004; Metaxakis et al. 2005). To control for genetic background, we used the parent stock 6599: y1 w67c23. For complementation at Dscam, we used stock 23162: w1118; Df(2R)BSC263/CyO (Cook et al. 2012). This stock has an engineered 300 kb deletion (i.e., deficiency) that eliminates approximately 45 genes including Dscam. As this deletion is homozygous lethal, it is maintained in a heterozygous state over a nonrecombining “balancer” chromosome, which is also homozygous lethal. We crossed this line to each of two wild type RAL lines with different natural alleles, and compared F1 genotypes with the balancer to the F1s with the deficiency. RAL line 365 contains the putative short-IPI allele at both genes, and line 555 contains the putative long allele at both genes, so these two stocks were used as the wild types in the complementation test.
Song Recording
Songs were recorded as previously described (Turner and Miller 2012). Briefly, we recorded songs from 10 males at a time: each male was placed with a single 1-day old virgin female from the RAL-380 line (a genotype randomly chosen to serve as a standardized courtship object) in a recording chamber. Ten recording chambers were each attached to 10 40PH array microphones (GRAS Sound and Vibration) in parallel, and background noise was removed by subtracting the median of all 10 recordings from each recording at each time point. A bandpass filter was used to filter out all frequencies above 1 kHz (as suggested by M. Ritchie, University of St. Andrews). The pulse song produced by males during courtship was distinguished from other sounds using filtering criteria: pulse length < 2 ms, cycles/pulse < 10, pulse amplitude > 1.0 mV, 5 or more consecutive pulses per pulse train, and IPI between 15 and 100 ms. Recording took place in an environmentally controlled room, 25 ± 0.5 °C. As IPI varies linearly with temperature (∼1.0 ms/°C; Ritchie and Kyriacou 1994), we used the temperature during each recording interval to standardize all measurements within the 1 °C of variation present. Some authors have reported that the IPI varies in a sinusoidal fashion during courtship (the Kyriacou and Hall cycle or KH cycle [Kyriacou and Hall 1980]). To minimize any possible effect of a KH cycle on measured IPI, we calculated the mean IPI per male from a 5-min courtship bout (average 192 IPI events). Males with fewer than 20 IPIs in a 5-min recording were discarded. The median IPI values for all lines are provided in supplementary table S1, Supplementary Material online.
GWAS
The median IPI per line was used to associate genotype and phenotype. No difference in IPI was found between lines infected with the Wolbachia symbiont (Mackay et al. 2012) and those that are uninfected (P = 0.66), so infection status was not considered further. Associations were tested at each marker using ANOVAs of the form Y = μ + M + ε, where M is the marker effect. These tests were implemented using the web utilities at dgrp.gnets.ncsu.edu (Mackay et al. 2012). Permutations were performed by randomizing the IPI values assigned to each line, then performing ANOVA in R using the lm() function. FDR was estimated by dividing the expected number of associations at any given P value threshold by the observed number of associations at that threshold, where the expected number is the average number of associations across 1,000 permutations.
Combining Data Sets
For each of the 2.46 million SNPs in the GWAS, we determined the allele frequencies in a previous data set (Turner and Miller 2012). In this data set, two populations were selected to have shorter IPI and two were selected to have longer IPI, and the allele frequency of variants across the genome were estimated from pooled sequencing. Details of this experiment are presented elsewhere. Briefly: selection populations were established by mixing the RAL inbred lines stochastically to the F5 generation. Selection strength was approximately 20%, with an average of 66 males selected to reproduce each generation with 200 females. To reduce variance in reproductive success that would reduce the effective population sizes, populations were reared in a set of vials, with only 2 males and 6 females per vial (offspring were mixed each generation). Selection was carried out for 14 generations. The 120 individuals with the most extreme trait values across generations 10–14 were pooled and pulped en masse for DNA extraction. The average sequence coverage of variable sites was 200-fold per population, or 800-fold experiment-wide. It should be noted that the RAL380 genotype used as the courtship object in the GWAS was not used in the E&R study. We have no evidence that the genotype of the female influences the male song, however, and no such evidence has been found by others.
