Abstract
Approximately 285 million people worldwide suffer from diabetes, with insulin supplementation as the most common treatment measure. Regenerative medicine approaches such as a bioengineered pancreas has been proposed as potential therapeutic alternatives. A bioengineered pancreas will benefit from the development of a bioscaffold that supports and enhances cellular function and tissue development. Perfusion-decellularized organs are a likely candidate for use in such scaffolds since they mimic compositional, architectural and biomechanical nature of a native organ. In this study, we investigate perfusion-decellularization of whole pancreas and the feasibility to recellularize the whole pancreas scaffold with pancreatic cell types. Our result demonstrates that perfusion-decellularization of whole pancreas effectively removes cellular and nuclear material while retaining intricate three-dimensional microarchitecture with perfusable vasculature and ductal network and crucial extracellular matrix (ECM) components. To mimic pancreatic cell composition, we recellularized the whole pancreas scaffold with acinar and beta cell lines and cultured up to 5 days. Our result shows successful cellular engraftment within the decellularized pancreas, and the resulting graft gave rise to strong up-regulation of insulin gene expression. These findings support biological utility of whole pancreas ECM as a biomaterials scaffold for supporting and enhancing pancreatic cell functionality and represent a step toward bioengineered pancreas using regenerative medicine approaches.
Keywords: Whole organ decellularization, Extracellular matrix scaffold, Tissue and organ engineering, Pancreatic β-cells
1. INTRODUCTION
Diabetes mellitus is a global disease with immense economic and social burden. According to the World Health Organization, at least 285 million people worldwide suffer from diabetes [1]. While pharmaceutical interventions and insulin supplementation are the most commonplace treatment of diabetes, these do not represent a cure and can potentially lead to long term complications [2]. Advances in tissue engineering have facilitated the development of replacement tissues or organs for the treatment of injured or degenerative soft tissue [3–5]. The development of a bioengineered pancreas by appropriate combination of cells, biomaterial scaffolds and biologically active molecules could provide an alternative avenue for diabetes therapy.
Three-dimensional (3D) scaffold plays a critical role in regenerative medicine and tissue engineering. For example, cell-scaffold interactions are important for the regulation of cellular behavior, including pancreatic islet-cell survival and insulin production through integrin-mediated activation and downstream signaling events [6, 7]. Native tissues and organs are comprised of unique extracellular matrix (ECM) compositions, microstructures and biomechanical properties, which maintain distinct signals for resident cells [8, 9]. Ideally, a tissue engineered scaffold would provide the same or similar microenvironmental niche to the seeded cells as that of a native ECM. Therefore, at present, the development of pancreatic tissue engineering scaffolds have focused on recreating milieu similar to the native islet and surrounding ECM (summarized in review by Cheng et al. [10] and Candiello et al. [11]). Various synthetic and natural biomaterials have been exploited in this field with the objective to restore the critical signal loss between native islet cells and their ECM environment following islet isolation. Most of these tissue engineering scaffolds, however, use a purified single ECM protein and do not mimic the intricacy of the pancreatic ECM composition and ultrastructure. Recently there has been a shift towards multiple-matrix systems of purified proteins, which have been shown to improve stem cell proliferation and differentiation [12, 13] and promote isolated islet and β-cell survival and function [14, 15]. Natural ECM derived from native tissues and organs can meet this requirement by providing a more physiologically relevant scaffold that recapitulates the complex in vivo microenvironment.
The novel concept of whole organ perfusion decellularization has been described recently to generate native ECM scaffolds from complex organs such as heart [16–19], liver [20–23], lung [24–29], kidney [30–32] and more recently pancreas [33–36]. Whole organ acellular matrices provide an attractive scaffold for the repopulation with cells for an engineered tissue/organ because of the physiological resemblance of the original tissue, including intact 3D anatomical architecture, preserved spatial array of ECM components, vascular network, and biomechanical properties. Prior studies have shown the positive effect of endothelial cell derived ECM for the enhancement of islet and β-cell attachment and proliferation [37, 38]. Other works have also shown improved islet functionality when cultured on decellularized matrices derived from small intestinal submucosa (SIS) [39, 40] and pancreatic slices [36, 41]. Decellularized pancreas work described by De Carlo et al. [41] and Mirmalek-Sani et al. [36] revealed the benefits of 2D decellularized pancreas slices to support and maintain islet functions. These initial studies clearly motivate the need and indicate the potential benefits that can be attained by a whole organ 3D reconstruction of pancreas. The importance of 3D scaffold is well recognized for a better representation of the complex in vivo microenvironment and is important for proper regulation of cell behavior [42], yet recellularization of whole pancreas 3D scaffold remains to be demonstrated. We hypothesize that the optimal scaffold to support pancreatic tissue engineering may be the natural 3D ECM scaffold of the whole pancreas. To this effect, we propose that perfusion-decellularization of whole pancreas yields an acellular scaffold suitable for pancreatic tissue and organ engineering.
The present study investigated the possibility of generating an acellular whole pancreas scaffold by a perfusion decellularization technique and utilizing this biomaterial as a scaffold to support pancreatic tissue engineering and whole organ regeneration. Toward this end, we characterized the resultant native pancreatic ECM-scaffold for preservation of ECM composition, 3D structural integrity, and biomechanical properties. We then repopulated the decellularized pancreas with endocrine and exocrine cell types to demonstrate the feasibility of recellularization. Furthermore, the effects of pancreas derived ECM scaffold on modulating cellular functionality such as insulin gene expression was determined.
2. METHODS
2.1 Mouse pancreas harvest and cannulation
All animal work performed was in accordance with animal welfare act, institutional guidelines and approved by Institutional Animal Care and Use Committee of the University of Pittsburgh). Female ICR mice (Taconic), between the ages of 6–12 weeks were euthanized by CO2 inhalation followed by cervical dislocation. Sterile conditions were observed when removing the pancreas. A laparotomy was performed and a 24 G catheter was inserted approximately 1 cm into the anterior hepatic portal vein and sutured in place. The distal end of the superior mesenteric vein and large branches of splenic arteries and veins were carefully ligated to prevent leakage. The pancreas was then carefully dissected free from all adjacent structures including the stomach, intestine, spleen and mesenteric tissue.
2.2 Perfusion decellularization of mouse pancreas
The isolated pancreas was connected to a perfusion system to allow retrograde perfusion at 8ml/min, in which solutions flowed through the hepatic portal vein, into the splenic vein, and throughout the vasculature of the pancreas. Next, ionic detergent, 0.5% SDS (Invitrogen) in deionized water was used as perfusate to rinse cells and cell debris out of the pancreas. After the tissues became translucent (about 325 min), subsequent steps of perfusion were 15 min of deionized water perfusion and 15 min of 1% Triton X-100 (Sigma Aldrich) in deionized water. A solution of benzonase (90U/ml, Sigma) was perfused for 15 min and a final washing step of 10% fetal bovine serum (FBS, Life Technologies) in PBS with Pen/Strep (100U/ml) perfused the pancreas for additional 48 hours to clear remaining cellular debris.
2.3 Cell culture
The AR42J acinar cell line (CRL-1492, ATCC, VA, USA) was used at passage 18–25. AR42J was cultured inF-12K medium (ATCC, VA, USA) supplemented with 20% FBS (Life Technologies) and 100 U/ml penicillin/streptomycin (Life Technologies) on T75 tissue culture flasks. The MIN-6 β-cell was used at passage 19–26. MIN-6 was cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 15% FBS (Life Technologies) and 100 U/ml penicillin/streptomycin (Life Technologies) on T75 tissue culture flasks. Both cell types were cultured at 37°C and in a 95% air/5% CO2 atmosphere.
2.4 In vivo implantation of decellularized pancreas
All animal work was approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. For preliminary biocompatibility studies, a 1 cm2 perfusion-decellularized pancreas construct was surgically placed within a dorsal subcutaneous pocket of an adult mouse. Female C57BL/6 mice, age 6–8 weeks, were purchased from Jackson Laboratories (Bar Harbor, ME). The mouse was anesthetized and maintained at a surgical plane of anesthesia with 1.5–2.5% isoflurane in oxygen and positioned in dorsal recumbency. The surgical site was prepared in sterile fashion using 70% isopropyl alcohol followed by the placement of sterile drapes. For the subcutaneous surgical implantation: A central longitudinal incision measuring approximately 1.5 cm in length was made in the epidermis, dermis, and fascia to expose the underlying muscle tissue on the dorsal side. Dermal layers and underlying connective tissue was undermined to create a pocket with similar size to the implant construct. The decellularized pancreas construct was sutured to the underlying muscle with four non-absorbable marking sutures (3-0 prolene) and the skin was closed over the implant site in uninterrupted fashion. A minimal amount of suture material was placed only at the construct corners to avoid eliciting a host response to the suture that would obscure the host response to the implanted material. The incision was covered in betadine ointment after closure and assessed for signs of infection for2 days postsurgery. The mouse received Buprenex (buprenorphine hydrochloride, 0.25 mg/kg) for analgesia, and Baytril (enro oxacin, 20 mg) an antibiotic, for 3 days postoperatively. The animal survived the surgical procedure and its predetermined study period without complications. After 14 days the mouse was euthanized with 5% iso urane in oxygen followed by an intracardiac injection of potassium chloride to induce cardiac arrest. Following euthanasia the implant along with the surrounding skin and muscle were harvested and fixed in 10% NBF for histologic evaluation.
