Abstract
The AlkB enzyme is an Fe(II)- and α-ketoglutarate-dependent dioxygenase that repairs DNA alkyl lesions by a direct reversal of damage mechanism as part of the adaptive response in E. coli. The reported substrate scope of AlkB includes simple DNA alkyl adducts, such as 1-methyladenine, 3-methylcytosine, 3-ethylcytosine, 1-methylguanine, 3-methylthymine, and N6-methyladenine, as well as more complex DNA adducts, such as 1,N6-ethenoadenine, 3,N4-ethenocytosine, and 1,N6-ethanoadenine. Previous studies have revealed, in a piecemeal way, that AlkB has an impressive repertoire of substrates. The present study makes two additions to this list, showing that alkyl adducts on the N2 position of guanine and N4 position of cytosine are also substrates for AlkB. Using high resolution ESI-TOF mass spectrometry, we show that AlkB has the biochemical capability to repair in vitroN2-methylguanine, N2-ethylguanine, N2-furan-2-yl-methylguanine, N2-tetrahydrofuran-2-yl-methylguanine, and N4-methylcytosine in ssDNA but not in dsDNA. When viewed together with previous work, the experimental data herein demonstrate that AlkB is able to repair all simple N-alkyl adducts occurring at the Watson–Crick base pairing interface of the four DNA bases, confirming AlkB as a versatile gatekeeper of genomic integrity under alkylation stress.
Introduction
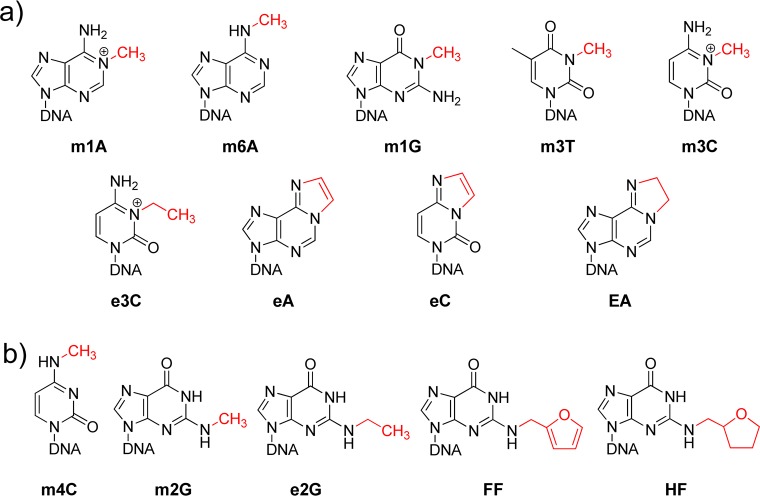
More than a decade ago, the unusual catalytic activity of the DNA repair enzyme AlkB was demonstrated by Sedgwick,1 Falnes,2 and co-workers, whereby oxygen is added to foreign, unnatural carbon atoms attached to nitrogen bases, leading to the liberation of an aldehyde and restoration of the canonical purine or pyrimidine base structures within DNA. Initially, the results pointed to a major role for AlkB3 in protecting cells against low molecular weight alkylating agents that react with nitrogen atoms on the Watson–Crick base-paring side of purine and pyrimidine DNA bases forming covalent adducts, such as the positively charged 1-methyladenine (m1A), 3-methylcytosine (m3C), and 3-ethylcytosine (e3C) lesions (Figure 1a).1,2,4−6 The methylated nucleobases 1-methylguanine (m1G), 3-methylthymine (m3T), and N6-methyladenine (m6A),4,7 uncharged at physiological pH, were also discovered to be substrates of AlkB. More complex DNA adducts formed either by oxidative stress-derived lipid byproducts or exogenous vinyl chloride exposure, such as 1,N6-ethenoadenine (eA) and 3,N4-ethenocytosine (eC),8,9 as well as the 1,N6-ethanoadenine (EA) adduct formed by the anticancer agent BCNU (bis-chloroethylnitrosourea), are also repaired by AlkB both in vitro and in vivo (Figure 1a).7,10 Although we explored the possibility of AlkB acting upon certain alkyl-adducts of oxygen (O6-methylguanine (O6mG)) and carbon (5-methylcytosine (m5C)), no such evidence was found.11 Several crystal structures of AlkB and homologues bound to alkyl substrates have provided detailed information on their substrate recognition and repair specificity.6,12−14 The substrate scope and properties of AlkB have been reviewed in several articles.15−20
Figure 1.
Chemical structures and abbreviations of DNA lesions investigated as possible repair substrates by the AlkB protein using ESI-TOF mass spectrometry. (a) Lesions in this panel are known substrates for AlkB; (b) lesions in this panel were tested as potential substrates for AlkB in this study. The repair target within each base is highlighted in red.
