Abstract
Background
Human rhinoviruses (HRVs) cause common colds, and the recently discovered HRV-C is increasingly associated with lower respiratory illness among populations such as children and asthmatic patients.
Objective
To determine how HRV-C is associated with respiratory illness and to evaluate changes in prevalence and species over 2 decades.
Methods
A prospective study of children younger than 5 years was performed at the Vanderbilt Vaccine Clinic over a 21-year period. Nasal-wash specimens from children presenting with upper or lower respiratory illness at acute care visits were tested for HRV and HRV-positives genotyped. Demographic and clinical features were compared between children with or without HRV, and with different HRV species.
Results
HRV was detected in 190 of 527 (36%) specimens from a population of 2009 children from 1982 through 2003. Of these, 36% were HRV-C. Age (P = .039) and month of illness (P < .001) were associated with HRV infection and HRV species. HRV-C was significantly associated with lower respiratory illness, compared with HRV-A (P = .014). HRV-A and HRV-C prevalence fluctuated throughout the 21-year period; HRV-C was more prevalent during winter (P = .058).
Conclusions
HRV-C is not a new virus but has been significantly associated with childhood lower respiratory illness in this population for several decades. Temporal changes in virus prevalence occur, and season may predict virus species. Our findings have implications for diagnostic, preventive, and treatment strategies due to the variation in disease season and severity based on species of HRV infection.
Key words: Rhinovirus, HRV-C, children, season, lower respiratory illness
Abbreviations used: HMPV, Human metapneumovirus; HRV, Human rhinovirus; LRI, Lower respiratory illness; PIV, Parainfluenza virus; RSV, Respiratory syncytial virus; URI, Upper respiratory illness; VVC, Vanderbilt Vaccine Clinic
Human rhinovirus (HRV), a small nonenveloped RNA virus in the Picornaviridae family, is the predominant cause of the common cold. Since its discovery in 1956, approximately 150 serotypes have been identified.1, 2, 3 Although the common cold is often clinically benign, HRV infections can be severe in certain populations such as infants, children with wheezing or asthma, and older adults. HRV is associated with both upper respiratory illness (URI) and lower respiratory illness (LRI), including pneumonia,4 bronchiolitis,5 and asthma.6 HRV is associated with the hospitalization of 5 of 1000 US children younger than 5 years and 18 of 1000 US children younger than 6 months.7 HRV-associated wheezing during infancy predicts the development of subsequent childhood asthma.6, 8 HRV infection also triggers many wheezing and asthma exacerbations.7, 9 Especially in the spring and fall seasons, HRV-associated asthma exacerbations10 cause missed school, hospitalizations, economic loss, and suffering in those affected.
The recently discovered HRV-C species is genetically distinct from the classic species HRV-A and HRV-B.11, 12, 13 Multiple HRV strains and species may circulate concurrently even within small populations.14 Studies have suggested that HRV-C and HRV-A may exhibit different seasonality patterns15, 16, 17 and that HRV-C may be associated with increased disease severity and asthma risk.18, 19, 20, 21, 22 However, most of these studies were limited to 1 or 2 seasons, and thus did not account for random variation by year, which may influence a given seasonal trend. In the current study, we examined 21 years of data and specimens that were collected prospectively year-round from children with acute respiratory illness, allowing us to measure the impact and seasonal trends of HRV species over a prolonged period.
Methods
Study design
Nasal-wash specimens were prospectively collected from 1982 through 2003 at the Vanderbilt Vaccine Clinic (VVC) in Nashville, Tennessee.23, 24, 25, 26, 27 The VVC was originally established to create a cohort of children for the evaluation of new vaccines, as well as determine the etiology of acute respiratory disease in early childhood in otherwise healthy young children. Thus, children with comorbid conditions other than mild asthma were excluded. Healthy, full-term infants were enrolled at birth and followed up to age 5 years. The VVC was the primary care provider for these children, and children were evaluated in the VVC for all well and acute care visits. All visits were conducted within the General Clinical Research Center, with care provided by the pediatric infectious diseases faculty and research nurse practitioners. Comprehensive care was offered to these children with doctors on call 24 hours a day; this increased the likelihood of capturing all illnesses in these children. Questions involving whether the child had atopy and/or food allergies were not specifically asked by providers, and so children with those conditions were not explicitly excluded from the study. Thirteen children were older than 5 years at their last visit and were also included in this study analysis. Four children were between 60 and 70 months, 4 between 71 and 80 months, 3 between 81 and 100 months, and 2 between 100 and 133 months old. During visits for illness, symptoms were recorded and nasal-wash samples were obtained and cultured for viruses. All studies were approved by the Vanderbilt University Medical Center Institutional Review Board. Parents provided written informed consent.
Study groups
Over the course of the study from 1982 through 2003, 2009 children were followed in the clinic, with approximately 200 children enrolled at any given time. Diagnoses were grouped into LRI (bronchiolitis, pneumonia, croup, and asthma) and URI (coryza, pharyngitis, and acute otitis media).28 Nasal-wash samples collected from children with URI or LRI were cultured for virus.29
Samples that were culture negative for parainfluenza virus (PIV), respiratory syncytial virus (RSV), influenza, adenovirus, and samples that were culture positive for HRV were tested in our study. Samples that were culture positive for other viruses were not retested for HRV. All samples that were culture negative for PIV, RSV, adenovirus, influenza, as well as those associated with LRI were tested in our study. A random sample of available culture-negative URI specimens was drawn and tested alongside the LRI samples. Culture-negative URI specimens were randomly drawn from each quarter of each year on the basis of the number of samples available for a given quarter to yield approximately 25% of all specimens in each quarter. Of the 1735 URI culture-negative samples available, we tested 407. Thus, there were a total of 527 specimens with aliquots available for testing in our study (either culture-negative or HRV culture-positive), 190 from patients diagnosed with LRI and 407 from patients diagnosed with URI. On average, there were 24 (SE ±2.33) samples each year available for testing. Figure E1 (in the Online Repository available at www.jacionline.org) depicts the number of samples tested by year. Data from children who were infected with human metapneumovirus (HMPV), PIV, RSV, influenza, or adenovirus were previously published23, 24, 25, 26, 30 and were analyzed in this study by using our models for comparison with HRV.
