SUMMARY
Filopodia are slender cellular protrusions that dynamically extend and retract to facilitate directional cell migration, pathogen sensing, and cell-cell adhesion [1–4]. Each filopodium contains a rigid and organized bundle of parallel actin filaments, which are elongated at filopodial tips by formins and Ena/VASP proteins [5–10]. However, relatively little is known about how the actin filaments in the filopodial shaft are spatially organized to form a bundle with appropriate dimensions and mechanical properties. Here, we report that the mammalian formin Daam1 (Disheveled associated activator of morphogenesis 1) is a potent actin bundling protein and localizes all along the filopodial shaft, which differs from other formins that localize specifically to the tips. Silencing of Daam1 led to severe defects in filopodial number, integrity and architecture, similar to silencing of the bundling protein fascin. This led us to investigate the potential relationship between Daam1 and fascin. Fascin and Daam1 co-immunoprecipitated from cell extracts, and silencing of fascin led to a striking loss of Daam1 localization to filopodial shafts but not tips. Further, purified fascin bound directly to Daam1, and multi-color single molecule TIRF imaging revealed that fascin recruited Daam1 to and stabilized Daam1 on actin bundles in vitro. Our results reveal an unanticipated and direct collaboration between Daam1 and fascin in bundling actin, which is required for proper filopodial formation.
RESULTS AND DISCUSSION
Endogenous Daam1 localizes all along the filopodial shafts
Filopodia contain unbranched actin filaments that are organized into parallel arrays and cross-linked along their lengths in the filopodial shaft [11, 12]. There is little understanding of how the filaments in filopodia are organized, but the actin bundling protein fascin plays a critical role in keeping filopodia rigid and straight [11–13]. Silencing of fascin leads to filopodia with a “wavy” appearance, in which the actin bundle is bent, buckled, and loosely organized [12]. Importantly, fascin depletion does not abolish bundling of filaments in filopodia, suggesting that other bundling proteins may contribute to this process [12]. Here, we report a role for the formin protein Daam1 in bundling actin to drive filopodia formation.
In a variety of cell types, formins localize to the tips of filopodial and lamellipodial protrusions, and have been demonstrated to be critical for actin nucleation and elongation at these sites [5, 7, 8, 14, 15]. However, many formins have the additional biochemical activity of bundling actin filaments, even though the potential relevance of this activity in vivo has remained largely unexplored. We performed immunostaining of endogenous Daam1 in B16F1 mouse melanoma cells and found that, unlike other reported formins, Daam1 localizes along the entire filopodial shaft and throughout the lamellipodium (Figure 1A and 1B). Line scan analysis revealed a strong co-localization of Daam1 with F-actin in these structures (Figure 1C). This localization to filopodia was verified with a second anti-Daam1 antibody (Figure S1A). Similar colocalization of Daam1 and F-actin in filopodia was observed in other cell lines, including NIH3T3 filbroblasts, N2A neuroblastoma cells, and Raw 264.7 macrophages (Figure S1B). Further, EGFP-tagged full-length Daam1 (EGFP-FL-Daam1) and constitutively active Daam1 (EGFP-Daam1ΔDAD) both localized to filopodia in B16F1 cells (Figure S1C).
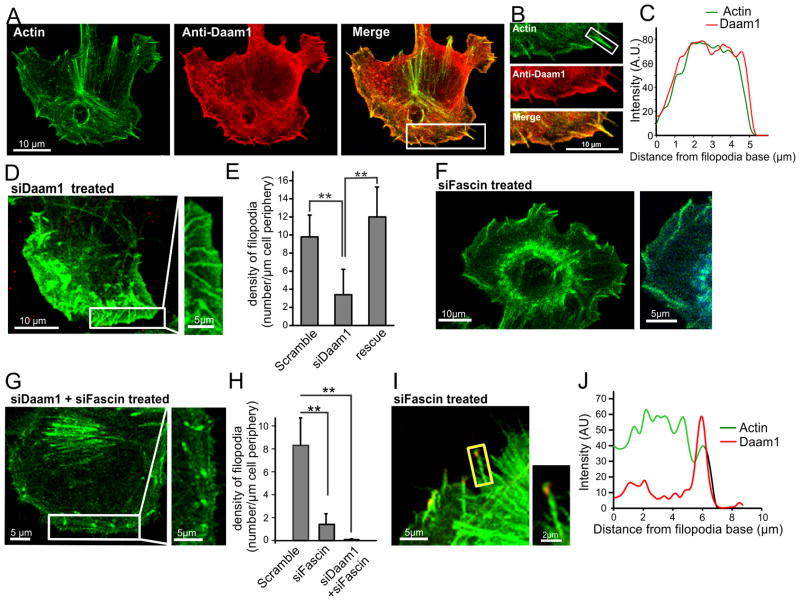
Figure 1. Daam1 localizes to filopodia shafts and is required for filopodia integrity.