2.5 Recellularization and culture of seeded-pancreatic construct
MIN-6 cells (30×106) were trypsinized and diluted in 3mL of medium. The cell suspension was introduced into the decellularized pancreas by means of retrograde gravity perfusion via the hepatic portal vein in 3 steps, 1mL each, with 20 min interval between each step. MIN-6 seeded pancreas was immersed and cultured in the same MIN-6 cell culture medium described above for 5 days. For co-recellularization strategy, AR42J was used as the second cell type for the recellularization. Similarly, AR42J (30×106 cells) were trypsinized and diluted in 3mL of medium. The AR42J cell suspension was inoculated into the pancreas by retrograde perfusion through the pancreatic duct but the MIN-6 cells were introduced through the vasculature. The two cell types were seeded in sequence, with the same multi-step repopulation mentioned for single-cell type seeding. Cells were allowed to attach for 2 hours before introducing a gentle rinse through both the hepatic portal vein and pancreatic duct in order to wash out any unattached cells. The cell-seeded pancreatic construct was left undisturbed and submerged in co-recell culture medium (DMEM/F12 supplemented with 15% FBS and 100 U/ml Pen/Strep). Both cannulation to the portal vein and pancreatic duct were kept intact to allow perfusion feeding of media every day. The seeded pancreas was cultured under static conditions at 37°C with a 95% air/5% CO2 atmosphere for 5 days.
2.6 MIN-6 seeded on different ECM substrates and 2D thin slices of decellularized pancreas
Growth factor-reduced Matrigel (BD) was used as a thin coating according to manufacturer’s instruction. Collagen IV (BD) and fibronectin (Milipore) were used at a concentration of 5 mg/ml. These ECM substrates were used for coating non-tissue culture-treated 12-well plates (BD). Collagen type I (BD) was used as 5 mm thick gels. Coated and untreated (UT) plates were blocked with 1% BSA (Sigma). Control culture was conducted on UT plates blocked with 1% BSA (Sigma). To prepare thin decellularized pancreata slices, decellularized pancreata were filled with 1.1% alginate solution, and submerged in 100mM Calcium Chloride solution to crosslink the alginate. The solidified pancreas was sliced with a tissue slicer (Thomas Scientific) to yield thin, uniform, transverse sections. The alginate within the thin slices of pancreas were dissolved in a sterile dissolution buffer consisting of 50 mM tri-sodium citrate dihydrate (Sigma), 77 mM sodium chloride (VWR International, Leicestershire, UK), and 10 mM HEPES (Sigma) in PBS for 15–20 min while stirring gently at room temperature. Finally, the thin slices of pancreas were rinsed and covered with sterile PBS with Pen/Strep (100U/ml), and placed at 4°C until use. Cloning cylinder (Fisher) was applied and sealed with sterile silicone grease (Dow Corning) to the pancreatic slice to consolidate the seeded cells only to the pancreatic slice. MIN-6 cells (25,000 cells/cm2) were cultured on these various different ECM substrates for 5 days before harvest for qPCR analysis.
2.7 Two-photon microscopy
Two-photon microscopy was performed with an upright Olympus FV1000 MPE multi-photon microscope (Olympus, Central Valley, PA, USA) and a Mai Tai DeepSee femtosecond-pulsed laser (Spectra-Physics, Santa Clara, CA, USA) tuned at 800 nm. The pancreatic slice was line-scanned and fluorescence emission was captured by three non-descanned external photomultiplier tube (PMT) detectors coupled to the following long-pass dichroic mirrors and band pass emission filters: 505 nm mirror and 460–500 nm filter (blue channel), 570 nm mirror and 520–560 nm filter (green channel) and 575–630 nm filter (red channel). The pancreatic slice was placed on an imaging dish having a #1.5 coverslip and immersed in PBS. Fixed xy planes spanning 505 × 375 μm at a resolution of 0.994 μm/pixel and depth of 1–50 μm from the surface of scaffold were imaged using a high numerical aperture (NA = 1.05), water-immersion 25X objective.
2.8 Immunohistochemistry (IHC)
Native, decellularized and recellularized pancreas were fixed, sectioned and stained following protocol previously described [43]. Briefly, tissue samples were fixed with 4% formaldehyde (ThermoFisher), cryoprotected with 30% sucrose and cut into 7 μm thick sections. For immunostaining, the following primary antibodies were used: rabbit anti-laminin, rabbit anti-collagen I, rabbit anti-collagen IV, rabbit anti-fibronectin (Abcam, 1:200), rabbit anti-Cpeptide (Cell signaling, 1:100) and rabbit anti-α-Amylase (Sigma Aldrich, 1:200). Secondary antibodies used were: donkey anti-rabbit Alexafluor 488 (1:500, Invitrogen) and donkey anti-rabbit Alexafluor 555 (1:500, Invitrogen). For co-labeling using antibodies from the same host species, sequential staining was conducted. After the first primary antibody staining, an additional blocking step was included prior to the addition of a second primary antibody. For actin staining, phalloidin rhodamine (Cytoskeleton, 1:200) was implemented and stained for 30 min in the dark. The slides were washed again three times with 1× PBS (5–10 min) each before being mounted with ProLong® Gold Anti-fade Reagent with DAPI (Invitrogen). Images were recorded with Metamorph 7.5.6.0 (Molecular Device) on an Olympus IX81 inverted microscope (Olympus, Central Valley, PA, USA).
2.9 ECM digestion and proteomics analysis
The ECM pellet was digested for 2h with each of the following enzymes: 0.15 mU/μl heparan sulfate lyase and chondroitin ABC lyase from Flavobacterium heparinum and Proteus vulgaris, respectively (Seikagaku Corporation, Japan) and washed in PBS. The ECM proteins were dissolved in 8M urea and 1x reducing SDS sample buffer. The proteins from the ECM preparation were separated on 3.5–15% gradient SDS PAGE under reducing conditions [44]. The gel lane was divided into 11 slices and taken through a typical in-gel digestion procedure. Briefly, gel bands were destained, reduced with 2.5mM tris (2-carboxyethyl) phosphine and alkylated with 3.75mM iodoacetamide, followed by in-gel digestion with 13ng/μl of trypsin (Promega) overnight at 37°C. Peptide digests were analyzed by nano Liquid Chromatography-tandem mass spectrometry (LC-MS/MS), on a Thermo Fisher LTQ OrbitrapVelos connected to a Waters Acquity UPLC system (Waters Corp., Milford, MA), using a 90 minute gradient. Protein identification was performed with Proteome Discoverer 1.3 using the Sequest search engine. Database searches used the Uniprot complete mouse database (downloaded in December 2012 merged with a contaminant database from ABSciex, 50838 sequences, 24435643 residues). Settings were for a full trypsin digest, with two missed cleavages, one static modification (cysteine carbamidomethylation), two dynamic modifications (oxidized methionines and hydroxyproline), mass tolerance of 10 ppm for precursor mass and 0.5 Da for fragment masses. Percolator, a post-processing software using a target/decoy database approach, was used to evaluate the accuracy of peptide identifications. Peptide identifications were filtered with a q-value cutoff of 0.01 (1% global False Discovery Rate, FDR). Proteins were grouped using the maximum parsimony principle and this list was imported to ProteinCenter (Thermo) and compared with the IPI mouse database for statistical analysis to identify over-represented Gene Ontology terms and functional classification.
2.10 DNA quantification
The decellularized pancreata (n=3) were digested with papain solution at 60°C for 6 h. The native pancreata (n=3) were digested in papain solution as controls. Papain (Sigma Aldrich) was dissolved at 400 mg/ml in 0.1 M phosphate buffer (pH 6.0), with 5 mM cysteine hydrochloride (Sigma Aldrich), and 5 mM EDTA (Sigma Aldrich). The lysates were used for detection of the DNA and sulfated glycosaminoglycan (sGAG) content. A DNA quantification kit - Quant-iT™ PicoGreen® dsDNA Assay kit (Invitrogen) was used to measure DNA content according to manufacturer’s instruction. The fluorescence reading (excitation: 485 nm and emission: 528 nm) was taken on a plate reader (Synergy 2, Biotek), and the absolute amount of DNA (ng/mL) was quantified against a lambda DNA standard curve (0 ng/mL– 1000 ng/mL).
2.11 sGAG content characterization
For qualitative determination of presence of GAG, Alcian Blue staining was done according to manufacturer’s instruction (Newcomer Supply). Briefly, slides were deparaffinized and hydrated through graded ethyl alcohols to distilled water. Next, slides were placed in 3% acetic acid for 3 min and then directly into 1% Alcian Blue solution pH 2.5 and stained for 30 min in room temperature. The slides were washed in running tap water for 10 min followed by a distilled water rinse. The counterstaining was done in Nuclear Fast Red stain for 5 min before rinsing, dehydrated through graded ethyl alcohol and cleared in xylene. The slides were mounted with xylene mounting medium. For qualitative measure of sGAG, the method to produce lysate for DNA quantification was used here as well. A sGAG quantification kit–Blyscan Sulfated Glycoaminoglycan Assay kit (Biocolor) was used to measure sGAG according to manufacturer’s instruction. Briefly, the specimen lysate was mixed with Blyscan dye to bind the GAG. The GAG-dye complex was then collected by centrifugation. After the supernatant was removed and the tube drained, the dissociation reagent was added. 100 μl solutions were transferred into a 96-well plate. Absorbance against the background control was obtained at a wavelength of 656 nm on a microplate spectrophotometer and the GAG amount was calculated based on a standard curve obtained with the standard GAG supplied with the kit.