Molecular analysis
RNA was extracted from nasal-wash specimens, and real-time RT-PCR was used to detect HRV.31 Conventional RT-PCR was performed on HRV-positive samples by using primers that amplified a 548 nt fragment encompassing the VP4/VP2 region and the hypervariable region in the 5′ noncoding region.32 Amplified fragments were gel-purified, cloned, and then sequenced at the Vanderbilt Sequencing Core. Sequences were aligned with each other and with published HRV sequences by using MacVector 11.0 (MacVector, Inc, Cary, NC), and phylogenetic analysis was conducted by using Mega 5.05.33
Statistical analysis
Poisson regression was used to model the prevalence of HRV over time as a function of age, race (white/black/other), sex, fever (yes/no), and season when the visit occurred. Generalized estimating equations methods were used34 to account for a small number of repeated measures. A similar model was used to compare the incidence of HRV species A versus C, adjusting for race, sex, and season. Both HRV-B and untypeable HRVs were excluded from analysis because of the small sample size. Month, year, and season were also factored into the analysis. Season was grouped into 4 categories: winter (December through February), spring (March through May), summer (June through August), and fall (September through November). Poisson regression was used to model the prevalence of diagnosis (bronchiolitis, pneumonia, croup, asthma, coryza, pharyngitis, and acute otitis media) separately as a function of HRV. Poisson regression with generalized estimating equations method was used to assess the association between HRV species (A, C) and classification of URI or LRI adjusting for race. A Poisson regression model was used to assess the seasonality of other respiratory viruses (HMPV, PIV, RSV, influenza, and adenovirus). Estimated relative risks with 95% CIs and/or P values are reported.
Results
All HRV infections
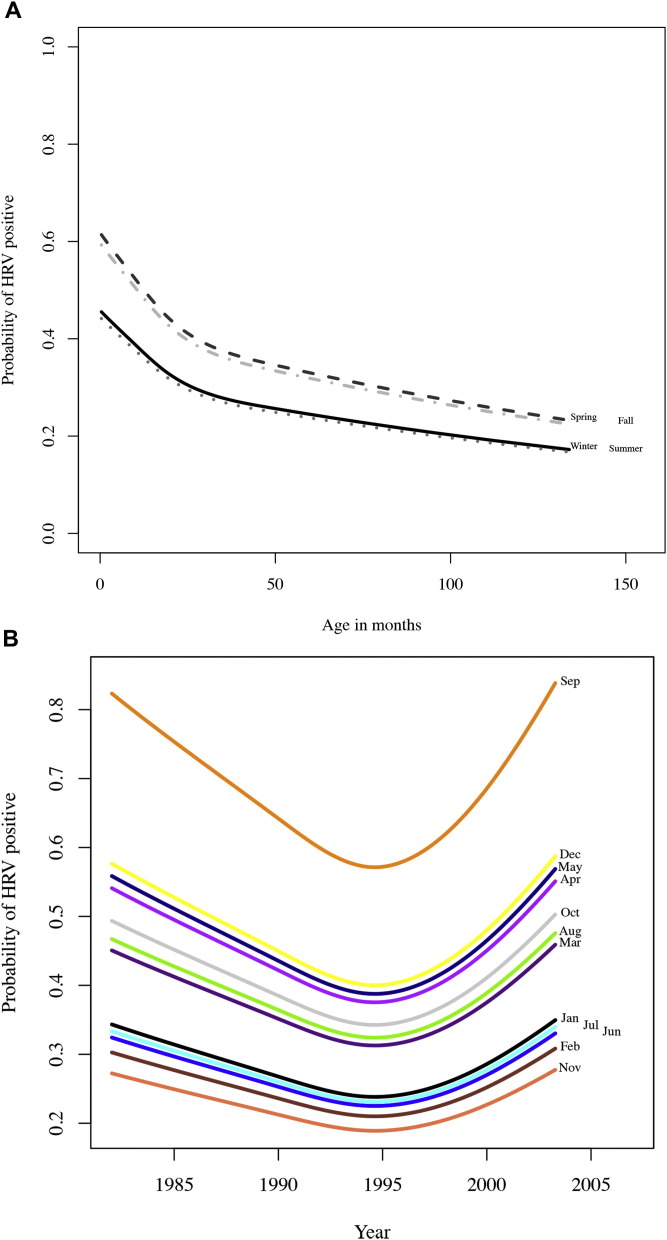
From a population of 2009 children, 527 samples were available (either culture-negative or HRV culture-positive) and tested for HRV and 190 (36%) were positive. Of the HRV-positive samples, 91 (48%) were HRV-A, 5 (3%) HRV-B, 69 (36%) HRV-C, and 25 (13%) untypeable (Table I ). Forty-eight of the 190 HRV-positive samples were collected from children diagnosed with LRI (25%) and 142 from those diagnosed with URI (75%). Detection of any HRV in the subpopulation was significantly correlated with the age of the child (P = .039), with younger children more likely to be infected with HRV (Fig 1 , A). Any HRV detection marginally correlated with race (P = .054) and season (P = .08; Fig 1, A). Black children had a lower relative risk of being diagnosed with HRV than white (0.73; 95% CI, 0.58-0.93). Month was also a significant predictor of HRV detection (P < .001; Fig 1, B). Samples from children who were sick during the month of September had a relative risk for HRV diagnosis of 2.47 (95% CI, 1.40-4.36) compared with those samples from sick children during July. Sex of the child, any contact with a smoker, whether they attended daycare, and diagnosis of fever did not alter the likelihood of any HRV infection. Sample sizes for partial and total breast-feeding were too low to analyze. Only 11 of 527 subjects partially breast-fed their children (2.1%) and 12 of 527 (2.3%) exclusively breast-fed their children.
Table I.