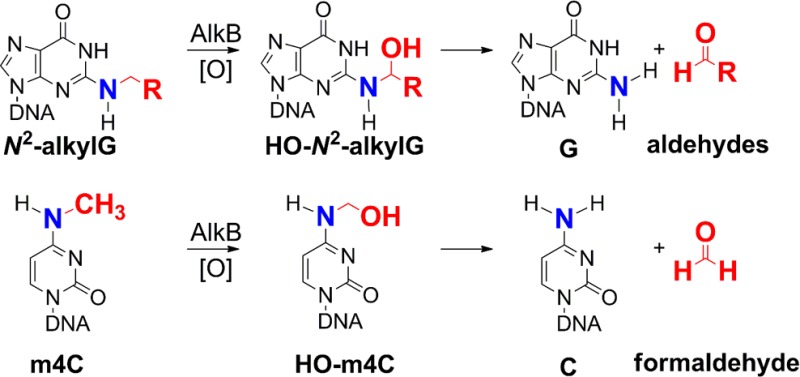
Many scenarios of alkylation damage have been explored in order to find possible substrates for AlkB with respect to base, position of alkylation, and type of alkyl group damage (Figure 1a). However, there are still two positions on the Watson–Crick base-paring interface of the four bases that, if alkylated, have not yet been investigated as possible substrates for AlkB, namely, the N2 position of guanine and the N4 atom of cytosine. To provide a complete picture of the substrate range of AlkB, we examined the possibility that this enzyme can act upon the following series of N2-alkylguanine adducts: N2-methylguanine (m2G), N2-ethylguanine (e2G), N2-furan-2-yl-methylguanine (FF), N2-tetrahydrofuran-2-yl-methylguanine (HF), and N4-methylcytosine (m4C) (Figure 1b). The spectrum of lesions chosen for this analysis was motivated by biological relevance gleaned from several studies. The m2G lesion is formed by a reaction of the N2-amino group of guanine with formaldehyde, a human and rodent carcinogen.21 The e2G lesion is formed by a similar reaction of deoxyguanosine with acetaldehyde, a chemical that occurs both endogenously from the metabolic oxidation of ethanol and from lipid peroxidation products, as well as from environmental sources such as tobacco smoke, vehicle exhaust, and food flavorings.22 Importantly, acetaldehyde has been shown to be carcinogenic in rodents and can elicit a variety of DNA lesions in human cells.23,24 The FF lesion is a structural model of the principal DNA adduct formed by nitrofurazone (NFZ),25 a topical antibacterial agent used for treating burns and skin grafts in patients and animals.26,27 This study also explores HF, the saturated analogue of FF. Postreplicative modification of cytosine by DNA methylases produces m4C, which is an epigenetic marker for bacteria.28−31 In this study, we used high resolution electrospray ionization time-of-flight (ESI-TOF) mass spectrometry to analyze the reaction products of AlkB and the five alkyl adducts mentioned above (Figure 1b). We demonstrate that these lesions are substrates for AlkB, leading to the conclusion that all simple N-alkyl lesions, such as methyl adducts, at the Watson–Crick base-pairing interface on the four bases can be dealkylated by AlkB in single stranded DNA.
Experimental Procedures
Oligonucleotide Synthesis
Sixteen-mer oligonucleotides of the sequence 5′-GAAGACCTXGGCGTCC-3′, where X is the lesion of interest (m2G, e2G, FF, HF, and m4C), were synthesized and purified as described.4,8,25,32−34 The calculated monoisotopic molecular weight (MW) for each oligonucleotide is shown in Table 1.
Table 1. Calculated and Observed Monoisotopic Molecular Weight of Oligonucleotides and Intermediates Expected and Found in AlkB Incubations.
| lesion or base | MW (calculated)
of neutral species |
m/z (calculated) –4 charge peak |
m/z (observed) –4 charge peak |
|---|---|---|---|
| G | 4904.86 | 1225.21 | 1225.11/24 |
| m2G | 4918.87 | 1228.71 | 1228.62 |
| e2G | 4932.89 | 1232.21 | 1232.25 |
| FF | 4984.88 | 1245.21 | 1245.25 |
| FF-2H | 4982.87 | 1244.71 | 1244.74 |
| HO-FF | 5000.88 | 1249.21 | 1249.25 |
| HF | 4988.91 | 1246.22 | 1246.26 |
| HF-2H | 4986.90 | 1245.72 | 1245.75 |
| HO-HF | 5004.91 | 1250.22 | 1250.26 |
| C | 4864.85 | 1215.20 | 1215.20 |
| U | 4865.83 | 1215.45 | 1215.45 |
| m4C | 4878.87 | 1218.71 | 1218.70 |
To create double-stranded substrates, complementary 17-mer oligonucleotides of the sequence 5′-TGGACGCCYAGGTCTTC-3′ were synthesized and purified. These were one base longer to avoid overlap with the MS signal from 16-mer lesion-containing oligonucleotides. Y denotes the base placed opposite the lesion X (C for N2-alkylG lesions and G for m4C). DNA concentration was measured by UV absorbance using a calculated extinction coefficient (ε) at 260 nm. The extinction coefficient used for an alkylated base was that of its unmodified counterpart, due to the negligible difference between the values in the context of 16-mer DNA.