(A) Co-localization of Daam1 and actin. B16F1 cells were fixed and stained with rhodamine phalloidin and anti-Daam1 antibody. (B) Magnified view of boxed region in ‘A’. (C) Line scan analysis of Daam1 and actin fluorescence intensities along the filopodium boxed in ‘B’. (D) Daam1 silencing leads to a decrease in filopodia density and altered filopodial morphology. (E) Average filopodia density (number of filopodia per 20 μm of cell periphery) in siDaam1-treated cells, and cells rescued by expression of EGFP-CDaam1 (n=10 cells, p<0.001). Error bars represent SD. (F) Silencing of fascin leads to a decrease in filopodia density and altered filopodial morphology, reminiscent of Daam1 silencing. (G) F-actin staining in cells where fascin and Daam1 were co-silenced. (H) Quantification of average filopodia densities. Error bars represent SD (n = 10 cells; p<0.001). (I) Silencing of fascin reduces Daam1-localization to filopodia shafts but not tips. (J) Line scan analysis of fluorescence intensities of Daam1 and actin along the filopodium boxed in ‘I’.
Daam1 silencing causes severe defects in filopodia formation
To analyze the potential role of Daam1 in filopodia formation, we silenced Daam1 in B16F1 cells using si-oligonucleotides (siDaam1) directed against its N-terminus. Silencing was confirmed by Western blot analysis (Figure S1D) and by loss of Daam1 immunostaining in siDaam1-treated cells (Figure S1E). Daam1 silencing led to pronounced defects, including filpodia with a wavy appearance (Figure 1D), reminiscent of the defects caused by fascin silencing in B16F1 cells [12, 16]. This observation suggested that Daam1, like fascin, is required for proper structural integrity of filopodia. In addition, there was a ~2.5 fold decrease in the density of filopodia in siDaam1-treated cells compared to cells treated with control scrambled oligos (Figure 1E), again similar to defects caused by fascin silencing and consistent with a destabilization of filopodia structure. These phenotypes were also rescued by expression of an RNAi-resistant EGFP-CDaam1 (encompassing FH1-FH2-COOH) (Figure 1E), demonstrating that the filopodial phenotypes arise from loss of Daam1, and that the C-terminal half of Daam1 (used in all of the biochemical experiments below) is sufficient to perform Daam1’s critical role(s) in filopodia formation. A similar striking loss of filopodia was observed upon Daam1 silencing in three additional cell lines (NIH3T3, N2A and Raw264.7; Figure S1B), indicating that Daam1 is critical for filopodia formation in diverse cells types.
A number of formins, including Drosophila DAAM, are capable of bundling actin filaments in vitro [16–23], but there have been few studies addressing their in vivo roles as bundlers. We considered whether mouse Daam1 might function in this capacity within the filopodia shaft. Therefore, we next silenced fascin to directly compare the effects to Daam1 silencing. Silencing of fascin alone caused defects similar to those previously reported [12], which included a drastic reduction in filopodia density and the appearance of aberrant wavy filopodia (Figure 1F and 1H). Co-silencing of Daam1 and fascin led to even stronger defects in filopodia density, although some remaining short F-actin-rich foci or nubs were observed (Figure 1G and 1H). These results indicate that fascin and Daam1 are each critical for maintaining the normal density and appearance of filopodia. In addition, we observed in fascin-silenced cells a drastic reduction in Daam1 localization along filopodia shafts, although Daam1 still localized to filopodia tips (Figure 1I, S1F and 1J). These observations show that fascin is required for proper localization of Daam1 to filopodial actin bundles, and suggest that Daam1 may perform separate roles in the filopodia shafts versus tips.