Frequency (and percentage) of specimens positive for HRV and type of HRV
| Feature | n (%) |
|||||
|---|---|---|---|---|---|---|
| HRV-A | HRV-B | HRV-C | Untypeable | Total + | Negative | |
| No. of patients | 91 | 5 | 69 | 25 | 190 | 336 |
| Median age (mo) | 17.7 | 15.2 | 15.4 | 14.76 | 11.47 | 14.45 |
| Female | 34 (37) | 1 (20) | 37 (54) | 11 (44) | 83 (44) | 146 (43) |
| White | 62 (67) | 1 (20) | 44 (64) | 18 (72) | 125 (66) | 194 (58) |
| Black | 19 (21) | 4 (80) | 24 (35) | 7 (28) | 54 (28) | 125 (37) |
| Other ethnicity | 10 (11) | 0 (0) | 1 (1) | 0 (0) | 11 (6) | 17 (5) |
| Winter | 21 (23) | 0 (0) | 26 (38) | 8 (32) | 55 (29) | 121 (36) |
| Spring | 28 (31) | 1 (20) | 18 (26) | 8 (32) | 55 (29) | 75 (22) |
| Summer | 17 (19) | 1 (20) | 9 (13) | 1 (4) | 28 (15) | 62 (18) |
| Fall | 25 (27) | 3 (60) | 16 (23) | 8 (32) | 52 (27) | 78 (23) |
Fig 1.
Age and month are significantly correlated with HRV infection. A, HRV is more common in younger children in fall and spring months. Winter, solid line; spring, dashed line; summer, gray dot; fall, gray dash/dot. Graph shown for white males. B, In September, children had a 2.47-fold increased risk of positive HRV diagnosis than in July. Lines represent modeled negative binomial regression.
Sixteen of 171 subjects (9.3%) were infected with HRV more than once. Time between infections ranged between 2 months and 29 months, with an average of 9.4 months (SE, 2.4 months). Of those 16, 9 (56%) were homologous infections (the same species of HRV, both HRV-A, eg). Four of these subjects were infected with HRV-A twice, 4 with HRV-C twice, and 1 had an unknown type of HRV both times. Of those 7 (44%) infections with different HRV species, 5 subjects were infected with HRV-A first, one with HRV-B first and one with HRV-C first. One of the 16 subjects was infected a total of 3 times, all HRV-As, and 1 was infected a total of 4 times, once with HRV-A and 3 times with HRV-C.
HRV-A compared with HRV-C
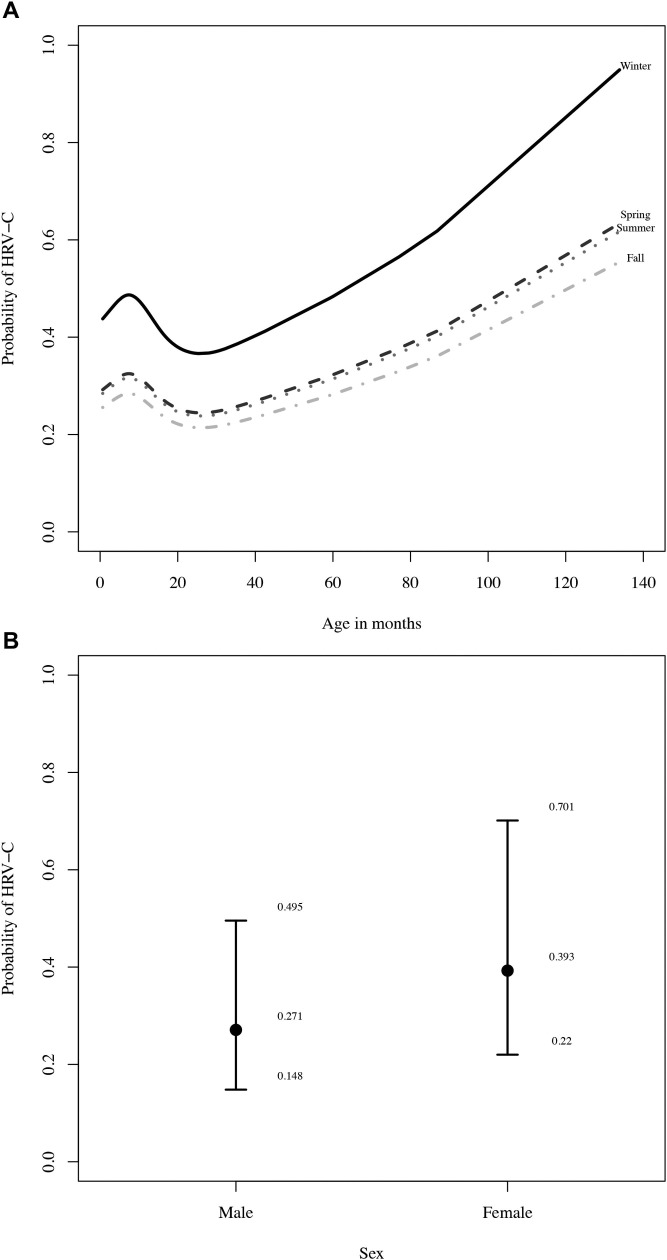
To examine more closely how the species of HRV may affect morbidity in young children, we compared HRV-A with HRV-C, controlling for race and season. As with the detection of any HRV, we found that the age of the child was a significant predictor of HRV species (P = .005), where older children were more likely to be diagnosed with HRV-C (Fig 2 , A, shows the modeled negative binomial regression derived from the data). There was also a significant difference between males and females in the likelihood of having HRV-A versus HRV-C detected (P = .02; Fig 2, B). Female children had a 1.4-fold relative risk of contracting HRV-C compared with male children (95% CI, 1-2.1). As expected, the trend was reversed with HRV-A infection, where females had a 0.69-fold (95% CI, 0.48-1.0) relative risk of contracting HRV-A compared with males. Again, season was marginally significant (P = .056), where children were more likely to be diagnosed with HRV-C in the winter months (Fig 2, A). The relative risk of HRV-C in the winter months was 1.7 (95% CI, 1.0-2.9) compared with the fall months (P = .058; Table I).
Fig 2.
Older females in the winter months were more likely to present with HRV-C. A, Age and season predicted HRV species (shown for white males). The negative binomial regression modeled here predicts that HRV-C is more common with increasing age. B, Sex was also a significant factor in the likelihood of HRV-C (shown for 11.2-month-old white child in the fall).