2.12 Scanning electron microscopy (SEM) and transmission electron microscopy (TEM)
Native and decellularized pancreata were fixed in 2.5% glutaraldehyde in 0.1 M PBS (pH 7.4) for 60 minutes. The samples were washed thoroughly in 3 changes 0.1 M PBS for 15 minutes each. Next, the samples were fixed in 1% OsO4 in 0.1 M PBS for 60 minutes. This was followed by another 3 changes of PBS washing steps for 15 minutes each. The samples were then dehydrated in gradient series of alcohol for 15 min each. Additionally, samples were critical point dried and coated with Au/Pd using a Cressington Coater 108A sputter coater. Electron microscope images were taken using a Jeol JSM-6335F field emission SEM. For TEM, the tissue sample was fixed in 2.5% glutaraldehyde in PBS. The tissue sample were post-fixed in 1% osmium tetroxide in PBS, dehydrated through a graded series of alcohols and embedded in Epon (Energy Beam Sciences, Agawam, MA). Thin (60-nm) sections were cut using a Reichert Ultracut S (Leica, Deerborn, MI), mounted on 200 mesh copper grids and counterstained with 2% aqueous uranyl acetate for 7 min and 1% aqueous lead citrate for 2 min. Observation was with a JEOL 1011 transmission electron microscope (Peabody, MA)
2.13 Preparation of pancreatic tissue for atomic force microscopy (AFM) measurement
Both native and decellularized unfixed samples from murine pancreas were cryoprotected by soaking in 30% sucrose solution in PBS for 24 hours. The entire murine pancreas was then set in a mold and covered in OCT cold embedding media and frozen by exposure to dry ice overnight. The OCT block containing the pancreas was cryosectioned to a thickness of 20 microns and mounted on SuperFrostPlus glass slides. All samples were thawed in PBS for 30 minutes prior to testing.
2.14 AFM measurement of pancreatic tissue stiffness
AFM force indentation measurements were performed using the MFP-3D Atomic Force Microscope (Asylum Research, CA, USA), mounted on top of an Olympus IX-71 fluorescence microscope (Olympus, Tokyo, Japan). All force measurements and analysis was done using the MFP3D software (Asylum Research) built on IgorPro 6 (Wavemetrics) as previously described [45]. For all measurements a glass silica sphere (radius 3.5 micron) was attached to the tip of a 100 micron silicon nitride cantilever (Veeco Systems) with a spring constant of ~0.6 N/m. The stiffness of each alginate gel was measured at n= 16 random locations on each sample. The Sneddon model was used to determine the gel stiffness from the force indentation plots.
2.15 Quantitative RT-PCR
RNA was extracted using NucleoSpin kit according to the manufacturer’s protocol. The sample absorbance at 280 nm and 260 nm was measured using a BioRad Smart Spec spectrophotometer to obtain RNA concentration and quality. Reverse transcription was performed using ImProm II Promega reverse transcription kit following the manufacturer’s recommendation. qRT-PCR analysis was performed for insulin markers, ins1 and ins2.
The cycle number at the threshold level of log-based fluorescence is defined as Ct number, which is the observed value in most real-time PCR experiments, and therefore the primary statistical metric of interest. ΔCt is equal to the difference in threshold cycle for target and reference or control (ΔCt = Cttarget−Ctreference). ΔΔCt is equal to the difference between ΔCtsample and ΔCtcontrol (ΔΔCt = ΔCtsample−ΔCtcontrol). The fold change of a target gene is defined by, fold change = 2−ΔΔCt. qRT-PCR analysis was repeated in triplicate.
2.16 Statistical analysis
Quantification data were expressed as mean ± SD. Significant differences among groups were determined by Wilcoxon rank-sum test for two-group comparisons or ANOVA followed by post-hoc analysis for multiple group comparisons. Probability values at P < 0.05 (*) indicated statistical significance.
3. RESULTS
3.1 Perfusion-decellularization of whole organ pancreas
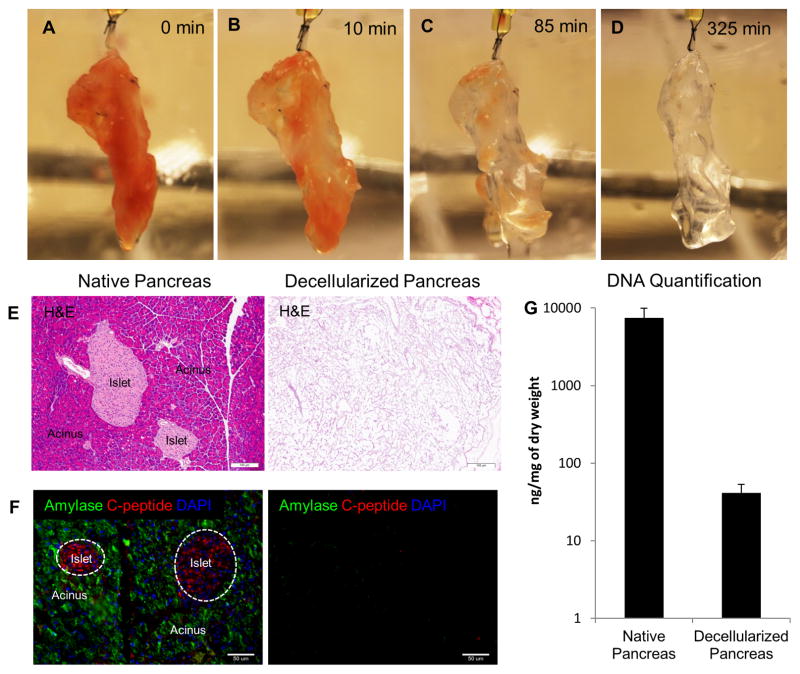
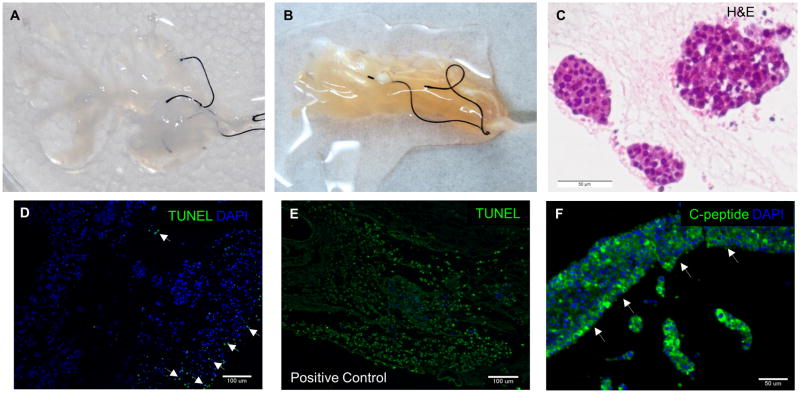
Isolated mouse pancreata were cannulated and retrograde perfused via hepatic portal vein with anionic based detergent, 0.5% SDS to remove cellular content. Alternately, retrograde perfusion via pancreatic duct was also attempted for pancreas decellularization, however the process took longer and was also less effective in cell removal (data not shown). Macroscopically, a gradual change of color was observed during the perfusion decellularization process (Fig. 1A–D). Perfusion was continued until whole pancreas turned completely translucent (about 325 min) as depicted in Fig. 1D. This generated an acellular pancreas scaffold while retaining the gross anatomical structure of the pancreas. Histological examination by H&E staining showed no remnant cells after the completion of decellularization (Fig. 1E). Immunostaining of two major constituents in native pancreas–acinar cells (Amylase) and β-islet cells (C-peptide) confirmed no residual presence of major pancreatic markers in the decellularized pancreas (Fig. 1F). To further assess the efficacy of decellularization, DNA quantification was performed using Picogreen assay. Picogreen analysis showed that DNA content decreased from 7442.3 ± 2450.2 ng/mg dry weight in normal pancreas to 41.3 ± 11.9 ng/mg dry weight in decellularized pancreas (P < 0.01) (Fig. 1G). These results suggest that perfusion mediated decellularization of pancreas efficiently removes pancreatic cellular components. Feasibility of this perfusion decellularization technique in human-sized organ was also demonstrated by decellularization of bovine pancreas (supplemental 1).
Figure 1. Perfusion-decellularization of murine pancreas.
(A–D) Panel images depict gradual change of color from perfusion-decellularization of mouse pancreas with 0.5% SDS. Resulting decellularized pancreas after 325 min (D) appeared translucent. (E) Histological comparison of native and decellularized pancreas by H&E staining showed removal of cells. (F) IHC evaluation of native and decellularized pancreas depict presence of major pancreatic constituents (C-peptide–red, and Amylase–green) in native pancreas (left) but absence in decellularized pancreas (right). (G) DNA quantification via picogreen analysis demonstrated less than 50ng/mg dry weight of DNA material present in the decellularized pancreas.
3.2 ECM characterization: immunohistochemistry and mass spectrometry-based proteomics analysis
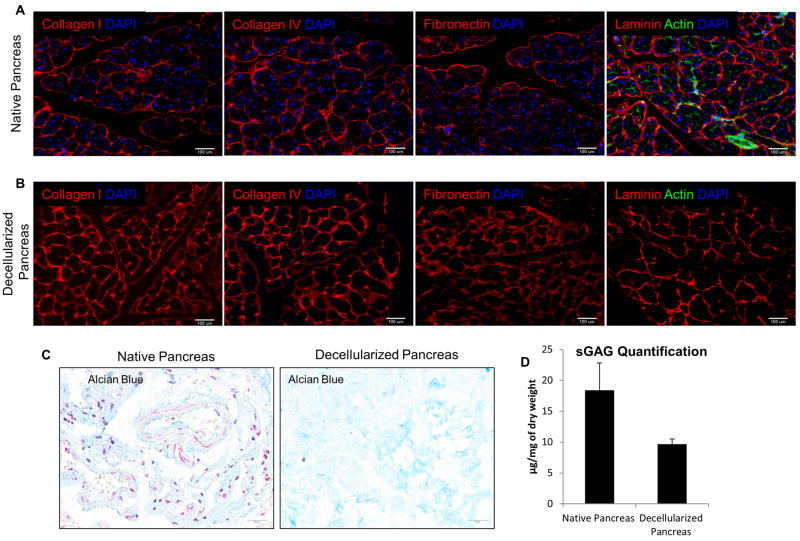
To first characterize the decellularized pancreas, immunohistochemical (IHC) staining was performed for the evaluation of major ECM components’ spatial presence relative to that of native pancreas. In native pancreas, the ECM is composed primarily of networks of collagens and other structural proteins filled with a hydrogel of proteoglycans [46, 47]. IHC staining showed that collagen I, IV, fibronectin and laminin were detected in the native pancreas (Fig. 2A). After decellularization, all four major ECM components were preserved, without any detectable DAPI or actin staining (Fig. 2B). This finding suggests complete removal of cellular and cytoskeletal elements while maintaining the ECM composition. In addition, the resulting ECM structure and fibril orientation looked identical to that of the native organ (Fig. 2B). The preservation of sulfated glycosaminoglycan (sGAG) proteins was evaluated qualitatively by alcian blue staining (Fig. 2C), which demonstrated that sGAG proteins were retained after decellularization process. In addition, sGAG content was also assessed quantitatively using a blyscan assay which demonstrated considerable retention of sGAG content in the decellularized pancreas, amounting to 52% of that in native pancreas (P<0.01) (Fig. 2D).