Daam1 is a potent actin filament bundler
To better understand the activities of Daam1 that underlie its in vivo role(s) in filopodia formation, we purified the C-terminal half of human Daam1, CDaam1 (FH1-FH2-COOH), and analyzed its effects on actin filament dynamics and organization in vitro. Similar to many other formins, in pyrene-actin assembly assays CDaam1 directly stimulated actin polymerization and protected growing barbed ends of filaments from inhibition by capping protein (Figure 2A). Further, actin nucleation by CDaam1 was suppressed by profilin (Figure 2A) and an I698A mutation in CDaam1 abolished actin nucleation activity (Figure 2B) [24–26].
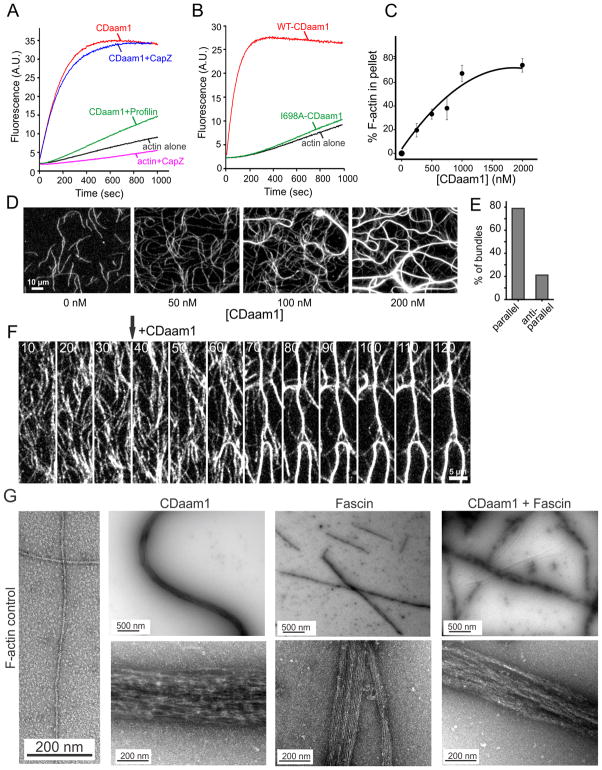
Figure 2. CDaam1 promotes actin filament assembly and bundling in vitro.
(A) Assembly of 2 μM G-actin (5% pyrene-labeled) in the presence of 20 nM CDaam1 (red curve) or 20 nM CDaam1 and 2 nM CapZ (blue curve). CDaam1-induced actin assembly is suppressed by 3 μM profilin (green curve). (B) Comparison of actin assembly stimulated by WT-CDaam1 (50 nM, red curve) and I698A-CDaam1 (200 nM, green curve). (C) Low-speed pelleting of F-actin in the presence of different concentrations of CDaam1. Gels of pellets and supernatants from one experiment are shown in Figure S2A. Error bars represent SD; n=3 experiments. (D) TIRF microscopy images (taken 10 min into reactions) of F-actin bundle formation (1 μM actin, 10% Oregon-Green (OG)-labeled) in the presence of different concentrations of CDaam1. (E) Quantification of the polarity of actin filaments in CDaam1-induced bundles. (F) TIRF microscopy time-lapse imaging of CDaam1 bundling effects on pre-formed actin filaments. Arrow indicates when 300 nM CDaam1 was introduced into the reaction. (G) Electron micrographs of negatively stained actin filaments (far left) or bundles of filaments generated by CDaam1 (left), fascin (center) or CDaam1 and fascin together (right), imaged after 30 min (upper panels) or 18 hours (lower panels) of incubation.
In addition to its polymerization effects, CDaam1 bundled actin filaments in a concentration dependent manner in low-speed sedimentation assays (Figure 2C and S2A). To examine actin filament bundling by CDaam1 in real time, we used total internal reflection fluorescence (TIRF) microscopy to visualize OG (Oregon Green)-labeled actin filaments. Nanomolar concentrations of CDaam1 induced bundle formation, and the abundance of bundles scaled with increasing CDaam1 concentrations (Figure 2D). Further, we observed zippering of polymerizing filaments into bundles (Figure S2B and Movie S1), reminiscent of previously reported fascin- or fimbrin-induced filament zippering [27, 28]. Analysis of bundle polarity by identification of the fast-growing barbed ends of filaments in time lapse imaging revealed that approximately 80% of CDaam1-induced bundles consisted of filaments with their barbed ends pointing in the same direction (Figure 2E, S2C, and Movie S1). This parallel arrangement is similar to the filament organization observed in filopodia [27, 29, 30]. We then tested whether, in addition to being able to bundle filaments that are actively growing, Daam1 could bundle preformed filaments. After polymerizing filaments in TIRF chambers, we flowed in 300 nM CDaam1 in the absence of actin monomers and observed rapid bundling, which reached completion by ~50 seconds after flow-in (Figure 2F and Movie S2).