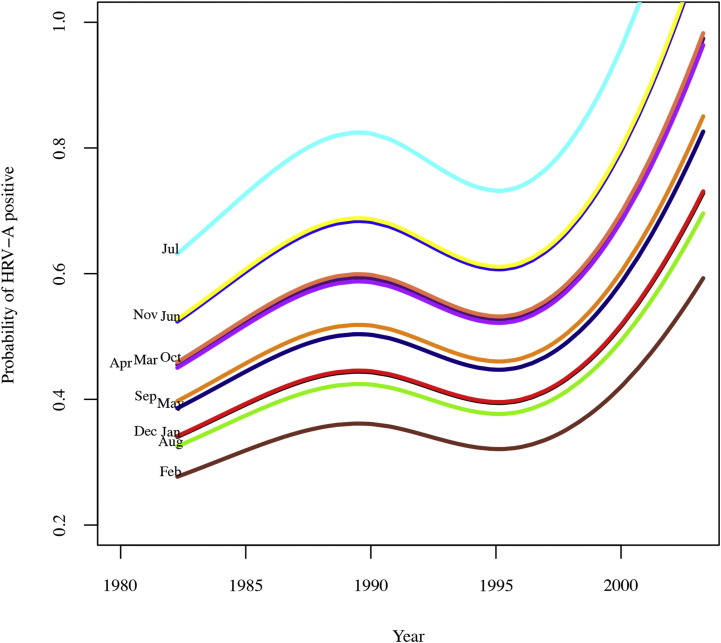
The probability of HRV-A diagnosis varied significantly over the 21-year course of this study (P = .03). Although no particular year dominated, there was a trend toward more HRV-A diagnoses in 1990 as well as in the most recent samples from the years 2000 to 2003. However, inference regarding estimates from recent years is limited because of small sample sizes. Fig 3 represents the negative binomial regression fitted to the data, suggesting that more recently there has been a rise in HRV-A-associated acute respiratory illness in this study population.
Fig 3.
HRV-A rate varies from year to year. HRV-A fluctuated significantly over the years; recently, there was more HRV-A present in children with respiratory illness. Lines represent the negative binomial regression model based on the data.
HRV compared with other viruses
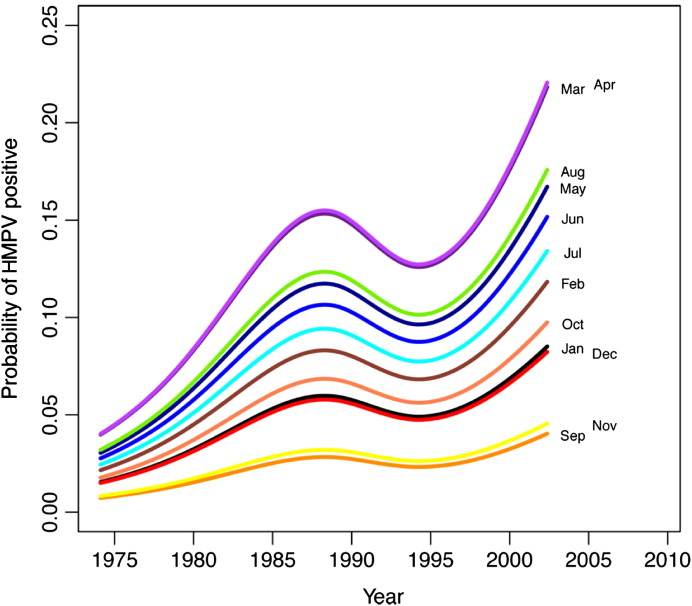
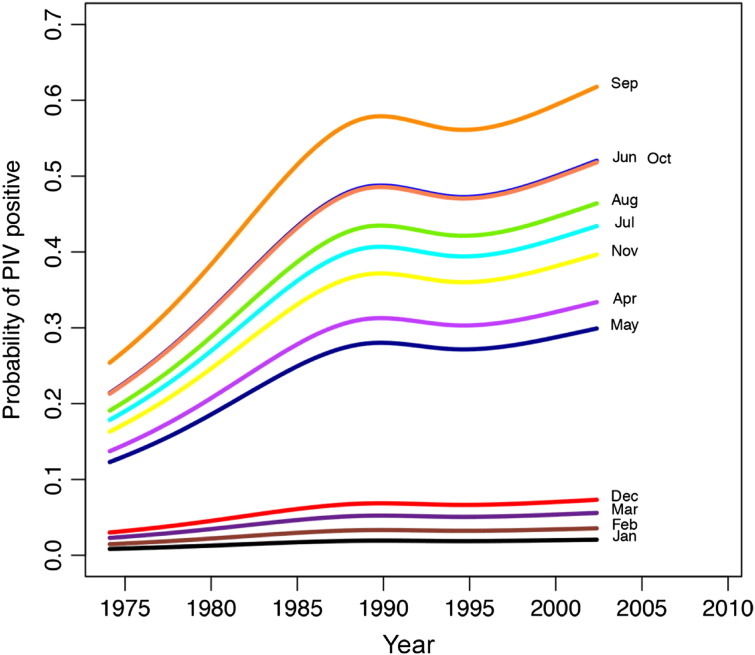
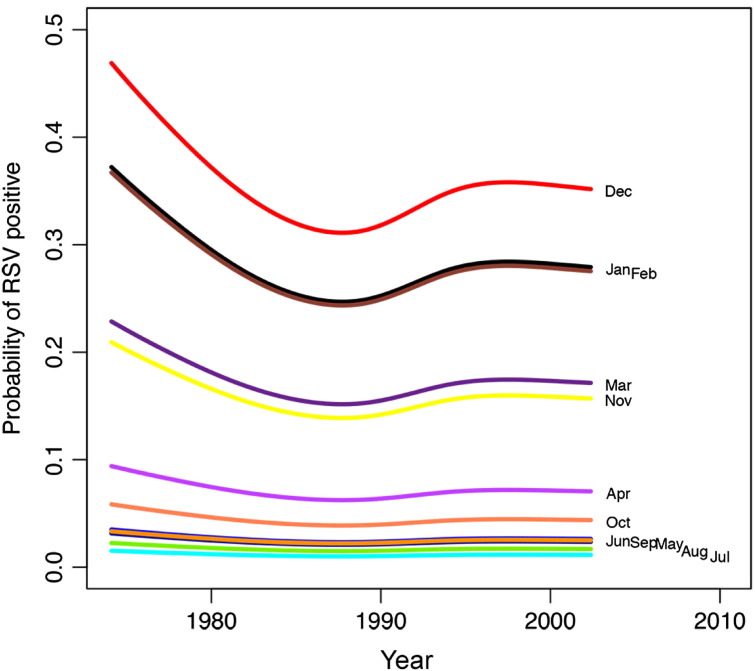
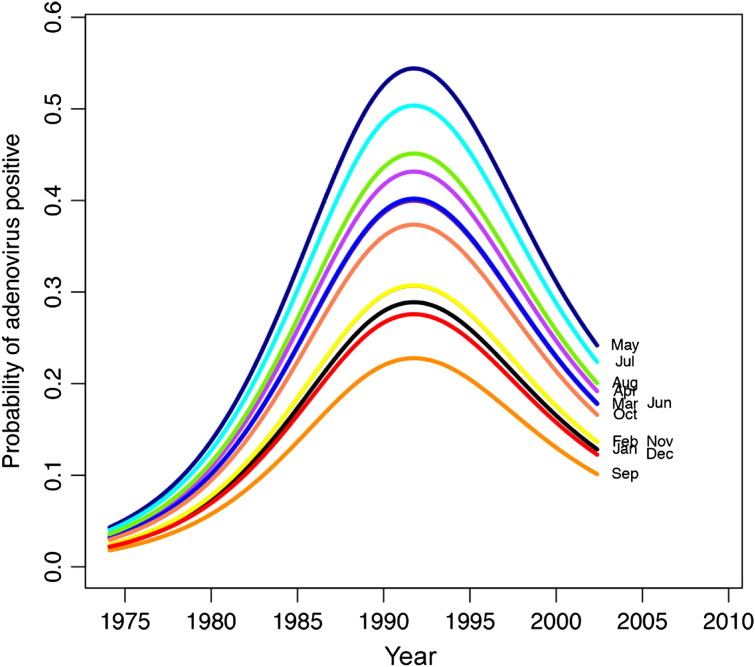
Detection of the other study viruses (HMPV, PIV, RSV, influenza, and adenovirus) varied over time (see Fig E2, Fig E3, Fig E4, Fig E5, Fig E6 in this article's Online Repository available at www.jacionline.org). HMPV varied significantly over both month (P < .001) and year (P = .012), with highest rates in April and March and lowest in September and November (Fig E2). PIV varied over both year (P = .02) and month (highest rate in September, lowest in January and February; Fig E3). RSV significantly varied over month (high in December, low in July and August; P < .001; Fig E4), as did influenza (binned months, because of small sample size: high in January/February, low in May through September; P < .001; Fig E5). Adenovirus showed significant variability for both year (P < .001) and month (high in May, low in September; P < .001; Fig E6).
Association of HRV species with URI or LRI
We next examined whether LRI or URI diagnosis was associated with species of HRV infection. There was a significant difference in the proportion of HRV-A and HRV-C in these grouped diagnoses. HRV-C was significantly more common among children with LRI (60%; relative risk C vs A = 2.152 [1.17-3.97]; P = .014; Fig 4 ). HRV-A was marginally more common among children with a diagnosis of URI (60%; relative risk of C vs A for URI = 0.81 [0.76-1.01]; P = .069).
