Abstract
The role and even the existence of myocyte proliferation in the adult heart remain controversial. Documentation of cell cycle regulators, DNA synthesis, and mitotic images has not modified the view that myocardial growth can only occur from hypertrophy of an irreplaceable population of differentiated myocytes. To improve understanding the biology of the heart and obtain supportive evidence of myocyte replication, three indices of cell proliferation were analyzed in dogs affected by a progressive deterioration of cardiac performance and dilated cardiomyopathy. The magnitude of cycling myocytes was evaluated by the expression of Ki67 in nuclei. Ki67 labeling of left ventricular myocytes increased 5-fold, 12-fold, and 17-fold with the onset of moderate and severe ventricular dysfunction and overt failure, respectively. Telomerase activity in vivo is present only in multiplying cells; this enzyme increased 2.4-fold and 3.1-fold in the decompensated heart, preserving telomeric length in myocytes. The contribution of cycling myocytes to telomerase activity was determined by the colocalization of Ki67 and telomerase in myocyte nuclei. More than 50% of Ki67-positive cells expressed telomerase in the overloaded myocardium, suggesting that these myocytes were the morphological counterpart of the biochemical assay of enzyme activity. Moreover, we report that 20–30% of canine myocytes were telomerase competent, and this value was not changed by cardiac failure. In conclusion, the enhanced expression of Ki67 and telomerase activity, in combination with Ki67-telomerase labeling of myocyte nuclei, support the notion that myocyte proliferation contributes to cardiac hypertrophy of the diseased heart.
The question of whether ventricular myocytes are terminally differentiated cells or retain the capacity to multiply in the adult heart remains controversial (1, 2). Results favoring the replicative potential of this cell population have been challenged, and the heart and the brain have been considered as the two typical organs unable to undergo physiologic repair. The discovery of neuronal regeneration in the human brain (3) and myocyte cytokinesis in the human heart (4, 5) has not changed the view of most investigators and clinicians in the cardiovascular field. To improve understanding the biology of the heart and obtain supportive evidence of myocyte division, three indices of cell proliferation were analyzed in dogs affected by a disease state, mimicking dilated cardiomyopathy in patients (6–8). These markers consisted of telomerase activity, Ki67 labeling, and identification of cycling telomerase competent cells. Moreover, the length of telomeres in myocyte nuclei was measured to establish whether telomerase activity preserved telomeric length in growing cells.
Chromosomes are protected by caps called telomeres, which are maintained by a specialized DNA polymerase. Telomeres constitute the physical ends of chromosomes, and telomerase is the enzyme capable of keeping intact the length of telomeres (9). Although telomerase-independent mechanisms of telomere elongation have been identified in vitro (10), their actual role in vivo has not been recognized. Conversely, the synthesis of telomeric repeats by telomerase allows complete replication of DNA in vivo (9,11). Telomerase-competent cells show telomerase activity only when they have entered the cell cycle; this enzyme prevents erosion of chromosomes postponing growth arrest (9, 11).
Proliferating cell nuclear antigen (PCNA), thymidine, and BrdUrd have been used for the detection of cycling myocytes (1, 2). However, PCNA cannot discriminate between cells traversing the cell cycle or undergoing DNA repair (1, 5). Thymidine and BrdUrd detect cells in S phase, but whether DNA synthesis is associated with repair processes, ploidy formation, or cytokinesis cannot be assessed (1, 2). Mitotic indices are complicated by the duration of mitosis, 30 min (12), and the difficulty of identifying nuclei in prophase (1); these factors lead to the underestimation of dividing myocytes. Ki67 is expressed in nuclei in G1, S, G2, prophase, and metaphase. Quiescent cells in G0 do not express Ki67 and this protein is not implicated in DNA repair; Ki67 strictly correlates with cell proliferation (13, 14). There is not a single evidence of Ki67 labeling in nonreplicating cells. Therefore, the parameters indicated above were analyzed in myocytes of control and failing dog hearts.
Materials and Methods
Ventricular Function.
Corkscrew electrodes attached to a pacemaker were inserted into young mongrel dogs (6–8, 15). Hearts were paced at 210 beats per min for 3 weeks and at 240 for another week. Animals were killed at 1 (n = 6), 3 (n = 6), and 4 (n = 11) weeks of pacing. Eight dogs were used as controls. After collection of hemodynamic data (6–8, 15), hearts were arrested in diastole, and each left ventricle was separated in two parts, one for myocyte isolation and biochemistry and the other for formalin fixation and cytochemistry.