In Vitro Incubations of Alkylated DNA with AlkB
All assays were performed with AlkBΔN11 protein, a truncated version of AlkB in which the first 11 residues were deleted. AlkBΔN11 protein was purified as described10 and shown previously to have activity similar to that of the wild-type protein.6,10,11 All AlkB incubation reactions used similar conditions as previously described.8,11 Reactions were performed at 37 °C in 45 mM HEPES (pH 8.0), 0.9 mM α-ketoglutarate, 67 μM Fe(NH4)2(SO4)2, and 1.8 mM ascorbate, followed by dry ice storage until ESI-TOF MS analysis. A typical reaction was performed with all the necessary cofactors, 5 μM lesion-containing DNA, and either 2.5 μM AlkB protein or protein purification buffer in a 10-μL volume for 1 h. Where indicated, complementary DNA (6 μM) was added and annealed at 65 °C for 5 min to form a dsDNA substrate for the AlkB reaction.
LC-ESI-TOF MS and MS/MS Analysis
Oligonucleotide analyses were performed on an Agilent 6510 ESI-TOF mass spectrometer (Palo Alto, CA). ESI was performed using a needle voltage of 3.5 kV. Nitrogen gas was set with drying at 10 L/min, with the heated capillary at 325 °C and the nebulizer set at 15 psig. Liquid chromatographic separations were performed using a Zorbax SB-Aq column (2.1 × 150 mm; 3.5 μm; Agilent Technologies, Palo Alto, CA) with a flow rate of 0.2 mL/min. Solvent A was 10 mM ammonium acetate in water (pH ∼7); solvent B was 100% acetonitrile. A linear gradient was carried out in the following steps: 98 to 70% A over 30 min, 70 to 98% A over 5 min, and 98 to 98% A over 10 min. Data were processed using Agilent MassHunter Workstation software v7.11
LC-MS/MS analyses were conducted on an Agilent 6510 QTOF system equipped with an ESI source. The MS/MS fragmentation was operated in the negative ion mode. Operating parameters were as follows: gas temperature, 350 °C; ESI capillary voltage, 4000 V; nebulizer pressure, 50 psi; drying gas flow, 10 L/min; fragmentation energy, 20 V. The theoretical m/z values, as illustrated in Figure S29 (Supporting Information), were calculated using Mongo Oligo Mass Calculator, v2.06 (http://library.med.utah.edu/masspec/mongo.htm).
Results
The ability of AlkB to modify the five alkylated bases was tested in vitro using a previously described experimental procedure.8,10,11 Briefly, N2-dG and N4-dC adducts were incorporated into 16-mer oligonucleotides in a site-specific manner. The modified oligonucleotides were then incubated with purified AlkB for 1 h at 37 °C, and the reaction products and intermediates (Scheme 1) were analyzed by high resolution ESI-TOF mass spectrometry. For each lesion, experiments were conducted both in the presence and absence of AlkB with all necessary cofactors. Additionally, the repair experiments were also carried out in the presence of a 17-mer complementary strand; the N2G lesions were placed opposite a cytosine, and the m4C lesion was placed opposite a guanine. The 16-mer oligonucleotides demonstrated a good signal in the −4 charge envelope of the mass spectra (Figure 2). To illustrate the method of analysis, the molecular weight (MW) of the 16-mer carrying an m2G lesion is calculated as 4918.87 Da (Table 1); its monoisotopic peak (all 12C, 14N, 16O, etc.) in the −4 charge state has a calculated m/z of 1228.71. Experimentally, we observed a peak at an m/z of 1228.62 (Figure 2a and Figure S1 (Supporting Information)).