Fig 4.
LRI diagnosis is more often associated with HRV-C and URI with HRV-A. Percentage of patients diagnosed with infection type: LRI (bronchiolitis, croup, pneumonia, and asthma) and URI (coryza, pharyngitis, and acute otitis media). White bars represent HRV-C, and gray bars represent HRV-A.
We also examined how HRV and individual diagnosis were related. Because of the small sample size of HRV-positive specimens within each individual diagnosis category (bronchiolitis, croup, pneumonia, asthma, coryza, pharyngitis, or acute otitis media), corresponding tests likely suffered from low power and we were unable to detect significant differences among specific diagnoses, even when grouping all positive HRV species together. The original goals of the VCC were to conduct respiratory surveillance and investigational vaccine trials in otherwise healthy young children, and thus children with chronic conditions including moderate or severe asthma were excluded. Because of this, the sample size for patients with mild asthma was only 19, 10 of which were positive for any HRV. Even with this low number, we found a 2.86-fold relative risk of asthma diagnosis when patients were infected with HRV, but the precision of that estimate is wide (95% CI, 0.23-35.47). Two of these patients were diagnosed with HRV-A and 6 with HRV-C, and 2 were untypeable.
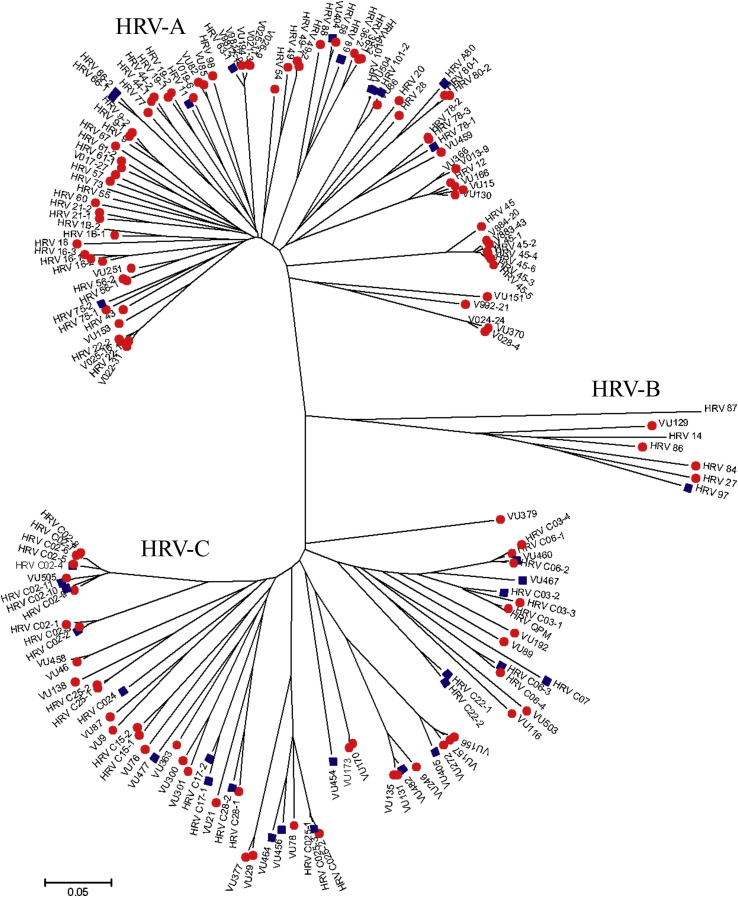
The HRV sequences were aligned and a phylogenetic tree constructed to determine evolutionary relationships. As expected, 3 distinct groups emerged comprising species HRV-A, HRV-B, and HRV-C (Fig 5 ). Sequences in this article have been deposited into Genbank under accession numbers JX560565- JX560730. Labeling of individual virus sequences with the clinical diagnosis of URI or LRI illustrated the association of HRV-A with URI (red circles) and HRV-C with LRI (blue squares). Furthermore, we compared the similarity of the sequences to known HRV-A, HRV-B, and HRV-C sequences. HRV-A sequences had on average a similarity of 78.0% (±0.27). HRV-B sequences had an average similarity of 79.7% (±0.42), and HRV-C sequences were 73.9% (±0.16) similar on average. This confirms the discovery of more new strains of HRV and suggests that HRV-C strains exhibited greater genetic diversity within the species compared with either HRV-A or HRV-B.