Figure 2. ECM characterization of decellularized pancreas.
(A) IHC staining showed the presence of major ECM composition–Col I, Col IV, Fibronectin, laminin and phalloidin actin staining (green) in native pancreas. (B) Comparison with decellularized pancreas, IHC staining demonstrated the corresponding ECM markers found in native pancreas also preserved in decellularized markers but DAPI and actin cytoskeletal element were absent. This indicates complete removal of cellular materials but preservation of important ECM proteins. (C) Alcian blue staining qualitatively determined the retention of sGAG protein after decellularization. (D) Blyscan assays quantitatively determined the sGAG retention in decellularized pancreas was on average 52% of that in native pancreas (P<0.05).
Insolubility of the ECM is a challenge to characterizing the pancreatic ECM proteome by mass spectrometry-based proteomics. In order to improve solubility, pancreatic ECM was deglycosylated to remove GAG side chains of proteoglycans followed by solubilization in high molar urea. A GeLC-MS/MS proteomics approach was used whereby the high molecular weight ECM proteins were separated by low percentage 1D gels, in-gel trypsin digested followed by LC-MS/MS analysis. Database searching used stringent 1% false discovery rate to report protein identifications. After protein grouping, a total of 114 unique proteins were identified in the pancreatic ECM (Supplemental table 1), the majority of which belonged to the ECM family based on Gene Ontology (GO) Cellular localization. The entire list of identified proteins was compared with the reference IPI mouse database to identify overrepresented GO cellular localization terms. The topmost GO terms included ECM and basement membrane (data not shown); suggesting that the decellularization process successfully removes cellular proteins yet preserves the ECM components. Table 1 is a categorized list of collagens, laminins, proteoglycans and ECM associated proteins.
Table 1.
Mass Spectrometry Composition Analysis
| Protein Family | Gene | Description |
|---|---|---|
| Collagens | Col6a3 | Collagen alpha-3(VI) chain |
| Col1a2 | Collagen alpha-2(I) chain | |
| Col4a2 | Collagen alpha-2(IV) chain | |
| Col4a1 | Collagen alpha-1(IV) chain | |
| Col1a1 | Collagen alpha-1(I) chain | |
| Col5a2 | Collagen alpha-2(V) | |
| Col6a2 | Collagen alpha-2(VI) | |
| Col6a1 | Collagen alpha-1(VI) | |
| Col5a1 | Collagen alpha-1(V) chain | |
| Col3a1 | Collagen alpha-1(III) | |
| Col14a1 | Collagen alpha-1(XIV) chain | |
| Col6a6 | Collagen alpha-6(VI) chain | |
| Col5a3 | Procollagen, type V, alpha 3 | |
| Col7a1 | Col7a1 Collagen alpha-1(VII) chain | |
| Gm7455 | Collagen alpha-5(VI) chain | |
| Col4a3 | Collagen alpha-3(IV) chain | |
| Col18a1 | Collagen alpha-1(XVIII) chain | |
| Laminin | Lamb2 | Laminin subunit beta-2 |
| Lamc1 | Laminin subunit gamma-1 | |
| Lama2 | Laminin subunit alpha-2 | |
| Lama5 | Laminin subunit alpha-5 | |
| Lamb1 | Laminin subunit beta-1 | |
| Lama1 | Laminin subunit alpha-1 | |
| Lama4 | Laminin subunit alpha-4 | |
| Proteoglycans | Hspg2 | Basement membrane-specific heparan sulfate proteoglycan core protein |
| Agrn | Agrin | |
| Nidogens | Nid1 | Nidogen-1 |
| Nid2 | Nidogen-2 | |
| Elastin associated | Fbn1 | Fibrillin-1 |
| Matricellular | Tnxb | Tenascin-X |
3.3 Biophysical characterization: structural and mechanical properties of decellularized pancreas
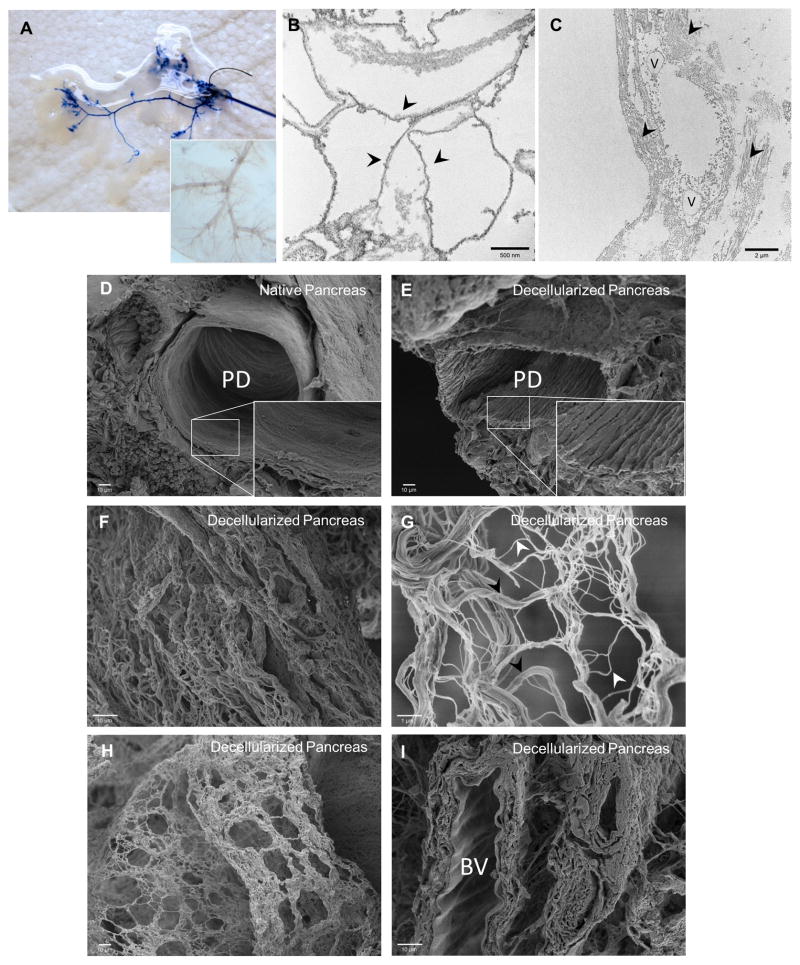
Integrity of the vasculature within the decellularized pancreas was analyzed by trypan blue dye retrograde injection through hepatic portal vein to visualize the continuity of the circulatory branching (Fig. 3A). Rapid diffusion through the vascular tree was also observed (Fig. 3A). Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were performed to evaluate the effect of the decellularization procedure on the 3D architecture and ECM micro-structures of the decellularized pancreas. TEM analysis revealed intact basement membrane and organized fibers of collagen preserved upon decellularization (Fig. 3B–C). SEM analysis of the cross-sectional images of the decellularized pancreatic constructs revealed that large ductal structures were retained whereas the microvilli cells lining the ducts were removed (Fig. 3D–E). Nanofibrous structures of the ECM were well retained within the parenchyma (Fig. 3F–G). Hollow and void spaces observed within the decellularized pancreas are likely attributes of the “footprints” left from pancreatic cells removed during the decellularization process (Fig. 3H). The recognizable scalloping pattern of arteries also indicates the preservation of vascular structure likely branched from the greater pancreatic artery (Fig. 3I). These results suggest that perfusion decellularization of the pancreas adequately retained the ultrastructure and architecture of the native pancreas. Biomechanical characteristics of the decellularized pancreas were analyzed using AFM. Representative cantilever displacement/sample deflection curves (Fig. 4A) shows that there was a discernible difference between the native and decellularized pancreas, with the stiffer decellularized sample having a much steeper slope. Young’s modulus as determined from the AFM curves was found to be 1210±77 Pa for native mouse pancreas, while the Young’s modulus of decellularized pancreas is on average 3 times greater (3736±1893 Pa) (Fig. 4B).
Figure 3. Ultrastructural characterization of decellularized pancreas.
(A) Trypan blue dye infusion visualized the progressive flow from large vessels to the fine blood vessel branches along the channels without leakage. Inset shows light microscopy image of vasculature branches remained intact after decellularization. (B) TEM of decellularized pancreas depicted the intact basement membrane (arrowhead points to the underlying basement membrane). (C) TEM image of decellularized pancreas showed the interstitial space with organized fibers of collagen (arrowhead). (D–E) SEM comparison of native and decellularized pancreas demonstrated preservation of 3D microstructure of pancreatic duct after decellularization. Inset shows the absence of lining ciliated microvilli cells from the luminal surface of the duct after decellularization process. (F) SEM image of decellularized pancreas showed ECM fibers within the parenchymal space. (G) Higher magnification of 3D meshwork showed a variety of fibers - large bundle of Type I collagen (black arrowhead) associated with a variety of smaller fibers (white arrowhead). (H) SEM image of decellularized pancreas demonstrated large/small fibers interlinked in a plane that forms a boundary such as to previously occupied pancreatic parenchymal cells. (I) SEM image of decellularized pancreas demonstrated scalloping appearance of the internal elastic lamina of an artery, indicating intact blood vessel (BV).