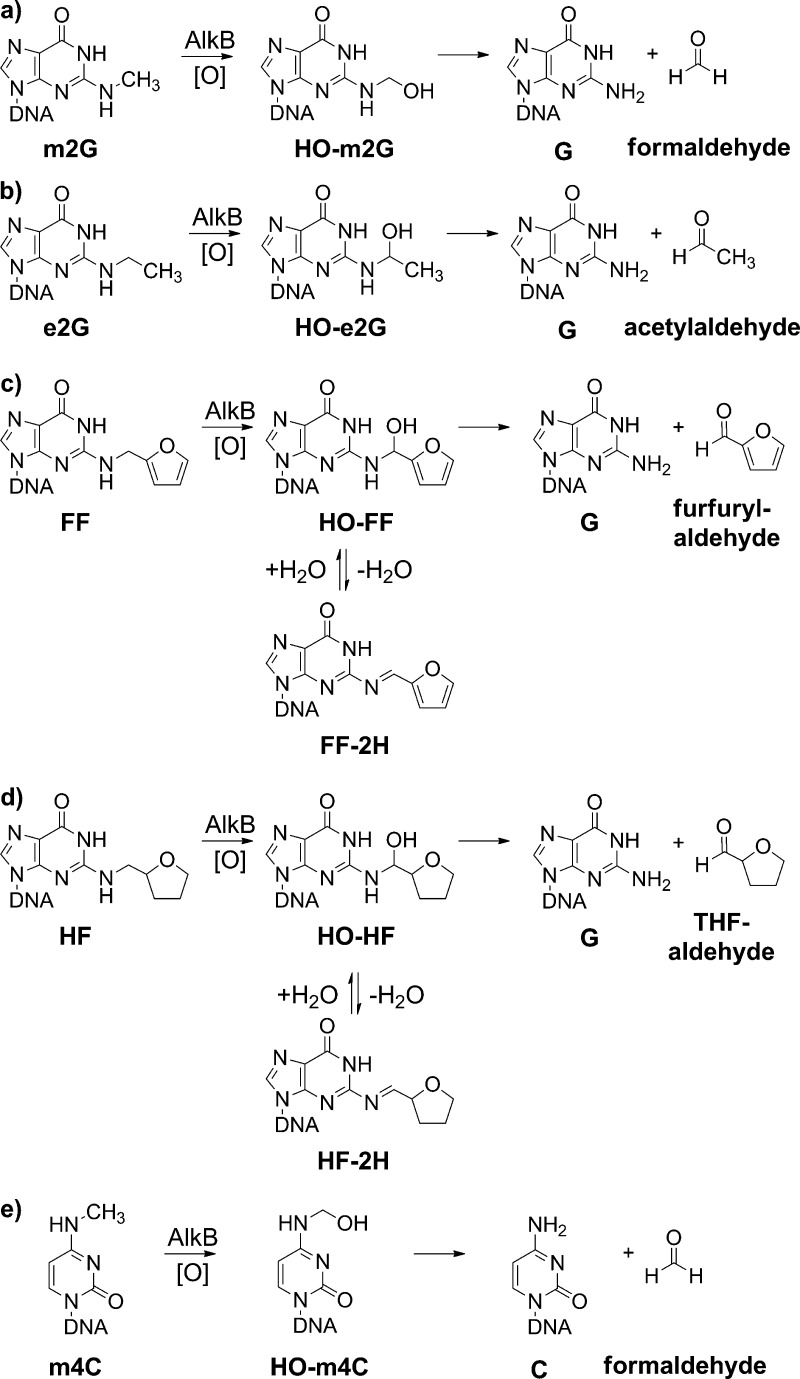
Scheme 1. Chemical Structures of the N-Alkyl Modified Bases Studied, with Proposed Chemical Transformations after AlkB-Mediated Substrate Oxidation for (a) m2G; (b) e2G; (c) FF; (d) HF; and (e) m4C.
Figure 2.
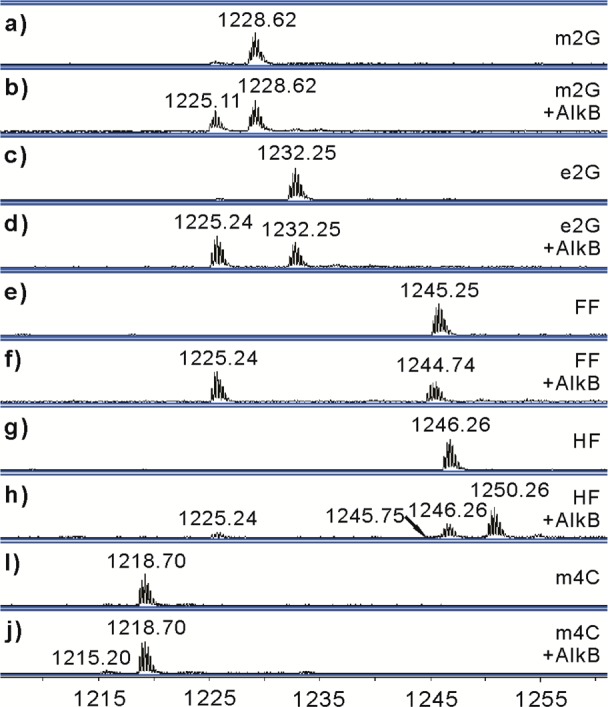
ESI-TOF mass spectra from incubations of N-alkyl DNA adducts with the AlkB protein. Data represent the intensity vs mass/charge (m/z) values for the −4 charge envelopes, with the monoisotopic (all 12C, 14N, 16O, etc.) peak labeled above each envelope. Calculated and observed monoisotopic molecular weights of oligonucleotides and intermediates present in the AlkB reactions are shown in Table 1. (a) m2G; (b) m2G + AlkB; (c) e2G; (d) e2G + AlkB; (e) FF; (f) FF + AlkB; (g) HF; (h) HF + AlkB; (i) m4C; and (j) m4C + AlkB.