Fig 5.
Phylogenetic tree of HRV species depicting HRV species and URI (red circle) or LRI (blue square) diagnosis. Grouping URI and LRI diagnoses suggests that HRV species is associated with certain clinical phenotypes (or respiratory disease severity). Novel sequences are designated by “VU” followed by the sample number, and sequences matching published HRV strains are marked as HRV followed by the strain number.
Discussion
We retrospectively analyzed data and samples from a prospective 21-year cohort study. Our findings show that the recently described HRV-C is not a new virus but has played an important role in childhood respiratory disease for at least 20 years. The data also confirm the growing evidence that HRV-C is associated with more severe lower respiratory disease, compared with HRV-A.19, 20, 21 Although other studies have examined HRV seasonality,15, 16 this is the first study to describe more than 20 years of data. This longitudinal cohort provides a unique ability to determine trends in HRV circulation over time without the risk of seasonal patterns attributable to random fluctuations. Interestingly, while HRV was prevalent in the fall and spring as expected,35, 36 HRV-C was found most commonly during winter months. Because these data encompass 21 years of prospective data, it is unlikely that the explanation is due to an atypical increase in HRV-C in a single winter.
In our study, HRV was more frequently detected in younger children and infants than in older children, in agreement with other studies.7, 37 However, we found that when older children were infected with HRV, it was more often HRV-C. Few studies have analyzed differences in HRV species in relationship to the age of the child; however, a study in Thailand detected HRV-A most often in children younger than 1 year and in adults and HRV-C more frequently in children aged 1 to 4 years.38 If HRV-C is more strongly associated with LRI, it is possible that parents of older children seek medical attention only for these more severe illnesses. Similarly, parents of younger children may take children to the doctor more often, even for less severe symptoms, than do parents of older children. Further studies are necessary to determine why age is associated with differential HRV species infections.
We also found that when examining both age and season HRV-C demonstrated different trends than did HRV overall. One possible explanation is that viral interference alters seasonal peaks and prevalence between HRV-C and other HRV species.39 It has been suggested that HRV may interfere with several other respiratory viruses, such as adenovirus, influenza virus A, PIV, HMPV, and coronavirus, based on the fact these viruses are significantly less likely to occur when coinfected with HRV.40 Wisdom et al39 found that though 10.7% of singly infected patients had RSV, only 2.4% had RSV when coinfected with HRV-C. In one of our previous studies of young hospitalized children, HRV-C codetection was 10% compared with 23% for HRV-A (P = .037).41 Another explanation may be that HRV-C is more likely to be communicable in winter months. A recent study on guinea pigs suggested that influenza B was more transmissible (both with and without contact) at colder temperatures.42 Although it is unknown why HRV-C peaks in the winter, recent work from Japan also supports this trend. During the 2008-2009 season, researchers found that HRV-C was most common in December.15 In our study population because we observed a marginally significant trend over a 20-year period, it is likely that in Nashville the HRV-C burden is higher in the winter months. This suggests that rapid viral detection, particularly in winter months when HRV-C, RSV, and influenza all peak, could modify treatment.
In our study, HRV was associated with a 2.86-fold increased relative risk (95% CI, 0.23-35.47) of asthma diagnosis, and the majority of these patients with asthma were infected with HRV-C. Although this result did not reach statistical significance and has a large data spread, it may be biologically relevant. Because of the experimental design, children with moderate or severe asthma were excluded from the original study, limiting the power to analyze the relationship between asthma and HRV. We had only 10 samples from patients with mild asthma that were positive for HRV; more data are needed to obtain a precise estimate of the true relative risk of asthma among those with different HRV species.
One limitation of the study was that we did not retest specimens that were positive for other viruses for HRV, thus likely underestimating HRV prevalence. In addition, we did not test specimens for bacterial coinfections, which may complicate acute respiratory viral infections.43 Another limitation was that study viruses other than coronaviruses30 and HMPV25 were identified by viral culture or rapid antigen tests, which are less sensitive than RT-PCR used to detect HRV. Thus, we could not directly compare HRV results with those previously obtained for some other viruses. Furthermore, rates of medical visits may have differed over time and among families in Nashville during the 21-year period. Breast-feeding rates were low in the study population, which limited power to detect any effect on infection; some studies show that breast-feeding may protect children from respiratory tract infections.44, 45 We also did not test healthy children for HRV, which has been shown to be present in asymptomatic individuals.46, 47 Strengths of our analysis include the prospective nature of the data and specimen collection, the use of molecular viral diagnostics with cloning and sequencing for HRV typing, and more than 2 decades of year-round recruitment. The demographics of children enrolled in this study were similar to those in the United States,48 and so our results should be largely applicable to the rest of the country. We did have slightly more black children in this study compared with the general makeup of the United States (34% vs 13% black in the United States48). However, race may not be a consistent risk factor for HRV infection. In a previous study from Nashville, Tennessee, HRV was associated more often with black children,17 the opposite of the trend we found.
In conclusion, HRV-C has circulated for many years, is prevalent in the winter, and is more strongly associated with LRI than is HRV-A. The temporal variation in virus detection we observed over 2 decades confirms that season is strongly linked with the likelihood of HRV infection and that HRV-C peaks during a different season than do other HRV species. These data suggest that HRV species can contribute to the severity of disease and asthma exacerbations in children and that certain HRV species are more common at different times of year. Further studies are required to better understand the pathogenesis of HRV species and their role in LRI and asthma to optimally target future diagnostic, preventive, and treatment strategies for specific HRV species and strains.