Our in vivo observations showing that two different bundling proteins, fascin and Daam1, are required to organize actin into a structure that can support normal filopodial protrusion prompted us to compare the ultrastructural effects of fascin and Daam1 on bundle formation in vitro using transmission electron microscopy (TEM) (Figure 2G and S2D). Fascin produced very straight bundles consisting of flattened arrays of regularly spaced filaments, enabling measurement of average number of filaments per bundle (17.7 ± 1.6) and average inter-filament distance (8.2 ± 0.5 nm) [16, 31] (Figure S2D). In contrast, CDaam1 produced wavy, densely packed, rounded bundles, precluding any analysis of the number of filaments per bundle or inter-filament distances. Interestingly, bundles formed in the combined presence of CDaam1 and fascin showed intermediate levels of bundle straightness, roundedness, and width, and intermediate levels of inter-filament spacing and organization. Thus, the composite bundles appear to acquire distinct properties from each bundler, which may help explain their co-requirement for filopodia formation in vivo.
Fascin directly binds and recruits Daam1 to actin filament bundles
Our in vivo observation that fascin is required for Daam1 localization to filopodial shafts prompted us to test whether CDaam1 and fascin physically associate. From B16F1 cells expressing GFP alone, GFP-CDaam1, or GFP-FL-Daam1 from plasmids, we found that endogenous fascin coimmmunoprecipitated with both GFP-Daam1 constructs but not with GFP alone, suggesting that fascin interacts with the C-terminal half of Daam1 (Figure 3A). Further, purified CDaam1 bound to immobilized GST-fascin but not GST (Figure 3B), demonstrating that fascin-Daam1 interactions are direct.
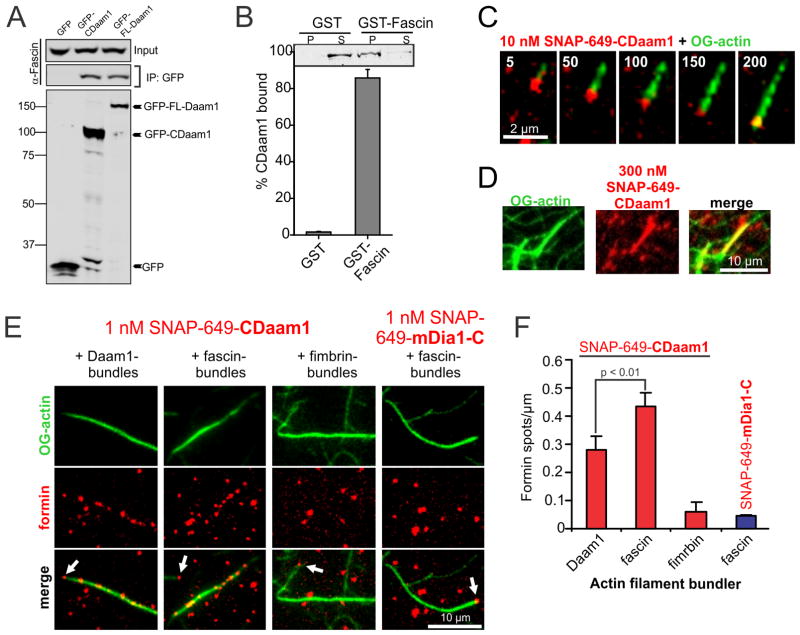
Figure 3. Fascin directly binds and recruits Daam1 to actin filament bundles.