Using the method described above, we found that in 1 h, 38% of m2G is converted to guanine in the presence of AlkB (Figure 2b, −4 charge m/z 1225.11), leaving 62% of the m2G unchanged (m/z 1228.62) (Figure 2b). Similarly, AlkB converted 64% of the e2G base to undamaged guanine (m/z 1225.24, Figure 2d) in 1 h. The oligonucleotide containing FF has an m/z of 1245.25 in the absence of AlkB (Figure 2e and Table 1). Incubation with AlkB resulted in the majority of the starting material being converted into two new species (Figure 2f), with masses being consistent with undamaged guanine at m/z 1225.24 and FF-2H at 1244.74 (Scheme 1c). The isotopic distribution pattern observed for the assigned FF-2H structure (Figure S9, Supporting Information) was similar to the FF starting material (Figure S7, Supporting Information), providing confidence that little to no FF starting material was buried underneath the FF-2H signal. A possible mechanism for FF transformation is that AlkB first oxidizes the furfuryl methylene group to generate an HO-FF intermediate (Scheme 1c), mirroring the oxidative hydroxylation mechanism AlkB uses on other DNA alkyl adducts, such as the formation of HO-m6A from m6A.7 The HO-FF intermediate can then either directly decompose to undamaged guanine or lose water to form the imine FF-2H. Because FF-2H is a Schiff’s base, it is susceptible to be hydrolyzed to the undamaged guanine and furfurylaldehyde. In the experiment with HF, an m/z of 1246.26 was observed for the starting material in the absence of AlkB (Figure 2g). Addition of AlkB resulted in four peak envelopes in the mass spectrum (Figure 2h). Unreacted starting material (35% remaining after 60 min incubation) was clearly identified at an m/z of 1246.26, while unmodified guanine (8%) was present at 1225.24 (Figure S11, Supporting Information). The peak envelope at 1250.26 (56%) matched the calculated MW of the HO-HF (1250.22, Scheme 1d and Figure S13, Supporting Information). For the HO-HF intermediate, the reaction could proceed either to form a dehydration product HF-2H or to decompose to guanine (Scheme 1d). The fourth peak, albeit a small one at ∼1%, corresponded to the Schiff’s base type intermediate HF-2H at m/z of 1245.75 (Figure 2h and Figure S12, Supporting Information), which can be hydrolyzed to the undamaged guanine and unconjugated THF-aldehyde (Scheme 1d). For the incubation of m4C without AlkB, an m/z of 1218.70 was observed for the 16-mer oligonucleotide containing m4C (Figure 2i), which corresponds well with the calculated m/z 1218.71 (Table 1). There was another small peak envelope at m/z of 1215.45 (Figure S15, Supporting Information) in the spectrum, which corresponds to the 16-mer containing a uracil base at the lesion site (calculated m/z 1215.45, Table 1). The uracil base in the oligonucleotide could be generated from the deamination reaction of m4C. In the presence of AlkB, about 10% of m4C is converted to cytosine after 1 h of incubation (Figure 2j and Figure S16, Supporting Information, −4 charge m/z 1215.20), with approximately 90% of m4C remaining unchanged (Figure 2j). The MW of the oligonucleotide containing cytosine (4864.85) is 1 Da less than that of the oligonucleotide containing uracil (4865.83). The MW difference between the two oligonucleotides at −4 charge state corresponds to an m/z difference of 0.25 unit, which is clearly demonstrated by the observed m/z of 1215.45 for uracil and 1215.20 for cytosine. These observations strongly support the conclusion that AlkB indeed repairs the m4C adduct to unmodified cytosine. To confirm the identity of the oligonucleotides containing lesions, repair intermediates, and products, MS/MS analysis was performed on each of the 5 AlkB reactions. In each case, both the unreacted oligonucleotide and the product oligonucleotide were chosen for collision-induced-dissociation (CID). The results, presented in detail in the Supporting Information, confirmed the identity of the corresponding oligonucleotide species (Figures S18–S29 and Tables S2–S12, Supporting Information).