Myocyte Isolation.
Myocytes were dissociated as described (6–8, 16). The number of myocytes varied from 7.3 × 106 to 4.5 × 106/g of myocardium in control and paced hearts, respectively. Nonmyocytes accounted for 2–3% of the cell population (6–8, 16).
Telomeric Repeat Amplification Protocol.
Myocytes were homogenized in 3-[3- cholamidopropyl)dimethyl-ammonio]-1-propanesulfonate buffer and centrifuged at 4°C. Four and one micrograms of untreated and RNase-treated myocyte extracts were incubated with [γ32P]ATP-end-labeled telomerase substrate (TS oligonucleotide: 5′-AATCCGTCGAGCAGAGTT-3′), Taq polymerase and anchored reverse primer (5′-GCGCGC[CTTACC]3CTAACC-5′) for 30 min. Samples were exposed to 27 amplification cycles (17). PCR products were separated on 12% polyacrylamide gel. Telomerase-induced reaction generated a 6-bp ladder. The OD of the bands was measured and normalized for PCR efficiency (17).
Immunoprecipitation.
Nuclear extracts were obtained by incubations of myocytes in hypotonic and hypertonic buffers. Proteins were immunoprecipitated with human telomerase reverse transcriptase (TERT) antibody (Amgen, Toronto, ON; gift from L. Harrington). After separation on 10% SDS/PAGE, proteins were transferred onto nitrocellulose and exposed to rabbit anti-murine TERT (Ab 3, Alpha Diagnostics, San Antonio, TX). Telomerase was seen as a 120- to 125-kDa band (7, 8).
Telomerase and Ki67.
Three antibodies were used for telomerase identification in the heart: rabbit anti-human TERT (H-231, Santa Cruz Biotechnology), rabbit anti-murine TERT and rabbit anti-human TERT (NB 100–141, Novus Biologicals, Littleton, CO). Sections were incubated with each antibody and then with FITC-conjugated goat anti-rabbit IgG. Negative controls included omission of primary antibodies and the use of nonimmune rabbit serum and an irrelevant primary antibody, rabbit anti-green fluorescent protein (Molecular Probes). Myocyte cytoplasm was identified by cardiac myosin (mAb 1458, Chemicon), fibroblasts by vimentin (clone V9, Sigma), endothelial cells by factor VIII (Sigma), and smooth muscle cells by α-smooth muscle actin (clone 1A4, Sigma) antibodies. For Ki67, sections were exposed to Ki67 antibody (clone MIB-5, Immunotech, Luminy, France). Myocyte cytoplasm was stained with cardiac myosin and nuclei with propidium iodide (PI). For telomerase localization, 10,000 myocytes, fibroblasts, and endothelial cells and 500 smooth muscle cells were examined in each heart. For Ki67, 10,000 myocytes were sampled in each case (18).
Telomere Length.
This was evaluated by telomeric restriction fragment (TRF) analysis (Southern blot) and quantitative fluorescence in situ hybridization. In both cases, immortal cell lines, with known telomeric length (refs. 19 and 20; gift from Maria Blasco, Universidad Autonoma de Madrid), were used for comparison.
TRF analysis.
A total of 106 myocytes in each sample were incubated at 55°C in lysis buffer containing proteinase K. After centrifugation, supernatants were exposed to isopropanol and ethanol to precipitate genomic DNA. DNA was digested with RsaI and HinfI (21), separated on 0.8% agarose gel/1× 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3, and transferred to nylon membranes (Amersham Pharmacia). Hybridization was performed overnight at 65°C in a solution containing 32P-end-labeled (C3TA2)3 probe (19, 22, 23). Mean TRF length was determined by densitometry.
Quantitative fluorescence in situ hybridization.
Smears of myocyte nuclei were treated with 4% formaldehyde, digested with pepsin, and hybridized with FITC-labeled (C3TA2)3 peptide nucleic acid (PNA) probe (Perkin–Elmer). Slides were stained with PI. Total fluorescence of FITC-PNA probe was determined by laser Compucyte cytometer (Compucyte, Cambridge, MA). Software was used to deconvolute histograms and estimate nuclei with specific telomeric length (24, 25); 5,000 myocyte nuclei were evaluated in each ventricle. Compucyte cytometer is superior to flow cytometry; it allows the analysis of randomly sampled individual nuclei (24). In comparison with digital fluorescence microscopy, the cytometer increased sampling by 25- to 125-fold, from 40–200 to 5,000 interphase nuclei (26).
Data Analysis.