Figure 4. Biomechanical properties of decellularization pancreas.
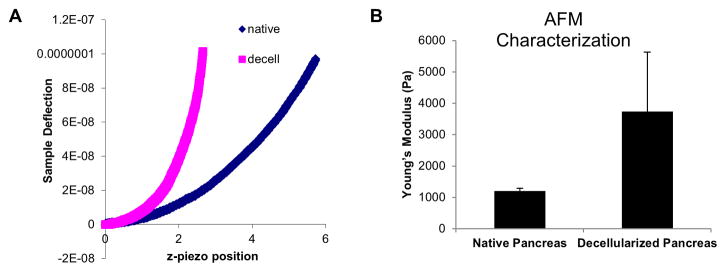
(A) Representative deflection plot from AFM evaluation of native and decellularized pancreas shows a steeper slope than force curve of decellularized pancreas from native pancreas, indicating higher Young’s modulus. (B) Bar graph representatives of AFM force curve shows on average 3 times higher stiffness in the decellularized pancreas than native pancreas.
3.4 In vivo response to decellularized pancreas
Pancreas ECM was implanted subcutaneously in order to preliminarily evaluate in vivo biocompatibility in a mouse model. At 14 days post-surgery, histological analysis of the subcutaneous implantation site showed the presence of mononuclear cells surrounding the partially degraded ECM (Fig. 5A) The pancreas ECM was non-cytotoxic after 14 days and like other efficiently decellularized ECM scaffolds [48, 49], was not associated with presence of multinucleate foreign body giant cells or other pathological signs of the foreign body response (Fig. 5B). Furthermore, active angiogenesis was a feature of the decellularized pancreas implantation site (Fig. 5B, arrows). These preliminary results suggest that the perfusion-decellularized pancreas ECM described herein represents a cytocompatible scaffold capable of integrating within host tissue.
Figure 5. Biocompatibility of perfusion-decellularized pancreas ECM.
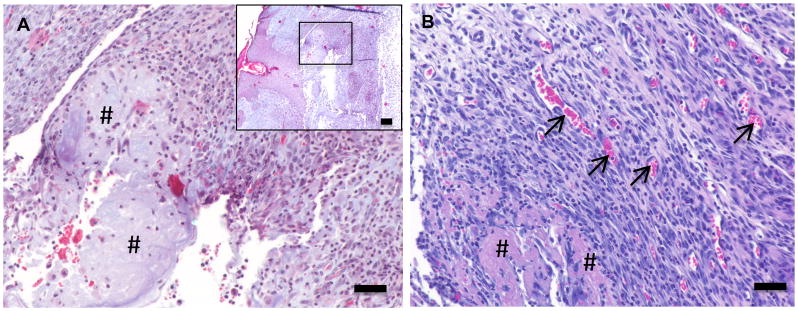
A perfusion-decellularized pancreas ECM construct was surgically implanted within a subcutaneous pocket in a C57Bl/6 mouse model. After 14 days Mason’s trichrome staining of the implant site (A) showed the ECM scaffold (#) infiltrated with host mononuclear cells. H&E staining (B) showed no signs of multinucleate giant cells and the presence of blood vessel (arrows). (Scale bars = 50μm, inset = low power magnification, inset scale bar = 200μm)
3.5 Recellularization of decellularized pancreatic scaffold
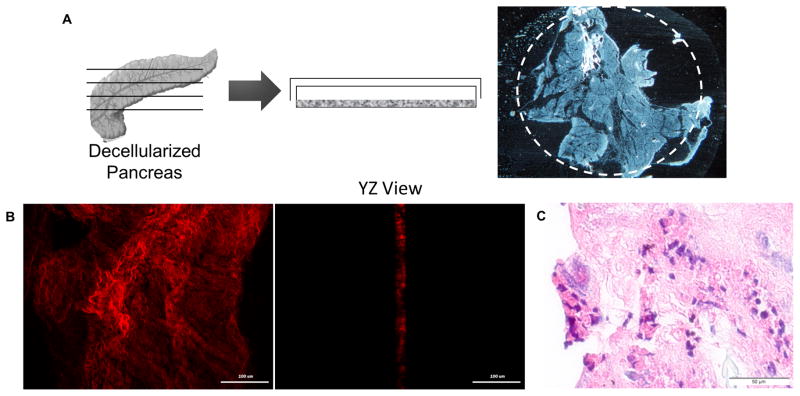
To evaluate the potential of decellularized pancreas as a scaffold for pancreatic tissue engineering, it was recellularized using relevant pancreatic cell lines. We first explored the effect of two dimensional (2D) slices of decellularized pancreatic matrix as a substrate for cell culture (Fig. 6A). Decellularized pancreatic matrix was cut into slices of about 10–20 μm thickness, as evaluated by multi-photon microscopy (Fig. 6B). MIN-6 cells (25,000 cells/cm2) were seeded on the 2D slice and cultured for 5 days. After 5 days, MIN-6 cells were found attached and spread on the surface of 2D slice (Fig. 6C).
Figure 6. MIN-6 culture on 2D slice pancreatic matrix.
(A) Diagram illustrates the generation of 2D pancreatic slice. White dashed circle depict the cloning cylinder applied to pancreatic slice to consolidate seeded cells to pancreatic substrate. (B) Multi-photon imaging on second harmonic generation (SHG) signal to visualize the collagen fibers demonstrated that thickness of the 2D slice pancreatic matrix is about 10–20 μm. (C) H&E staining demonstrated the attachment of MIN-6 cells on 2D pancreatic slice after 5 days of culture, and insulin expression (C-peptide: green) was maintained.
To evaluate whole organ recellularization potential, endocrine β cell line, MIN-6 cells (30×106) were first seeded into the decellularized pancreas via hepatic portal vein by multistep infusion technique following the seeding strategy described for liver recellularization [20, 21]. Approximately 10×106 cells were introduced at each step, for a total of 3 steps, with 20-min intervals between each step. Upon cell seeding, a uniform color change was observed throughout the whole decellularized pancreas, indicating a homogenous cell delivery (Fig. 7A–B). The repopulated pancreas was then maintained in static culture for 5 days with culture media changed by perfusion through the pancreas every day. After 5 days, the reconstructed pancreas was analyzed for engraftment, survival and functionality by IHC. From the histology, MIN-6 cells appeared to be engrafted both surrounding the parenchymal region and around the larger vessels (Fig. 7C). TUNEL staining detected only minimal apoptosis (<18%), suggesting that the 3D decellularized pancreas scaffold is cytocompatible and supportive of cell growth (Fig. 7D). Furthermore, these engrafted cells maintained their insulin expression over 5 days as evidenced by positive C-peptide staining (Fig. 7F).
Figure 7. Recellularization of decellularized pancreas.
(A) Decellularized whole pancreas before cell seeding. (B) Recellularization of the same pancreas via multi-step infusion of about 30 million cells via portal vein infusion. (C) H&E staining of the recellularized pancreas after 5 days showed the engraftment of the seeded cells at surrounding parenchymal region. (D) TUNEL staining of the recellularized construct showed less than 18% of apoptotic cells (arrowhead), demonstrating the cytocompatibility of the scaffold. (E) Positive control of TUNEL staining by adding DNAse to the consecutive sections of recellularized pancreas’s histological slide. (F) IHC of recellularized pancreas with MIN-6 only showed C-peptide+ cells engraftment around the larger vessel (arrowhead).