Clinical implications.
Since 1982, HRV-C has been associated with lower respiratory illness, particularly during the winter. Rapid viral detection during winter when HRV-C, RSV, and influenza peak is necessary to modify treatment.
Acknowledgments
We thank Dr Kathryn Edwards and the families who participated in the Vanderbilt Vaccine Clinic. Statistical analysis was conducted by Z.L. and B.R.S.
Footnotes
This study was supported by the National Institutes of Health (grant nos. RR-24977-03 and AI-091691-02 to E.K.M. and grant no. AI-085062 to J.V.W.) and Vanderbilt Institutional Clinical and Translational Research (grant nos. UL1 RR024975-01 and UL1 TR000445-06 to E.K.M.).
Disclosure of potential conflict of interest: P. F. Wright has received grants from the National Institute of Allergy and Infectious Diseases/Vaccine and Treatment Evaluation Units, the Gates Foundation, and the National Institutes of Health (NIH) and is employed by Dartmouth College and Vanderbilt University. J. V. Williams has received grants from the NIH and is on the scientific advisory board for Quidel. E. K. Miller has received grants from the NIH, the March of Dimes, Thrasher, MedImmune, and Vanderbilt Institutional Clinical and Translational Research; has consultant arrangements with the American Association for the Advancement of Science; and has received payment for lectures from the FOOCUS Asthma Group. The rest of the authors declare that they have no relevant conflicts of interest.
Appendix
Fig E1.
Number of samples analyzed for HRV by year. Black bars indicate the number of positive samples.
Fig E2.
The probability of HMPV varies significantly over month and year. Lines represent negative binomial regression model based on data.
Fig E3.
The probability of parainfluenza virus varies significantly over month and year. January and February were least likely to be associated with positive PIV. Lines represent negative binomial regression model based on data.
Fig E4.
The probability of RSV infection varies by month. December, January, and February were more likely to be associated with virus detection. Lines represent negative binomial regression model based on data.
Fig E5.
The probability of influenza varies by month. Because of low sample size, data were binned into 2-month intervals. January/February was most likely to be associated with positive influenza diagnosis. Lines represent negative binomial regression model based on data.
Fig E6.
The probability of adenovirus varies by both month and year. September was least likely to be associated with adenovirus, and May was most often associated with adenovirus. Lines represent negative binomial regression model based on data.
References
- 1.Pelon W., Mogabgab W., Phillips I., Pierce W. A cytopathogenic agent isolated from naval recruits with mild respiratory illnesses. Proc Soc Exp Biol Med. 1957;94:262–267. doi: 10.3181/00379727-94-22915. [DOI] [PubMed] [Google Scholar]
- 2.Hamparian V.V. A collaborative report—rhinoviruses, extensions of the numbering system from 89 to 100. Virology. 1987;159:191–192. doi: 10.1016/0042-6822(87)90367-9. [DOI] [PubMed] [Google Scholar]
- 3.Simmonds P., McIntyre C., Savolainen-Kopra C., Tapparel C., Mackay I.M., Hovi T. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J Gen Virol. 2010;91:2409–2419. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- 4.Louie J.K., Roy-Burman A., Guardia-LaBar L., Boston E.J., Kiang D., Padilla T. Rhinovirus associated with severe lower respiratory tract infections in children. Pediatr Infect Dis J. 2009;28:337–339. doi: 10.1097/INF.0b013e31818ffc1b. [DOI] [PubMed] [Google Scholar]
- 5.Jartti T., Lehtinen P., Vuorinen T., Osterback R., van den Hoogen B., Osterhaus A. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10:1095–1101. doi: 10.3201/eid1006.030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemanske R.F., Jackson D.J., Gangnon R.E., Evans M.D., Li Z.H., Shult P.A. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–577. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 7.Miller E.K., Lu X.Y., Erdman D.D., Poehling K.A., Zhu Y.W., Griffin M.R. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 2007;195:773–781. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson D.J., Lemanske R.F., Jr. The role of respiratory virus infections in childhood asthma inception. Immunol Allergy Clin North Am. 2010;30:513–522. doi: 10.1016/j.iac.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gern J.E. Rhinovirus respiratory infections and asthma. Am J Med. 2002;112:19S–27S. doi: 10.1016/s0002-9343(01)01060-9. [DOI] [PubMed] [Google Scholar]
- 10.Olenec J.P., Kim W.K., Lee W.-M., Vang F., Pappas T.E., Salazar L.E.P. Weekly monitoring of children with asthma for infections and illness during common cold seasons. J Allergy Clin Immunol. 2010;125:1001–1006. doi: 10.1016/j.jaci.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W.-M., Kiesner C., Pappas T., Lee I., Grindle K., Jartti T. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. Plos One. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ledford R.M., Patel N.R., Demenczuk T.M., Watanyar A., Herbertz T., Collett M.S. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau S.K.P., Yip C.C.Y., Tsoi H.-W., Lee R.A., So L.-Y., Lau Y.-L. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic previously undetected HRV cluster, probably representing a species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peltola V., Waris M., Osterback R., Susi P., Hyypia T., Ruuskanen O. Clinical effects of rhinovirus infections. J Clin Virol. 2008;43:411–414. doi: 10.1016/j.jcv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Kaida A., Kubo H., Takakura K.-I., Togawa M., Shiomi M., Kohdera U. Molecular epidemiology of human rhinovirus C in patients with acute respiratory tract infections in Osaka City, Japan. Jpn J Infect Dis. 2011;64:488–492. [PubMed] [Google Scholar]