Results are mean ±SD. Significance in multiple comparisons was determined by the Bonferroni method. P < 0.05 was considered significant.
Results
Telomerase Activity.
Hemodynamic changes associated with ventricular pacing have been well characterized (6–8, 15). Similar abnormalities in function have been found here. The increase in left ventricular end-diastolic pressure and the decrease in left ventricular dP/dt at 1 week of pacing worsened with time, resulting at 4 weeks in depressed systolic performance and overt failure with pulmonary edema and ascites (data not shown).
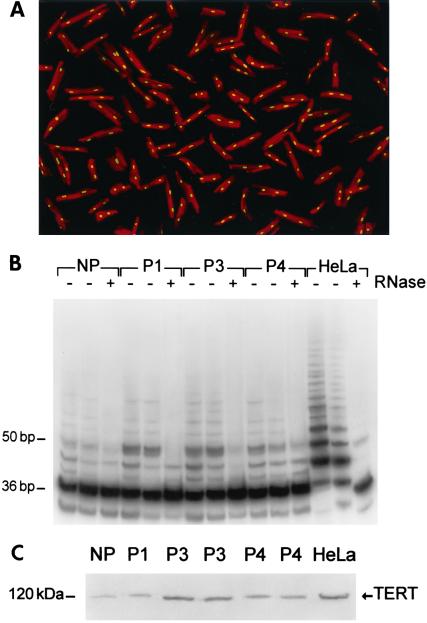
Telomerase activity was measured in isolated myocytes (Fig. 1A). Contamination from blood cells did not occur with this preparation; animals were injected with heparin and the coronary circulation was perfused extensively with buffer before collagenase digestion (6–8, 16). Residual blood cells have never been found. Formation of the 6-bp ladder was detected in myocytes from control and paced hearts at 1, 3, and 4 weeks (Fig. 1B). The OD of the bands reflected the number of TS oligonucleotides extended by telomerase. In comparison with controls (79 ± 19, n = 8), OD did not vary with 1 week (106 ± 24, n = 6), but increased 3.1-fold (248 ± 59, n = 6; P < 0.001) and 2.4-fold (186 ± 38, n = 11; P < 0.001) with 3 and 4 weeks of pacing. These results were complemented with measurements of TERT in myocyte nuclear proteins (Fig. 1C). OD data demonstrated that TERT in controls, 0.96 ± 0.59 (n = 8), remained constant after 1 week, 1.13 ± 0.48 (n = 6), and increased 6-fold (P < 0.001) and 3-fold (P < 0.001) after 3 weeks, 5.78 ± 1.4 (n = 6), and 4 weeks , 3.16 ± 1.31 (n = 11), of pacing. TERT protein decreased 45% (P < 0.001) from 3 to 4 weeks.
Figure 1.
(A) Myocytes isolated from a paced heart. Red fluorescence, α-sarcomeric actin staining of myocyte cytoplasm; yellow fluorescence, PI labeling of nuclei. Confocal microscopy, ×200. (B) Telomerase activity in myocytes from nonpaced (NP) and paced hearts for 1 (P1), 3 (P3), and 4 (P4) weeks. Products of telomerase activity start at 50 bp and display 6-bp periodicity. Myocytes treated with RNase (+) were used as negative control, and HeLa cells (3 and 1 μg) as positive control. (C) TERT in myocyte nuclear lysates (400 μg) and HeLa cells (80 μg).
Telomerase and Ki67 Localization.