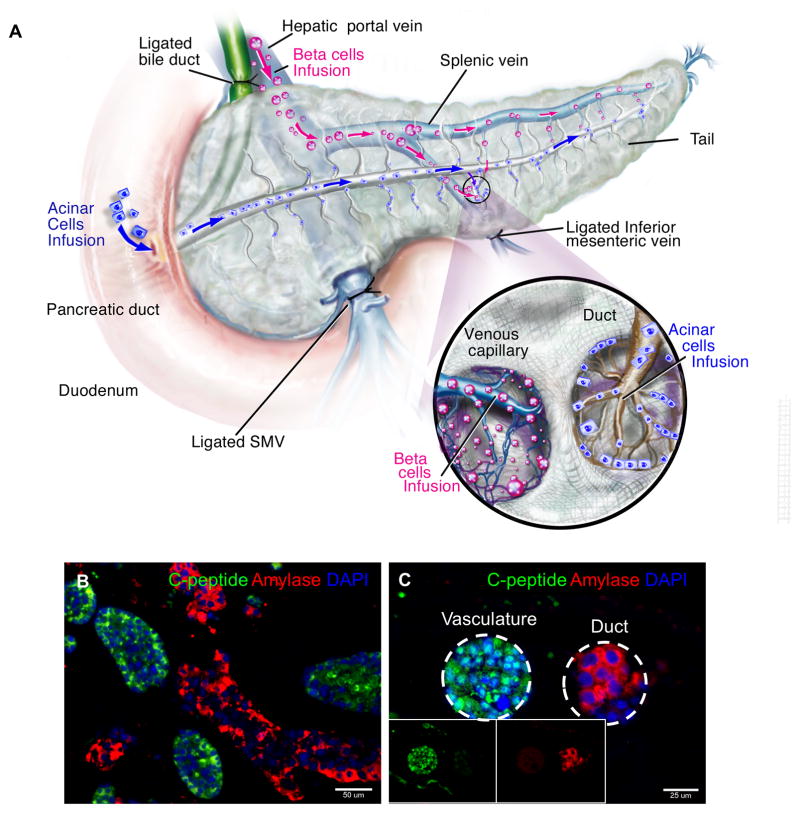
The native pancreas is a dual-functioning glandular organ comprised and capable of both endocrine and exocrine tissue and function, to this end, next the decellularized pancreas was reconstructed with both the endocrine β cell line (MIN-6) and exocrine acinar cell line (AR42J) by simultaneous seeding. The β cells were perfused through the hepatic portal vein as before, while the acinar cells were perfused through the pancreatic duct (Fig 8A). The two cell types were seeded in sequence, with the hepatic duct perfusion of the MIN6 cells preceding the pancreatic duct perfusion of acinar AR42J cells. This seeding strategy attempts to retain the cellular composition of native pancreas and the anatomical proximity of the cells to different secretory locations: (i.e.; endocrine β cells–vasculature; exocrine acinar cells–ductal system). As before, the dual cell-seeded pancreas constructs was maintained for up to 5 days in static culture. At day 5, IHC analysis showed that both cell types attached and localized to their specific topographical locations as a result of their different cell seeding route (Fig. 8B). Immunofluorescence staining confirmed the expression of C-peptide, as an analogue of insulin marker identifying endocrine β-cells and amylase as a digestive enzyme identifying exocrine acinar-cells (Fig. 8B, C). Most C-peptide+ cells were localized to the luminal spaces of large vessels and parenchymal regions similar to that observed in single cell reconstructed pancreas. Distinct from the localization of C-peptide+ cells, tubular ductal spaces lined with acinar cells expressing amylase were observed (Fig. 8C). No inter-dispersed population of the two cell types were observed within the engraftment region, suggesting the preservation of the distinct architecture of vasculature and ductal structure (Fig. 8C). These results indicate that the decellularized pancreas is capable of supporting multiple pancreatic cell types and this co-seeding strategy allows the reconstitution of major cellular constituents as in the native pancreas.
Figure 8. Dual recellularization strategy with ƒÀ- and acinar cell types.
(A) Two different cell types (MIN-6 and AR42J) were inoculated into the decellularized pancreas 3D scaffold utilizing two different seeding routes. (B) IHC of recellularization with both MIN-6 and AR42J cell lines showed engraftment of both cell types. Both cell types maintained their respective functional markers–MIN-6 (Cpeptide–green) and AR42J (Amylase–red). (C) Vasculature and pancreatic duct situated next to each other with their respective seeded different population of cells (vasculature-MIN6 and duct-AR42J). Inset shows single channel images. This indicates that preservation of architecture of vasculature and ductal structure, hence different cell population were not inter-dispersed after seeding.
3.6 Modulation of β-cell insulin gene expression by native pancreatic ECM
Both primary pancreatic islets and individual isolated pancreatic β-cells have been demonstrated to have better in vitro function when cultured on ECM-derived substrates [37, 39–41, 50] and purified ECM proteins [15, 38, 51, 52]. Having confirmed engraftment and survival of the relevant pancreatic cell types in the native pancreatic scaffold, the contribution of this pancreas-derived ECM scaffold on cell function was next evaluated by the insulin gene expression level. For comparative evaluation, control cultures of MIN-6 cells (25,000 cells/cm2) were plated on surfaces with the following ECM proteins: collagen I, collagen type IV, fibronectin, and growth factor-reduced (GFR) Matrigel. In addition, the MIN-6 cells were also cultured on 2D slice configurations of pancreatic ECM for comparison between the effects of 2D versus 3D culture configurations. Upon plating, the MIN-6 cells readily attached to all tested substrates and remained viable over the culture period (data not shown). The cells were cultured for 5 days, after which the cell lysate was analyzed for insulin gene expression (ins1 and ins2 expression) by qRT-PCR (Fig. 9). Quantitative RT-PCR of MIN-6 demonstrated significantly higher ins1 and ins2 gene expression when cultured on these ECM proteins and pancreatic ECM scaffold (P<0.05) compared to control (untreated plate) with the exception of collagen I gel. Among the ECM proteins tested, collagen IV and fibronectin substrates induced similar insulin gene expression levels. Matrigel promoted the highest insulin gene expression (6.9-fold ins1 and 4.1-fold ins2). MIN-6 seeded on 3D pancreatic ECM exhibited higher insulin gene expression than 2D pancreatic ECM. Furthermore, 3D pancreatic ECM also demonstrated higher insulin gene expression when compared to single ECM proteins such as collagen IV and fibronectin but displayed similar level of insulin gene expression with the Matrigel coated surface (no significant difference). These results clearly demonstrate that β-cells interact closely with the ECM substrates, and that interaction with specific substrates can significantly affect insulin gene expression. The native organ derived of decellularized pancreas was found to have a strong positive attribute to ins1 and ins2 gene expression, comparable to Matrigel coating and significantly higher than other tested single purified molecules.
Figure 9. Comparison of ECM mediated insulin gene expression of MIN-6 cells.
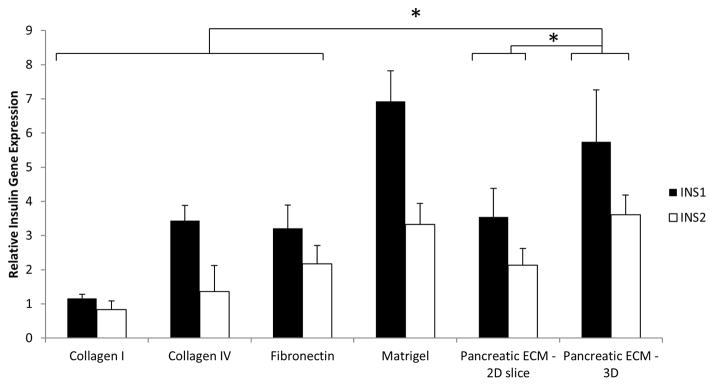
qPCR data demonstrated modulation of MIN-6 cell insulin gene expression by different types of ECM protein. 3D Pancreatic ECM demonstrated higher ins1 and ins2 genes expression than Collagen I, Collagen IV, Fibronectin and 2D pancreatic slice ECM but only comparable to Matrigel. Normalization was performed to MIN-6 cells seeded on control non-tissue culture treated plastic. * indicates P<0.05
4. DISCUSSION
The production of an intact acellular organ such as the pancreas by perfusion-decellularization offers a promising alternate approach for pancreatic tissue engineering and functional organ replacement. Herein, we demonstrate that perfusion-decellularization of whole pancreas results in the generation of a natural pancreas ECM scaffold with a perfusable vascular tree, ductal network and intact 3D architecture, which acts as a suitable template for pancreatic tissue engineering and whole organ regeneration. Comprehensive characterization of the decellularized pancreatic matrix showed the preservation of complex multi-faceted ECM proteins, 3D spatial orientation and microstructure, and relative stiffness of native pancreas. Decellularized pancreatic matrix was non-cytotoxic, and promoted angiogenesis when subcutaneously implanted in a mouse model. Repopulation of decellularized pancreas with pancreatic cells showed the cytocompatibility of the scaffold. The recellularized constructs maintained their respective phenotypic expression (β-cells: C-peptide; acinar cells: Amylase) after 5 days of culture. Finally, we demonstrated that the decellularized pancreas matrix is supportive of β-cell function, as evidenced by the strong up-regulation of insulin gene expression.
The objectives for successful decellularization are 1) complete or near complete removal of cellular material, and 2) preservation of ECM composition [53]. Commonly reported decellularization protocols require mechanical agitation or freezing and thawing which may take up to days or weeks to remove all cellular materials [54–56]. Such protocols do not guarantee preservation of the ECM microstructure, which has been shown to be instrumental for the generation of functional tissue engineered constructs [57]. In contrast, our approach of whole organ perfusion decellularization reduces the diffusion distance required for decellularization agents to reach the cells and facilitates removal of the cellular material from the tissue by convective transport [53, 58, 59]. This technique allowed the efficient generation of acellular scaffold with preserved ECM, 3D architecture and perfusable network resembling a native organ (approximately 325 min as shown in our data Fig. 1D). The resulting decellularized pancreas met the stringent requirement to define a successful decellularization [48] - absence of nuclear material with DAPI and H&E staining, and retained only less than 50ng dsDNA per mg ECM dry weight as shown by quantitative DNA measurement (Fig. 1E–G). This requirement is crucial because residual DNA fragments in decellularized ECM have shown to lead to cytocompatibility issues in vitro and adverse immunological response upon implantation [49, 60, 61]. We and other groups have tried decellularization using similar strategies on other circulatory networks to reduce diffusion distance (e.g., bile duct in liver, pancreatic duct in pancreas) but it was much less effective likely due to incomplete distribution of a conduit network to reach all the cells throughout the target organ.
Another important consideration for organ decellularization is minimizing the undesirable alteration and loss of biologically active ECM components. Decellularized pancreas scaffolds demonstrated maintenance of key ECM proteins in adult pancreas [46, 47], including collagen I, collagen IV, laminin, and fibronectin, as well as sulfated GAG. Most of the ECM proteins retained their physiological organization after decellularization as noted by IHC (Fig. 2A). This finding was further substantiated by mass spectrometry analysis to comprehensively characterize the proteome of the decellularized pancreatic ECM. Retention of ECM protein/glycan compositions is typically characterized by IHC; this approach is however limited by its robustness and only confined to a small number of ECM candidates studied. Proteomic analysis by mass spectrometry is unbiased and does not require specific probes. Though our analysis was qualitative, notably all the top scoring proteins belonged to the ECM family (supplemental table 1). These identifications have a high number of peptide spectral counts providing rough measures of protein abundance.