(A) Coimmunoprecipitation in B16F1 cells of endogenous fascin with plasmid-expressed GFP–CDaam1 and GFP-FL-Daam1 but not GFP. Blots were probed with antibodies to fascin (top) and GFP (bottom). (B) Direct binding between purified fascin and CDaam1. Immunoblot probed with anti-Daam1 antibodies and quantification of levels of soluble CDaam1 in pellet fractions after incubation with GST or GST–fascin beads. Error bars represent SD, n = 3. (C) Dual color TIRF microscopy of an elongating actin filament (green), conditions as above, processively capped by a SNAP-649-CDaam1 molecule (red). Time is given in sec. (D) F-actin bundling by SNAP-649-CDaam1. 1 μM actin (10% OG-labeled) was polymerized in the presence of 300 nM SNAP-649-CDaam1, producing bundles heavily decorated with SNAP-649-CDaam1. (E) Single-molecule binding of SNAP-649-CDaam1 to pre-formed F-actin bundles. 1 μM actin (10% OG-labeled) was polymerized in the presence of 300 nM CDaam1, fascin or fimbrin (Sac6) to generate actin filament bundles, then 1 nM SNAP-649-CDaam1 or SNAP-649-mDia1-C was introduced, and binding of formin molecules (red) to bundles (green) was monitored. Arrows mark formin molecules associated with filament/bundle barbed ends. (F) Quantification of the number of SNAP-649-CDaam1 or SNAP-649-mDia1-C spots per micron length of bundle.
To better understand the functional relationship between fascin and Daam1, we employed two-color TIRF microscopy to directly visualize labeled Daam1 molecules interacting with actin bundles. A purified SNAP-CDaam1 fusion protein was labeled with Benzylguanine-Dy649. SNAP-649-CDaam1 nucleated actin assembly in bulk assays indistinguishably from unlabeled CDaam1 (Figure S3A). Further, it enhanced filament elongation rates in TIRF assays in a profilin-dependent manner (Figure S3B). Single SNAP-649-CDaam1 molecules processively moved on growing barbed ends of actin filaments in the presence and absence of profilin (Figure 3C, S3C and Movies S3) as observed for SNAP-mDia1 [32]. At concentrations above 100 nM, SNAP-649-CDaam1 induced actin bundle formation, similar to untagged CDaam1, and decorated the bundles (Figure 3D). Thus, SNAP-649-CDaam1 was functionally equivalent to CDaam1 in all of its effects on actin dynamics and organization.
To analyze dynamics of SNAP-649-CDaam1 molecules on actin bundles, we used low nanomolar concentrations of SNAP-649-CDaam1, which reduced the background and enabled detection of single-molecule binding events. Since these concentrations of SNAP-649-CDaam1 do not bundle filaments, we pre-bundled filaments with 300 nM of unlabeled CDaam1, fascin, or fimbrin (as a control), then flowed-in lower concentrations (1–10 nM) of SNAP-649-CDaam1. SNAP-649-CDaam1 molecules rarely interacted with the sides of single (non-bundled) filaments but readily associated with the barbed ends (Figure 3C and R.J. unpublished observations). When filaments were bundled by fascin or unlabeled CDaam1, SNAP-649-CDaam1 molecules bound to the sides of bundles (Figure 3E). In contrast, when filaments were bundled by fimbrin, SNAP-649-CDaam1 showed no interactions with the sides of bundles (Figure 3E). Thus, SNAP-649-CDaam1 recruitment to bundles is highly specific, consistent with the direct interaction between fascin and CDaam1.
As an additional control, we asked whether a different formin, mDia1, which lacks bundling activity and is absent from filopodial shafts [33–35], was recruited to the sides of bundles. SNAP-tagged mDia1 (SNAP-649-mDia1-C, FH1-FH2-COOH) readily associated with barbed ends of filaments and remained processively attached during elongation as expected [32]. However, no specific interactions of SNAP-649-mDia1-C with the sides of fascin-generated bundles were detected (Figure 3E). Thus, interactions of SNAP-649-CDaam1 with the sides of bundles are highly specific. Quantification of the number of SNAP-649-CDaam1 and SNAP-649-mDia1-C spots per unit length of bundle further revealed that SNAP-649-CDaam1 interactions are more pronounced for fascin-induced bundles compared to CDaam1-induced bundles (Figure 3F). Taken together with our other observations, fascin appears to directly recruit Daam1 to bundles.
Finally, we analyzed the dynamics of single SNAP-649-CDaam1 molecules in CDaam1- and fascin-generated actin bundles to gain insights into the mechanism of Daam1 interactions with these structures. SNAP-649-CDaam1 molecules were observed to transiently bind CDaam1-bundled filaments, with the majority of dwell times being shorter than 50 seconds (Figure 4A, 4C, S3D and Movie S4). However, in fascin-induced bundles SNAP-649-CDaam1 dwell times greatly increased (Figure 4B, 4D, S3D and Movie S4), suggesting that fascin stabilizes CDaam1 associations with the bundles. SNAP-649-CDaam1 molecules displayed 2-dimensional diffusion along CDaam1-induced bundles, as determined by mean-square displacement analysis (Figure 4A, 4E and Movie S5). In contrast, diffusion was abrogated in fascin-induced bundles as indicated by the spatially static spots (Figure 4B, 4D and Movie S4). Thus, fascin confines SNAP-CDaam1 in the bundle, which is remarkably consistent with our in vivo observations showing that Daam1 localization to filopodial shafts is diminished after fascin silencing.