The results of the repair reactions of the five lesions carried out in the presence of the complementary strand are summarized in Table S1 (Supporting Information). A control experiment to test repair efficiency in both ssDNA and dsDNA was setup using the known AlkB substrate m1A, which was ∼20% converted to A in 1 h in both contexts. Unlike the reactions in ssDNA, the N2-aklyl-G and m4C lesions in dsDNA were left essentially untouched by AlkB.
Discussion
This study completes our analysis of the sites on DNA that AlkB can access in order to repair single stranded DNA. The data, taken together, show that the enzyme repairs alkylation damage at all possible nitrogens involved in the normal base-pairing of guanine, cytosine, adenine, and thymine. Here, we show that AlkB repairs the simple alkyl lesions m2G, e2G, and m4C, as well as the bulky lesions FF and HF when present in ssDNA, but not in dsDNA. The modified oligonucleotides were incubated with or without purified AlkB enzyme in vitro, with or without the complementary strand, and the reaction products were analyzed by ESI-TOF mass spectrometry and MS/MS. These assay conditions were not developed to provide kinetic constants for the AlkB reactions. Rather, they show qualitatively whether or not a given lesion has the chemical properties needed to be a substrate. If the lesion is chemically competent for repair, it is introduced into cells in subsequent studies to determine if AlkB acts upon the lesion in vivo.32
The finding that AlkB can act on the m4C DNA adduct suggests an additional biological function for AlkB, besides its normal role as a DNA repair enzyme. The m4C adduct, which is similar to m6A, is normally not considered a deleterious lesion. Rather, it is an important postreplicative DNA modification for certain bacteria.28,29 The results presented above suggest that AlkB might not only act as a repair enzyme to defend against DNA alkyl damage but also play a role in controlling gene expression and/or DNA replication and segregation.
The present study also highlights the versatility of AlkB in dealing with different sizes of alkyl adducts at the N2 position of guanine. The enzyme can modify not only the small alkyl lesions m2G or e2G but also the bulky FF and HF lesions. Interestingly, the FF and HF lesions, which are structurally similar to each other, displayed very different amounts of AlkB-induced repair intermediates (Figure 2). Conversion of FF to undamaged guanine was also much greater than the conversion for HF. Reasons for these observations may be that the FF lesion possesses an aromatic furan ring, which may aid in oxidation of the aliphatic carbon to form HO-FF (analogous to the oxidation of a chemically reactive benzylic position), as well as dehydration of HO-FF to FF-2H. Also, the furfurylaldehyde released enjoys conjugation between the aldehyde carbonyl group and the aromatic furan ring, which is lacking for the ring-saturated THF-aldehyde released from the abundant HO-HF intermediate. Interestingly, a major intermediate, FF-2H (Scheme 1c), consistent with being a Schiff’s base dehydration product, was much more prevalent for the FF AlkB incubation than was HF-2H for the analogous HF-derived incubation. Meanwhile, the amount of HO-FF was very small compared to the amount of HO-HF. One possible explanation is that the dehydration of HO-FF is driven by the formation of the intermediate FF-2H (Scheme 1c), a very stable extensively conjugated system throughout the entire guanine and furan rings. However, HF-2H (Scheme 1d) lacks such conjugation leading to the accumulation of larger amounts of HO-HF (Figure 2h).
AlkB is known to repair lesions, such as m1A and m3C, in both ssDNA and dsDNA, with ssDNA being the preferred substrate.1,2,12,15 These lesions feature alkyl groups that disrupt the Watson–Crick base-pairing interactions, thus facilitating their recognition by AlkB and its homologues in both ssDNA and dsDNA.13,14 Interestingly, while all the lesions tested in this work were repaired by AlkB in ssDNA, they were not good substrates for repair by AlkB when presented in dsDNA (Table S1, Supporting Information). One possible explanation for these observations relies on the fact that the N2-alkyl-dG and m4C lesions display alkyl-groups on the exocyclic amino groups of dG or dC; these alkyl groups can rotate in the minor or major groove to allow for unimpeded Watson–Crick base-pairing, which possibly avoids being detected by AlkB. Several crystal structures of AlkB with different DNA alkyl lesions in ssDNA and dsDNA have been published.6,12−14 We modeled the N2G lesions based on the structures of AlkB with m1A and eA and modeled the m4C lesion based on the m3C structure. Among the different conformers generated from the free-rotation of the exocyclic amino groups in of N2G and m4C lesions, we chose the conformers that directed the N-attached lesion carbon closest to the Fe(II) center. The analyses showed that the distance between this carbon and the Fe(II) center was about 5 Å, which was similar to the distance between the N-attached lesion carbon and Fe(II) center in the crystal structures of oligonucleotides containing m1A, m1G, eA, and m3C with AlkB.6,12−14 On the basis of the structural analyses, the five exocyclic lesions tested in this work should be able to adopt conformations that place the alkyl groups in the vicinity of the catalytic Fe(II) center of AlkB, allowing the enzyme to oxidize and dealkylate the modified bases.