Biomechanical evaluation of engineered scaffolds and decellularized constructs is an important assessment for the rate of preservation of functional integrity. Numerous studies have shown that biomechanical properties change after decellularization of tissues [28, 62, 63]. By using AFM, we measured the stiffness of the decellularized pancreas ECM. Our results demonstrated that Young’s modulus of decellularized pancreas is on average 3 times greater than the native pancreas (P<0.01, Fig. 4A, B). This is partially attributed to the loss of GAG content from the decellularization protocol (Fig. 2D). GAG proteins have a myriad of biological functions and some of them are associated with structural and biomechanical properties. GAGs possess a fixed negative charge that renders them hydrophilic and attracts water into the tissue causing an osmotic swelling, which in turn could contribute to tissue physical properties. Removal of GAG side chains by heparitinase/chondroitinase treatment has been shown to increase stiffness of basement membrane of tissue [11]. Although our result showed a higher Young’s modulus in decellularized pancreas than native pancreas, it is still well within the mechanical property range of “soft tissue” [64, 65]. While the magnitude of increase depends on the specific organ under study, some studies have shown that a slight alteration of tissue biomechanical properties was not associated with any detrimental effect on cellular functionality. For instance, successful repopulation of a cardiac matrix was demonstrated despite a higher tangential modulus was detected within the decellularized heart when compared to native rat ventricles [16], which is consistent with our findings.
Recent studies have suggested that the ECM composition and expression pattern are specific to a given anatomical location to direct or support site-appropriate cell attachment and function [25, 66]. In the native pancreas, the primary functional units of the pancreas divide easily into endocrine cells and exocrine cells. The endocrine cells are arranged mainly in groups as islets of Langerhans and secrete different polypeptides delivered to other parts of the body via the vasculature. In contrast, the secretions of acinar exocrine cells are carried away through the ductal system. To reconstitute the major functional processes in pancreas, we utilized these unique differential secretory channels to deliver two major pancreatic cell types–endocrine (β-cells) and exocrine (acinar cells)–in close proximity to their respective native niches. Our result showed that both cells types retained their functionality and distinct location upon recellularization (Fig. 8B, C).
Reestablishment of a critical ECM-β-cells interaction has been shown to improve β-cells survival and functions [15, 38, 40, 50–52]. In this study, we evaluated the effect of substrate on β-cell function. Insulin gene expression of MIN-6 cultured on Matrigel was found to be significantly higher than coated single purified proteins. This finding indicates that β-cells are responsive to ECM substrate differences. Hence it was expected that native pancreatic ECM scaffold with preserved native ECM milieu will also likely enhance insulin gene expression. Consistent to our hypothesis, ins1 and ins2 expression of MIN-6 cultured on decellularized pancreas 3D scaffold was higher than single purified molecules (P<0.05, Fig. 9) and comparable to Matrigel (no statistical difference). However, 2D substrates of pancreatic ECM only showed similar level of insulin gene expression with single purified molecules. This is likely due to the reason that pancreatic cell niche in vivo is a 3D environment and previous studies have shown that cell-matrix interaction in 3D matrices are more favorable from those on 2D substrates [67, 68]. Our result was supportive of this previous finding with the 3D pancreatic scaffold demonstrating better support of β-cell function. The high insulin gene expression observed when β-cells are cultured on Matrigel suggests the benefits of synergistic effects from multi-faceted ECM proteins. Similarly, decellularized pancreas scaffold also possesses multiplex ECM composition and the effects are proving to be similar when compared to Matrigel. Even though Matrigel is widely used in various tissue engineering studies, it is limited in its translational potential due to its tumor cell origin. In contrast, decellularized xenogenic sources of ECM scaffolds are typically regulated by the FDA as medical device and currently in wide clinical use [69]. Hence, the development of a bioscaffold using decellularized pancreatic ECM will provide a better clinical translation opportunity for pancreatic tissue engineering and regenerative medicine applications.
5. CONCLUSIONS
This report presents the first step toward successful decellularization and 3D reconstruction of whole pancreas. Thorough characterization revealed that the decellularized pancreas retained its native 3D architecture, vasculature and ductal channels along with important ECM compositions. The preserved pancreatic ECM scaffold was cytocompatible, supportive of representative pancreatic cell types, and enhanced insulin function when seeded with β-cells. Such native organ derived scaffold is likely to have a great impact in pancreatic tissue engineering, by providing a niche microenvironment for pancreatic cell types and even stem/progenitor cells.
Supplementary Material
Figure S1. Decellularization of bovine pancreas. Panel images show progressive decellularization of bovine pancreas over the course of 65 hours.
Acknowledgments
We acknowledge our funding source for the generous support: NIH New Innovator Award DP2 116520. We thank Jonathon Franks from Center of Biological Imaging University of Pittsburgh for the technical assistance with SEM and TEM, and Timothy Maul for his assistance with bovine pancreas procurement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Research and Clinical Practice. 2010;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Bloomgarden ZT. Diabetes complications. Diabetes Care. 2004;27:1506–14. doi: 10.2337/diacare.27.6.1506. [DOI] [PubMed] [Google Scholar]
- 3.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–6. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 4.Griffith LG, Naughton G. Tissue engineering--current challenges and expanding opportunities. Science. 2002;295:1009–14. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 5.Shin H, Jo S, Mikos AG. Biomimetic materials for tissue engineering. Biomaterials. 2003;24:4353–64. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 6.Stendahl JC, Kaufman DB, Stupp SI. Extracellular matrix in pancreatic islets: relevance to scaffold design and transplantation. Cell Transplant. 2009;18:1–12. doi: 10.3727/096368909788237195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaithilingam V, Tuch BE. Islet transplantation and encapsulation: an update on recent developments. Rev Diabet Stud. 2011;8:51–67. doi: 10.1900/RDS.2011.8.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown BN, Barnes CA, Kasick RT, Michel R, Gilbert TW, Beer-Stolz D, et al. Surface characterization of extracellular matrix scaffolds. Biomaterials. 2010;31:428–37. doi: 10.1016/j.biomaterials.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sellaro TL, Ravindra AK, Stolz DB, Badylak SF. Maintenance of hepatic sinusoidal endothelial cell phenotype in vitro using organ-specific extracellular matrix scaffolds. Tissue Eng. 2007;13:2301–10. doi: 10.1089/ten.2006.0437. [DOI] [PubMed] [Google Scholar]
- 10.Cheng JY, Raghunath M, Whitelock J, Poole-Warren L. Matrix components and scaffolds for sustained islet function. Tissue Eng Part B Rev. 2011;17:235–47. doi: 10.1089/ten.TEB.2011.0004. [DOI] [PubMed] [Google Scholar]
- 11.Candiello J, Cole GJ, Halfter W. Age-dependent changes in the structure, composition and biophysical properties of a human basement membrane. Matrix Biology. 2010;29:402–10. doi: 10.1016/j.matbio.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Brafman DA, Shah KD, Fellner T, Chien S, Willert K. Defining long-term maintenance conditions of human embryonic stem cells with arrayed cellular microenvironment technology. Stem Cells Dev. 2009;18:1141–54. doi: 10.1089/scd.2008.0410. [DOI] [PubMed] [Google Scholar]
- 13.Flaim CJ, Teng D, Chien S, Bhatia SN. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 2008;17:29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 14.Weber LM, Anseth KS. Hydrogel encapsulation environments functionalized with extracellular matrix interactions increase islet insulin secretion. Matrix Biol. 2008;27:667–73. doi: 10.1016/j.matbio.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber LM, Hayda KN, Anseth KS. Cell-matrix interactions improve beta-cell survival and insulin secretion in three-dimensional culture. Tissue Eng Part A. 2008;14:1959–68. doi: 10.1089/ten.tea.2007.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 17.Wainwright JM, Czajka CA, Patel UB, Freytes DO, Tobita K, Gilbert TW, et al. Preparation of cardiac extracellular matrix from an intact porcine heart. Tissue Eng Part C Methods. 2010;16:525–32. doi: 10.1089/ten.tec.2009.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remlinger NT, Wearden PD, Gilbert TW. Procedure for decellularization of porcine heart by retrograde coronary perfusion. J Vis Exp. 2012:e50059. doi: 10.3791/50059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SL, Narayanan K, Gao S, Wan AC. Lineage restricted progenitors for the repopulation of decellularized heart. Biomaterials. 2011;32:7571–80. doi: 10.1016/j.biomaterials.2011.06.065. [DOI] [PubMed] [Google Scholar]
- 20.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–20. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soto-Gutierrez A, Zhang L, Medberry C, Fukumitsu K, Faulk D, Jiang H, et al. A whole-organ regenerative medicine approach for liver replacement. Tissue Eng Part C Methods. 2011;17:677–86. doi: 10.1089/ten.tec.2010.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shupe T, Williams M, Brown A, Willenberg B, Petersen BE. Method for the decellularization of intact rat liver. Organogenesis. 2010;6:134–6. doi: 10.4161/org.6.2.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604–17. doi: 10.1002/hep.24067. [DOI] [PubMed] [Google Scholar]