Figure 4. Single molecule analysis of SNAP-649-CDaam1 dynamics on actin bundles generated by unlabeled CDaam1 or fascin.
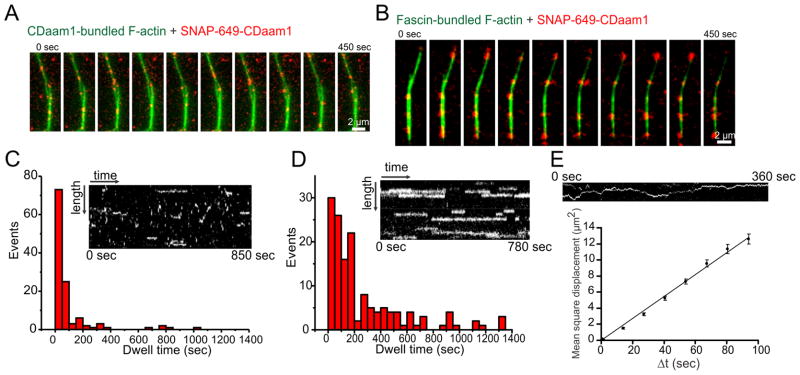
(A and B) Time-lapse imaging of SNAP-649-CDaam1 molecules binding to actin filament bundles produced by CDaam1 (A) or fascin (B). (C) and (D) Kymographs of SNAP-649-CDaam1 binding to bundles from experiments as in ‘A’ and ‘B’. Histograms show dwell-times of SNAP-649-CDaam1 molecules on CDaam1 (C) and fascin (D) bundles. (E) Mean-square displacement of the diffusion of a single SNAP-649-CDaam1 molecule (as shown in kymograph above) on a CDaam1-generated bundle plotted against the time interval. A linear curve fit yields a diffusion coefficient of D = 0.06825 μm2s−1.
Concluding Remarks
In summary, our results reveal a critical role for the formin Daam1 in organizing filaments in the filopodial shaft, and in maintaining the structural integrity of filopodia. Until now, fascin has been the only bundler shown to both localize all along the filopodia shaft and to be required for filopodia integrity. We found that Daam1 similarly localizes along the shafts, and demonstrated that both Daam1 and fascin are required for proper filopodia formation. Further, we found that fascin and Daam1 directly associate, and that fascin recruits Daam1 to filopodial shafts in vivo. In vitro, fascin also is sufficient to recruit Daam1 to actin bundles and to restrict Daam1 diffusion along bundles. Together, these data reveal an unanticipated and direct collaboration between these two actin bundlers, where each is critical in vivo for the proper formation of filopodia.
Finally, our results expand the in vivo roles of formins to include actin bundling. Until now, formin cellular functions in regulating the actin cytoskeleton have focused primarily on actin filament nucleation and elongation, and there have been few in vivo investigations into their potential roles as bundlers. Even though bundling activities have been described in vitro for many formins [17, 33], the in vivo relevance of these activities has remained in question. Our findings demonstrate a critical role for the formin Daam1 in bundling filaments and collaborating with fascin to give the filopodia shaft structural integrity. Formins also decorate a number of other actin arrays in vivo that contain bundled filaments (e.g. stress fibers, cytokinetic rings, stereocilia, invadopodia, bristles, and sarcomeres) [36–41], where their bundling activities may be relevant.
Supplementary Material
RESEARCH HIGHLIGHTS.
Daam1 is a potent actin bundling protein and localizes to filopodial shafts
Silencing of Daam1 disrupts filopodia formation and architecture
Fascin binds to and recruits Daam1 to filopodial shafts in vivo
Fascin stabilizes Daam1 association with actin bundles in vitro
Acknowledgments
We are grateful to Ray Habas, Jan Faix, and Jeff Gelles for sharing key reagents, to Ed Dougherty for assistance with confocal microscopy, to Jeff Gelles for advice on single molecule imaging, and to Chen Xu for his assistance in the Brandeis EM facility. We also thank Brian Graziano, Brooke McCartney, Avital Rodal, and Casey Ydenberg for comments on the manuscript. This work was funded by a fellowship to D.B. from the DFG (BR 4116-1/1) and by grants to B.L.G. from the NIH (GM083137) and NSF (DMR-MRSEC-0820429).