While AlkB may appear to be a panacea for wiping the nucleobase N-alkylation damage slate clean at all Watson–Crick base-pairing positions, the relative extents of alkylation removal will depend not only on the position of the alkylated nitrogen atom for each base but also on other factors, which include the AlkB binding specificity for a given lesion, the relative conformation of the alkyl group in the active site, and the rate of decomposition of the oxidized intermediates to products. Further kinetic studies are needed to establish the rank order of alkylation repair by AlkB and its homologues for the diverse constellation of their alkyl lesion substrates.
Acknowledgments
We thank the Tannenbaum/Wishnok Laboratory and the MIT Center for Environmental Health Sciences for providing the ESI-TOF mass spectrometry facility and Agilent Technologies for providing the UHPLC system. We also thank Professor Graham C. Walker, Dr. John. S. Wishnok, Dr. Vipender Singh, Dr. Lauren E. Frick, and Dr. James J. Foti for helpful discussions, and Ms. Charlotte M. Page for her assistance on the in vitro AlkB reactions.
Glossary
Abbreviations
- m1A
1-methyladenine
- m3C
3-methylcytosine
- e3C
3-ethylcytosine
- m1G
1-methylguanine
- m3T
3-methylthymine
- m6A
N6-methyladenine
- eA
1,N6-ethenoadenine
- eC
3,N4-ethenocytosine
- EA
1,N6-ethanoadenine
- O6mG
O6-methylguanine
- m5C
5-methylcytosine
- m2G
N2-methylguanine
- e2G
N2-ethylguanine
- FF
N2-furan-2-yl-methylguanine
- HF
N2-tetrahydrofuran-2-yl-methylguanine
- m4C
N4-methylcytosine
- ESI-TOF
electrospray ionization time-of-flight
- NFZ
nitrofurazone
- BCNU
bis-chloroethylnitrosourea
- CID
collision-induced dissociation
Supporting Information Available
ESI-TOF mass spectra of the starting materials, intermediates, and products of the AlkB repair reactions; MS/MS fragmentation spectra of the species involved in the AlkB repair reactions; predicted collision-induced dissociation fragmentation patterns of the 16-mer oligonucleotides used in the repair reactions; percentage of lesion repair by AlkB in ssDNA and dsDNA; and the predicted and observed m/z for MS/MS fragmentation patterns. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# McKinsey & Company, 2929 Arch Street, Suite 1400, Philadelphia, PA 19104, United States.
Author Present Address
▽ Visterra Inc., Cambridge, MA 02139, United States.
Author Present Address
○ School of Pharmaceutical Science and Technology, Tianjin University, Tianjin, P. R. China 300072.
Author Present Address
¶ Johnson & Johnson Pharmaceutical Research & Development, 930 Route 202 South, Raritan, NJ 08869, United States.