As a cumulative measure of differentiation under selection for each variant, we used the sum of allele frequency differences between the two replicate comparisons (abs[short-IPI population 1 − long-IPI population 1 + short-IPI population 2 − long-IPI population 2]). The maximum value would therefore be 2, achieved if a variant reciprocally fixes twice. We compared the distribution of this statistic with the distribution of GWAS P values by binning the data and comparing bins with a Kolmogorov–Smirnov test. We compared statistics for different frequency classes of SNPs: all comparisons were quantitatively similar, so we present only the most common and most rare frequency classes here (these comparisons are the most qualitatively different).
Functional Annotations
FlyAtlas (Chintapalli et al. 2007) data were accessed using FlyMine (Lyne et al. 2007). The expression profiles of candidate genes were compared with a list of all genes in the 5.43 D. melanogaster genome annotation, and to a list of the 37 most brain-biased genes in the genome available at FlyMine.com (last accessed February 27, 2012). DAVID (Huang et al. 2009) was also used to conduct a functional clustering of gene-ontology terms (supplementary table S4, Supplementary Material online).
Quantitative Complementation
For each quantitative complementation test, we measured the trait values of four genotypes: F1 offspring resulting from crosses between each of two natural isolates with both a mutant strain and a control strain. ANOVA was not used to test for significant interactions because the distributions of IPI values within each genotype were found to be significantly nonnormal using a Shapiro–Wilk test (Shapiro and Wilk 1965). Instead, we used the nonparametric adjusted rank transform (ART) test (Leys and Schumann 2010). Like the rank transform test (Conover and Iman 1981), the ART test replaces phenotypic values with ranks. In addition, the ART allows for tests of interaction terms by eliminating main effects before transformation. We initially quantified a total of 100–200 flies per complementation test. Data at Dscam were almost exactly parallel (no interaction), so further data were not collected. Data at Syn1 were consistent with an interaction term of the expected magnitude, so data were collected in batches of several hundred flies we reached the sample size required for this interaction to become significant. The median phenotypes of each genotype were very consistent throughout this process.
Supplementary Material
Supplementary data, figures S1–S4, and tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank many UCSB undergraduates for recording tens of thousands of flies singing and Alison Pischedda for editing the manuscript. This work would not have been possible without the community supported resources available at Flybase.org and the Bloomington Drosophila Stock Center. This work was supported by the University of California Santa Barbara, the National Institute of Health grant R01 GM098614, and the National Science Foundation support to the Center for Scientific Computing at UCSB (NSF MRSEC DMR-1121053 and NSF CNS-0960316).
References
- Allen HL, Estrada K, Lettre G, et al. (291 co-authors) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, et al. (11 co-authors) Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, et al. (12 co-authors) A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 2010;6:e1001193. doi: 10.1371/journal.pgen.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, et al. (12 co-authors) The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennet-Clark HC, Ewing AW. Pulse interval as a critical parameter in the courtship song of Drosophila melanogaster. Anim Behav. 1969;17:755–759. [Google Scholar]
- Chan YF, Marks ME, Jones FC, et al. (16 co-authors) Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2009;327:303–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Conover WJ, Iman RL. Rank transformations as a bridge between parametric and nonparametric statistics. Am Statist. 1981;35:124–129. [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling DE, Burnet B. Courtship songs and genetic control of their acoustic characteristics in sibling species of the Drosophila melanogaster subgroup. Anim Behav. 1981;29:924–935. [Google Scholar]
- Ewing AW, Miyan JA. Sexual selection, sexual isolation and the evolution of song in the Drosophila repleta group of species. Anim Behav. 1986;34:421–429. [Google Scholar]