Footnotes
Supplemental Information, which includes three figures and corresponding figure legends, Supplemental Experimental Procedures, and five movies can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: Complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Faix J, Rottner K. The making of filopodia. Current opinion in cell biology. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nature reviews Molecular cell biology. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Block J, Stradal TE, Hanisch J, Geffers R, Kostler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. Journal of microscopy. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- 6.Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nature cell biology. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS biology. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. The EMBO journal. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallavarapu A, Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. The Journal of cell biology. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeRosier DJ, Edds KT. Evidence for fascin cross-links between the actin filaments in coelomocyte filopodia. Experimental cell research. 1980;126:490–494. doi: 10.1016/0014-4827(80)90295-5. [DOI] [PubMed] [Google Scholar]
- 12.Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. The Journal of cell biology. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto JJ, Kane RE, Bryan J. Formation of filopodia in coelomocytes: localization of fascin, a 58,000 dalton actin cross-linking protein. Cell. 1979;17:285–293. doi: 10.1016/0092-8674(79)90154-5. [DOI] [PubMed] [Google Scholar]
- 14.Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Current biology : CB. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris ES, Gauvin TJ, Heimsath EG, Higgs HN. Assembly of filopodia by the formin FRL2 (FMNL3) Cytoskeleton. 2010;67:755–772. doi: 10.1002/cm.20485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. Mechanism of actin filament bundling by fascin. The Journal of biological chemistry. 2011;286:30087–30096. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barko S, Bugyi B, Carlier MF, Gombos R, Matusek T, Mihaly J, Nyitrai M. Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. The Journal of biological chemistry. 2010;285:13154–13169. doi: 10.1074/jbc.M109.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esue O, Harris ES, Higgs HN, Wirtz D. The filamentous actin cross-linking/bundling activity of mammalian formins. Journal of molecular biology. 2008;384:324–334. doi: 10.1016/j.jmb.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 19.Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006;16:1924–1930. doi: 10.1016/j.cub.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 20.Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- 21.Machaidze G, Sokoll A, Shimada A, Lustig A, Mazur A, Wittinghofer A, Aebi U, Mannherz HG. Actin filament bundling and different nucleating effects of mouse Diaphanous-related formin FH2 domains on actin/ADF and actin/cofilin complexes. J Mol Biol. 2010;403:529–545. doi: 10.1016/j.jmb.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 23.Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 24.Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breitsprecher D, Koestler SA, Chizhov I, Nemethova M, Mueller J, Goode BL, Small JV, Rottner K, Faix J. Cofilin cooperates with fascin to disassemble filopodial actin filaments. Journal of cell science. 2011;124:3305–3318. doi: 10.1242/jcs.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Current biology : CB. 2010;20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. The Journal of cell biology. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Small JV. The actin cytoskeleton. Electron microscopy reviews. 1988;1:155–174. doi: 10.1016/s0892-0354(98)90010-7. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa R, Sakamoto T, Ando T, Higashi-Fujime S, Kohama K. Polarized actin bundles formed by human fascin-1: their sliding and disassembly on myosin II and myosin V in vitro. Journal of neurochemistry. 2003;87:676–685. doi: 10.1046/j.1471-4159.2003.02058.x. [DOI] [PubMed] [Google Scholar]
- 32.Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. The Journal of biological chemistry. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 34.Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. The Journal of cell biology. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, Desmarais V, Holman HA, Kitchen S, Backer JM, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. The Journal of cell biology. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. The Journal of cell biology. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D, Hallett MA, Zhu W, Rubart M, Liu Y, Yang Z, Chen H, Haneline LS, Chan RJ, Schwartz RJ, Field LJ, Atkinson SJ, Shou W. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development. 2011;138:303–315. doi: 10.1242/dev.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato A, Khadka DK, Liu W, Bharti R, Runnels LW, Dawid IB, Habas R. Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development. 2006;133:4219–4231. doi: 10.1242/dev.02590. [DOI] [PubMed] [Google Scholar]
- 39.Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Current biology : CB. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 40.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. The EMBO journal. 2008;27:618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Molecular biology of the cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.