This work was supported by National Institutes of Health Grants CA080024, CA26731, and ES002109 (to J.M.E.). C.L.D. is a Howard Hughes Medical Institute Investigator.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Trewick S. C.; Henshaw T. F.; Hausinger R. P.; Lindahl T.; Sedgwick B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178. [DOI] [PubMed] [Google Scholar]
- Falnes P. Ø.; Johansen R. F.; Seeberg E. (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419, 178–182. [DOI] [PubMed] [Google Scholar]
- Aravind L., and Koonin E. V. (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. (2004) Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 14051–14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas P. A.; Otterlei M.; Falnes P. O.; Vågbø C. B.; Skorpen F.; Akbari M.; Sundheim O.; Bjørås M.; Slupphaug G.; Seeberg E.; Krokan H. E. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421, 859–863. [DOI] [PubMed] [Google Scholar]
- Yu B.; Edstrom W. C.; Benach J.; Hamuro Y.; Weber P. C.; Gibney B. R.; Hunt J. F. (2006) Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 439, 879–884. [DOI] [PubMed] [Google Scholar]
- Li D.; Delaney J. C.; Page C. M.; Yang X.; Chen A. S.; Wong C.; Drennan C. L.; Essigmann J. M. (2012) Exocyclic carbons adjacent to the N6 of adenine are targets for oxidation by the Escherichia coli adaptive response protein AlkB. J. Am. Chem. Soc. 134, 8896–8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J. C.; Smeester L.; Wong C.; Frick L. E.; Taghizadeh K.; Wishnok J. S.; Drennan C. L.; Samson L. D.; Essigmann J. M. (2005) AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 12, 855–860. [DOI] [PubMed] [Google Scholar]
- Mishina Y.; Yang C.-G.; He C. (2005) Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 127, 14594–14595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick L. E.; Delaney J. C.; Wong C.; Drennan C. L.; Essigmann J. M. (2007) Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc. Natl. Acad. Sci. U.S.A. 104, 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Delaney J. C.; Page C. M.; Chen A. S.; Wong C.; Drennan C. L.; Essigmann J. M. (2010) Repair of DNA alkylation damage by the Escherichia coli adaptive response protein AlkB as studied by ESI-TOF mass spectrometry. J. Nucleic Acids 2010, 369434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-G.; Yi C.; Duguid E. M.; Sullivan C. T.; Jian X.; Rice P. A.; He C. (2008) Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C.; Jia G.; Hou G.; Dai Q.; Zhang W.; Zheng G.; Jian X.; Yang C.-G.; Cui Q.; He C. (2010) Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature 468, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. J.; Hollis T. (2010) Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching. PLoS One 5, e8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B.; Bates P. A.; Paik J.; Jacobs S. C.; Lindahl T. (2007) Repair of alkylated DNA: recent advances. DNA Repair 6, 429–442. [DOI] [PubMed] [Google Scholar]
- Yi C.; Yang C.-G.; He C. (2009) A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 42, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav N.; Li D.; Essigmann J. M. (2010) Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis 31, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundheim O.; Talstad V. A.; Vågbø C. B.; Slupphaug G.; Krokan H. E. (2008) AlkB demethylases flip out in different ways. DNA Repair 7, 1916–1923. [DOI] [PubMed] [Google Scholar]
- Sedgwick B. (2004) Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5, 148–157. [DOI] [PubMed] [Google Scholar]
- Yi C.; He C. (2013) DNA repair by reversal of DNA damage. Cold Spring Harb. Perspect. Biol. 5, a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2006) Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monogr. Eval. Carcinog. Risks Hum. 88, 1–478. [PMC free article] [PubMed] [Google Scholar]
- Nakao L. S.; Fonseca E.; Augusto O. (2002) Detection of C8-(1-Hydroxyethyl)guanine in liver RNA and DNA from control and ethanol-treated rats. Chem. Res. Toxicol. 15, 1248–1253. [DOI] [PubMed] [Google Scholar]
- Abraham J.; Balbo S.; Crabb D.; Brooks P. J. (2011) Alcohol metabolism in human cells causes DNA damage and activates the Fanconi anemia-breast cancer susceptibility (FA-BRCA) DNA damage response network. Alcohol.: Clin. Exp. Res. 35, 2113–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P. J.; Theruvathu J. A. (2005) DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol 35, 187–193. [DOI] [PubMed] [Google Scholar]
- Jarosz D. F.; Godoy V. G.; Delaney J. C.; Essigmann J. M.; Walker G. C. (2006) A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439, 225–228. [DOI] [PubMed] [Google Scholar]
- Hiraku Y.; Sekine A.; Nabeshi H.; Midorikawa K.; Murata M.; Kumagai Y.; Kawanishi S. (2004) Mechanism of carcinogenesis induced by a veterinary antimicrobial drug, nitrofurazone, via oxidative DNA damage and cell proliferation. Cancer Lett. 215, 141–150. [DOI] [PubMed] [Google Scholar]
- Rodgers G. L.; Mortensen J. E.; Fisher M. C.; Long S. S. (1997) In vitro susceptibility testing of topical antimicrobial agents used in pediatric burn patients: comparison of two methods. J. Burn Care Rehabil. 18, 406–410. [DOI] [PubMed] [Google Scholar]
- Ehrlich M.; Gama-Sosa M. A.; Carreira L. H.; Ljungdahl L. G.; Kuo K. C.; Gehrke C. W. (1985) DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 13, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X. (1995) Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 24, 293–318. [DOI] [PubMed] [Google Scholar]
- Ratel D.; Ravanat J.-L.; Berger F.; Wion D. (2006) N6-methyladenine: the other methylated base of DNA. BioEssays 28, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wion D.; Casadesús J. (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. (2006) Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 408, 1–15. [DOI] [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. (1999) Context-dependent mutagenesis by DNA lesions. Chem. Biol. 6, 743–753. [DOI] [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. (2001) Effect of sequence context on O6-methylguanine repair and replication in vivo. Biochemistry 40, 14968–14975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.