- 24.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–33. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 25.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–41. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AP, England KA, Matson AM, Blazar BR, Panoskaltsis-Mortari A. Development of a decellularized lung bioreactor system for bioengineering the lung: the matrix reloaded. Tissue Eng Part A. 2010;16:2581–91. doi: 10.1089/ten.tea.2009.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daly AB, Wallis JM, Borg ZD, Bonvillain RW, Deng B, Ballif BA, et al. Initial binding and recellularization of decellularized mouse lung scaffolds with bone marrow-derived mesenchymal stromal cells. Tissue Eng Part A. 2012;18:1–16. doi: 10.1089/ten.tea.2011.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallis JM, Borg ZD, Daly AB, Deng B, Ballif BA, Allen GB, et al. Comparative assessment of detergent-based protocols for mouse lung de-cellularization and re-cellularization. Tissue Eng Part C Methods. 2012;18:420–32. doi: 10.1089/ten.tec.2011.0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonvillain RW, Danchuk S, Sullivan DE, Betancourt AM, Semon JA, Eagle ME, et al. A nonhuman primate model of lung regeneration: detergent-mediated decellularization and initial in vitro recellularization with mesenchymal stem cells. Tissue Eng Part A. 2012;18:2437–52. doi: 10.1089/ten.tea.2011.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan DC, Mirmalek-Sani SH, Deegan DB, Baptista PM, Aboushwareb T, Atala A, et al. Decellularization methods of porcine kidneys for whole organ engineering using a high-throughput system. Biomaterials. 2012;33:7756–64. doi: 10.1016/j.biomaterials.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Ross EA, Williams MJ, Hamazaki T, Terada N, Clapp WL, Adin C, et al. Embryonic stem cells proliferate and differentiate when seeded into kidney scaffolds. J Am Soc Nephrol. 2009;20:2338–47. doi: 10.1681/ASN.2008111196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song JJ, Guyette JP, Gilpin SE, Gonzalez G, Vacanti JP, Ott HC. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19:646–51. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conrad C, Schuetz C, Clippinger B, Vacanti JP, Markmann JF, Ott HC. Bio-engineered endocrine pancreas based on decellularized pancreatic matrix and mesenchymal stem cell/islet cell coculture. Journal of the American College of Surgeons. 2010;211:S62. [Google Scholar]
- 34.Goh S-K, Bertera S, Banerjee I. AIChE. Minneapolis Convention Center; Minnesota: 2011. Perfusion-decellularization of pancreatic matrix - a scaffold for bio-engineered pancreas. [Google Scholar]
- 35.Goh SK, Bertera S, Banerjee I. Perfusion-decellularization of pancreas as a scaffold for the differentiation of human embryonic stem cells into insulin-producing cells. 10th Annual Scientific Meeting of Int Soc Stem Cell Res; Yokohama, Japan. 2012. p. 67. [Google Scholar]
- 36.Mirmalek-Sani S-H, Orlando G, McQuilling JP, Pareta R, Mack DL, Salvatori M, et al. Porcine pancreas extracellular matrix as a platform for endocrine pancreas bioengineering. Biomaterials. 2013;34:5488–95. doi: 10.1016/j.biomaterials.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thivolet CH, Chatelain P, Nicoloso H, Durand A, Bertrand J. Morphological and functional effects of extracellular matrix on pancreatic islet cell cultures. Exp Cell Res. 1985;159:313–22. doi: 10.1016/s0014-4827(85)80005-7. [DOI] [PubMed] [Google Scholar]
- 38.Nikolova G, Jabs N, Konstantinova I, Domogatskaya A, Tryggvason K, Sorokin L, et al. The vascular basement membrane: a niche for insulin gene expression and beta cell proliferation. Dev Cell. 2006;10:397–405. doi: 10.1016/j.devcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Tian XH, Xue WJ, Ding XM, Pang XL, Teng Y, Tian PX, et al. Small intestinal submucosa improves islet survival and function during in vitro culture. World J Gastroenterol. 2005;11:7378–83. doi: 10.3748/wjg.v11.i46.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods EJ, Walsh CM, Sidner RA, Zieger MAJ, Lakey JRT, Ricordi C, et al. Improved in vitro function of islets using small intestinal submucosa. Transplantation Proceedings. 2004;36:1175–7. doi: 10.1016/j.transproceed.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 41.De Carlo E, Baiguera S, Conconi MT, Vigolo S, Grandi C, Lora S, et al. Pancreatic acellular matrix supports islet survival and function in a synthetic tubular device: in vitro and in vivo studies. Int J Mol Med. 2010;25:195–202. [PubMed] [Google Scholar]
- 42.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 43.Goh S-K, Olsen P, Banerjee I. Extracellular matrix aggregates from differentiating embryoid bodies as a scaffold to support ESC proliferation and differentiation. PLoS One. 2013;8:e61856. doi: 10.1371/journal.pone.0061856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balasubramani M, Schreiber EM, Candiello J, Balasubramani GK, Kurtz J, Halfter W. Molecular interactions in the retinal basement membrane system: a proteomic approach. Matrix Biol. 2010;29:471–83. doi: 10.1016/j.matbio.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Candiello J, Balasubramani M, Schreiber EM, Cole GJ, Mayer U, Halfter W, et al. Biomechanical properties of native basement membranes. FEBS J. 2007;274:2897–908. doi: 10.1111/j.1742-4658.2007.05823.x. [DOI] [PubMed] [Google Scholar]
- 46.Meyer T, Chodnewska I, Czub S, Hamelmann W, Beutner U, Otto C, et al. Extracellular matrix proteins in the porcine pancreas: a structural analysis for directed pancreatic islet isolation. Transplant Proc. 1998;30:354. doi: 10.1016/s0041-1345(97)01302-x. [DOI] [PubMed] [Google Scholar]
- 47.Meyer T, Czub S, Chodnewska I, Beutner U, Hamelmann W, Klock G, et al. Expression pattern of extracellular matrix proteins in the pancreas of various domestic pig breeds, the goettingen minipig and the wild boar. Ann Transplant. 1997;2:17–26. [PubMed] [Google Scholar]
- 48.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–43. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keane TJ, Londono R, Turner NJ, Badylak SF. Consequences of ineffective decellularization of biologic scaffolds on the host response. Biomaterials. 2012;33:1771–81. doi: 10.1016/j.biomaterials.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 50.Schuppin GT, Bonner-Weir S, Montana E, Kaiser N, Weir GC. Replication of adult pancreatic-beta cells cultured on bovine corneal endothelial cell extracellular matrix. In Vitro Cell Dev Biol Anim. 1993;29A:339–44. doi: 10.1007/BF02633963. [DOI] [PubMed] [Google Scholar]
- 51.Daoud J, Petropavlovskaia M, Rosenberg L, Tabrizian M. The effect of extracellular matrix components on the preservation of human islet function in vitro. Biomaterials. 2010;31:1676–82. doi: 10.1016/j.biomaterials.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 52.Wang RN, Rosenberg L. Maintenance of beta-cell function and survival following islet isolation requires re-establishment of the islet-matrix relationship. J Endocrinol. 1999;163:181–90. doi: 10.1677/joe.0.1630181. [DOI] [PubMed] [Google Scholar]
- 53.Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortiella J, Niles J, Cantu A, Brettler A, Pham A, Vargas G, et al. Influence of acellular natural lung matrix on murine embryonic stem cell differentiation and tissue formation. Tissue Eng Part A. 2010;16:2565–80. doi: 10.1089/ten.tea.2009.0730. [DOI] [PubMed] [Google Scholar]
- 55.Shamis Y, Hasson E, Soroker A, Bassat E, Shimoni Y, Ziv T, et al. Organ-specific scaffolds for in vitro expansion, differentiation, and organization of primary lung cells. Tissue Eng Part C Methods. 2011;17:861–70. doi: 10.1089/ten.tec.2010.0717. [DOI] [PubMed] [Google Scholar]
- 56.Nakayama KH, Batchelder CA, Lee CI, Tarantal AF. Decellularized rhesus monkey kidney as a three-dimensional scaffold for renal tissue engineering. Tissue Eng Part A. 2010;16:2207–16. doi: 10.1089/ten.tea.2009.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mauck RL, Baker BM, Nerurkar NL, Burdick JA, Li WJ, Tuan RS, et al. Engineering on the straight and narrow: the mechanics of nanofibrous assemblies for fiber-reinforced tissue regeneration. Tissue Eng Part B Rev. 2009;15:171–93. doi: 10.1089/ten.teb.2008.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soto-Gutierrez A, Wertheim JA, Ott HC, Gilbert TW. Perspectives on whole-organ assembly: moving toward transplantation on demand. J Clin Invest. 2012;122:3817–23. doi: 10.1172/JCI61974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song JJ, Ott HC. Organ engineering based on decellularized matrix scaffolds. Trends Mol Med. 2011;17:424–32. doi: 10.1016/j.molmed.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–30. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 61.Brown BN, Valentin JE, Stewart-Akers AM, McCabe GP, Badylak SF. Macrophage phenotype and remodeling outcomes in response to biologic scaffolds with and without a cellular component. Biomaterials. 2009;30:1482–91. doi: 10.1016/j.biomaterials.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams C, Liao J, Joyce EM, Wang B, Leach JB, Sacks MS, et al. Altered structural and mechanical properties in decellularized rabbit carotid arteries. Acta Biomater. 2009;5:993–1005. doi: 10.1016/j.actbio.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gratzer PF, Harrison RD, Woods T. Matrix alteration and not residual sodium dodecyl sulfate cytotoxicity affects the cellular repopulation of a decellularized matrix. Tissue Eng. 2006;12:2975–83. doi: 10.1089/ten.2006.12.2975. [DOI] [PubMed] [Google Scholar]
- 64.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 65.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Cui CB, Yamauchi M, Miguez P, Roach M, Malavarca R, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology. 2011;53:293–305. doi: 10.1002/hep.24012. [DOI] [PubMed] [Google Scholar]
- 67.Chun T-H, Hotary KB, Sabeh F, Saltiel AR, Allen ED, Weiss SJ. A pericellular collagenase directs the 3-dimensional development of white adipose tissue. Cell. 2006;125:577–91. doi: 10.1016/j.cell.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 68.Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 69.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: structure and function. Acta Biomaterialia. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Decellularization of bovine pancreas. Panel images show progressive decellularization of bovine pancreas over the course of 65 hours.