- Gleason JM. Mutations and natural genetic variation in the courtship song of Drosophila. Behav Genet. 2005;35:265–277. doi: 10.1007/s10519-005-3219-y. [DOI] [PubMed] [Google Scholar]
- Gleason JM, Ritchie MG. Evolution of courtship song and reproductive isolation in the Drosophila willistoni species complex: do sexual signals diverge the most quickly? Evolution. 1998;52:1493–1500. doi: 10.1111/j.1558-5646.1998.tb02031.x. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, et al. (25 co-authors) Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A. 2012;109:15553–15559. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen E, Ritchie MG. The genomic response to courtship song stimulation in female Drosophila melanogaster. Proc Biol Sci. 2012;279:1359–1365. doi: 10.1098/rspb.2011.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Greenspan RJ. The nature of genetic influences on behavior: lessons from “simpler” organisms. Am J Psychiatry. 2006;163:1683–1694. doi: 10.1176/ajp.2006.163.10.1683. [DOI] [PubMed] [Google Scholar]
- Kopp A. Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution. 2009;63:2771–2789. doi: 10.1111/j.1558-5646.2009.00761.x. [DOI] [PubMed] [Google Scholar]
- Kyriacou CP, Hall JC. Circadian rhythm mutations in Drosophila melanogaster affect short-term fluctuations in the male’s courtship song. Proc Natl Acad Sci U S A. 1980;77:6729–6733. doi: 10.1073/pnas.77.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. The Genetic Basis of Evolutionary Change. New York: Columbia University Press; 1974. [Google Scholar]
- Leys C, Schumann S. A nonparametric method to analyze interactions: the adjusted rank transform test. J Exp Social Psychol. 2010;46:684–688. [Google Scholar]
- Lyne R, Smith R, Rutherford K, et al. (21 co-authors) FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, et al. (52 co-authors) The Drosophila melanogaster genetic reference panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowsky R, Pajewski NM, Klimentidis YC, Vazquez AI, Duarte CW, Allison DB, de los Campos G. Beyond missing heritability: prediction of complex traits. PLoS Genet. 2011;7:e1002051. doi: 10.1371/journal.pgen.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- McQuilton P, Pierre SES, Thurmond J, Consortium F. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai R, Hashimoto R, Yamaguchi M. Drosophila syntrophins are involved in locomotion and regulation of synaptic morphology. Exp Cell Res. 2010;316:2313–2321. doi: 10.1016/j.yexcr.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Ober U, Ayroles JF, Stone EA, et al. (11 co-authors) Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 2012;8:e1002685. doi: 10.1371/journal.pgen.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova EG, Vieira C, Mackay TFC. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics. 2000;156:1129–1146. doi: 10.1093/genetics/156.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MG, Halsey EJ, Gleason JM. Drosophila song as a species-specific mating signal, and the behavioural importance of Kyriacou & Hall cycles in D. melanogaster song. Anim Behav. 1999;58:649–657. doi: 10.1006/anbe.1999.1167. [DOI] [PubMed] [Google Scholar]
- Ritchie MG, Kyriacou CP. Genetic variability of courtship song in a population of Drosophila melanogaster. Anim Behav. 1994;48:425–434. [Google Scholar]
- Service PM. How good are quantitative complementation tests? Sci Aging Knowledge Environ. 2004;2004:pe13. doi: 10.1126/sageke.2004.12.pe13. [DOI] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talyn BC, Dowse HB. The role of courtship song in sexual selection and species recognition by female Drosophila melanogaster. Anim Behav. 2004;68:1165–1180. [Google Scholar]
- Thomas A, Lee P-J, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS One. 2012;7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Miller PM. Investigating natural variation in Drosophila courtship song by the evolve and resequence approach. Genetics. 2012;191:633–642. doi: 10.1534/genetics.112.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Stewart EE, Arnold LL, Neidert AH, Haerum BK, Thompson EM, Akhras S, Smith-Winberry G, Shefner L. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science. 2009;326:540–544. doi: 10.1126/science.1176980. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. (12 co-authors) Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.