- 16.Savolainen-Kopra C., Blomqvist S., Kaijalainen S., Jounio U., Juvonen R., Peitso A. All known human rhinovirus species are present in sputum specimens of military recruits during respiratory infection. Viruses-Basel. 2009;1:1178–1189. doi: 10.3390/v1031178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller E.K., Williams J.V., Gebretsadik T., Carroll K.N., Dupont W.D., Mohamed Y.A. Host and viral factors associated with severity of human rhinovirus-associated infant respiratory tract illness. J Allergy Clin Immunol. 2011;127:883–891. doi: 10.1016/j.jaci.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denlinger L.C., Sorkness R.L., Lee W.-M., Evans M.D., Wolff M.J., Mathur S.K. Lower airway rhinovirus burden and the seasonal risk of asthma exacerbation. Am J Respir Crit Care Med. 2011;184:1007–1014. doi: 10.1164/rccm.201103-0585OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller E.K., Khuri-Bulos N., Williams J.V., Shehabi A.A., Faouri S., Al Jundi I. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol. 2009;46:85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bizzintino J., Lee W.M., Laing I.A., Vang F., Pappas T., Zhang G. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McErlean P., Shackelton L.A., Lambert S.B., Nissen M.D., Sjoots T.P., Mackay I.M. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards K.M., Thompson J., Paolini J., Wright P.F. Adenovirus infections in young children. Pediatrics. 1985;76:420–424. [PubMed] [Google Scholar]
- 24.Neuzil K.M., Zhu Y.W., Griffin M.R., Edwards K.M., Thompson J.M., Tollefson S.J. Burden of interpandemic influenza in children younger than 5 years: a 25-year prospective study. J Infect Dis. 2002;185:147–152. doi: 10.1086/338363. [DOI] [PubMed] [Google Scholar]
- 25.Williams J.V., Harris P.A., Tollefson S.J., Halburnt-Rush L.L., Pingsterhaus J.M., Edwards K.M. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. New Engl J Med. 2004;350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams J.V., Wang C.Y.K., Yang C.F., Tollefson S.J., House F.S., Heck J.M. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J Infect Dis. 2006;193:387–395. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed G., Jewett P.H., Thompson J., Tollefson S., Wright P.F. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children <5 years old. J Infect Dis. 1997;175:807–813. doi: 10.1086/513975. [DOI] [PubMed] [Google Scholar]
- 28.Jamjoom G.A., al-Semrani A.M., Board A., al-Frayh A.R., Artz F., al-Mobaireek K.F. Respiratory syncytial virus infection in young children hospitalized with respiratory illness in Riyadh. J Trop Pediatr. 1993;39:346–349. doi: 10.1093/tropej/39.6.346. [DOI] [PubMed] [Google Scholar]
- 29.Wright P.F., Ross K.B., Thompson J., Karzon D.T. Influenza A infections in young children. New Engl J Med. 1977;296:829–834. doi: 10.1056/NEJM197704142961501. [DOI] [PubMed] [Google Scholar]
- 30.Talbot H.K., Shepherd B.E., Crowe J.E., Jr., Griffin M.R., Edwards K.M., Podsiad A.B. The pediatric burden of human coronaviruses evaluated for twenty years. Pediatr Infect Dis J. 2009;28:682–687. doi: 10.1097/INF.0b013e31819d0d27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu X., Holloway B., Dare R.K., Kuypers J., Yagi S., Williams J.V. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savolainen C., Mulders M.N., Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardin J., Hilbe J. Chapman and Hall/CRC; London: 2003. Generalized estimating equations. [Google Scholar]
- 35.Brittain-Long R., Andersson L.-M., Olofsson S., Lindh M., Westin J. Seasonal variations of 15 respiratory agents illustrated by the application of a multiplex polymerase chain reaction assay. Scand J Infect Dis. 2012;44:9–17. doi: 10.3109/00365548.2011.598876. [DOI] [PubMed] [Google Scholar]
- 36.Turner R.B. Rhinovirus: more than just a common cold virus. J Infect Dis. 2007;195:765–766. doi: 10.1086/511829. [DOI] [PubMed] [Google Scholar]
- 37.van Piggelen R.O., van Loon A.M., Krediet T.G., Verboon-Maciolek M.A. Human rhinovirus causes severe infection in preterm infants. Pediatr Infect Dis J. 2010;29:364–365. doi: 10.1097/INF.0b013e3181c6e60f. [DOI] [PubMed] [Google Scholar]
- 38.Fry A.M., Lu X.Y., Olsen S.J., Chittaganpitch M., Sawatwong P., Chantra S. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. Plos One. 2011;6:e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisdom A., Kutkowska A.E., Leitch E.C.M., Gaunt E., Templeton K., Harvala H. Genetics, recombination and clinical features of human rhinovirus species C (HRV-C) infections: interactions of HRV-C with other respiratory viruses. Plos One. 2009;4:e8518. doi: 10.1371/journal.pone.0008518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greer R.M., McErlean P., Arden K.E., Faux C.E., Nitsche A., Lambert S.B. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol. 2009;45:10–15. doi: 10.1016/j.jcv.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller E.K., Edwards K.M., Weinberg G.A., Lwane M.K., Griffin M.R., Hall C.B. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol. 2009;123:98–104. doi: 10.1016/j.jaci.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pica N., Chou Y., Bouvier N., Palese P. Transmission of influenza B viruses in the guinea pig. J Virol. 2012;86:4279–4287. doi: 10.1128/JVI.06645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bezerra P.G.M., Britto M.C.A., Correia J.B., Duarte MdCMB., Fonceca A.M., Rose K. Viral and atypical bacterial detection in acute respiratory infection in children under five years. Plos One. 2011;6:e18928. doi: 10.1371/journal.pone.0018928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller E.K., Bugna J., Libster R., Shepherd B.E., Scalzo P.M., Acosta P.L. Human rhinoviruses in severe respiratory disease in very low birth weight infants. Pediatrics. 2012;129:E60–E67. doi: 10.1542/peds.2011-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein M.I., Bergel E., Gibbons L., Coviello S., Bauer G., Benitez A. Differential gender response to respiratory infections and to the protective effect of breast milk in preterm infants. Pediatrics. 2008;121:E1510–E1516. doi: 10.1542/peds.2007-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright P.F., Deatly A.M., Karron R.A., Belshe R.B., Shi J.R., Gruber W.C. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J Clin Microbiol. 2007;45:2126–2129. doi: 10.1128/JCM.02553-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato M., Li H., Ikizler M.R., Werkhaven J.A., Williams J.V., Chappell J.D. Detection of viruses in human adenoid tissues by use of multiplex PCR. J Clin Microbiol. 2009;47:771–773. doi: 10.1128/JCM.02331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.United States Census Bureau. Summary File 1. United States Census, 2010. Washington, DC. http://2010.census.gov/2010census/data.