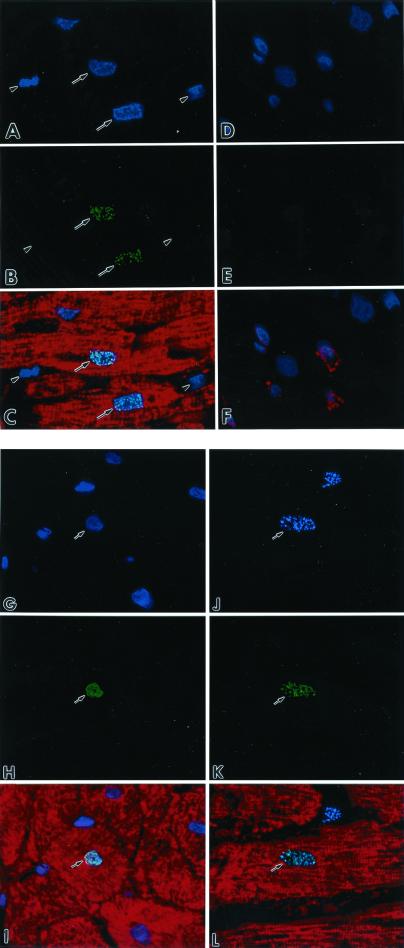
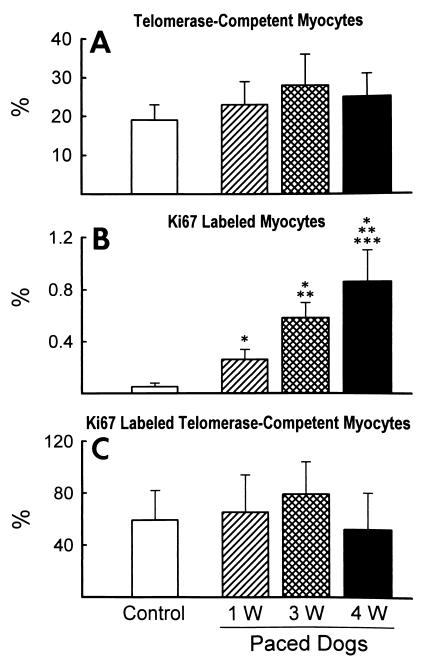
The three antibodies used for the identification of telomerase-competent cells in the myocardium showed no differences in their nuclear pattern and magnitude of labeling. Myocytes were the only cells expressing telomerase (Fig. 2 A–C). Telomerase was not seen in endothelial cells, smooth muscle cells, and fibroblasts (Fig. 2 D–F). The localization of Ki67 (Fig. 2 G–I) and the presence of Ki67 and telomerase together (Fig. 2 J–L) in myocyte nuclei also were observed. The percentage of telomerase-positive myocyte nuclei remained relatively constant with pacing (Fig. 3A). Conversely, pacing increased the number of Ki67-positive myocyte nuclei by 5-fold (P < 0.05) at 1 week, 12-fold (P < 0.001) at 3 weeks, and 17-fold (P < 0.001) at 4 weeks (Fig. 3B). The number of cycling myocytes at 4 weeks was 1.5-fold (P < 0.01) and 3.3-fold (P < 0.001) higher than at 3 weeks and 1 week, respectively.
Figure 2.
(A and D) Nuclei by the blue fluorescence of PI. (B) Green fluorescence telomerase staining (arrows) of nuclei. (E) The absence of telomerase labeling. Red fluorescence cardiac myosin staining of myocyte cytoplasm (C) and factor VIII labeling of endothelial cells (F). Bright fluorescence in C corresponds to the combination of PI and telomerase staining of nuclei. Arrowheads, interstitial nuclei negative for telomerase. (G) Nuclei by the blue fluorescence of PI. (H) Green fluorescence Ki67 staining of a nucleus. (I) Myocyte cytoplasm by the red fluorescence of cardiac myosin. Bright fluorescence (I) corresponds to the combination of PI and Ki67 localization in a myocyte nucleus. (J) Blue fluorescence Ki67 labeling of a nucleus. (K) Green fluorescence telomerase staining in the same nucleus. (L) The myocyte cytoplasm by the red fluorescence of cardiac myosin. Bright fluorescence (L) corresponds to the colocalization of Ki67 and telomerase in the myocyte nucleus. Confocal microscopy, ×1,000.
Figure 3.
Effects of pacing on telomerase-competent myocytes (A), Ki67-labeled myocytes (B), and Ki67-labeled telomerase-competent myocytes (C). Results are mean ± SD. *, **, and ***, P < 0.05 vs. control dogs (n = 8), dogs paced for 1 week (1w; n = 6) and for 3 weeks (3w, n = 6), respectively. Dogs paced for 4 weeks: 4w, n = 8.
The in vivo expression of Ki67 was assessed in myocardial sections; this approach did not allow the analysis of the distribution of Ki67 in the nuclear compartments. The centromeric, telomeric, or nucleolar localization of this protein is more apparent in highly proliferating cells in vitro (14). Quantitatively, Ki67 indicated that nearly 1% of myocytes reentered the cell cycle and were proliferating in the failing heart at 4 weeks. On this basis, the fraction of cycling telomerase-competent myocytes was determined. The colocalization of Ki67 and telomerase represented the morphological counterpart of the biochemically measured enzyme activity. The percentage of myocyte nuclei labeled by Ki67 that also had telomerase exceeded 50% in all cases (Fig. 3C). Although the values among groups were not significant, their relative differences were similar to those in telomerase activity shown in Fig. 1B.
Length of Telomeres.
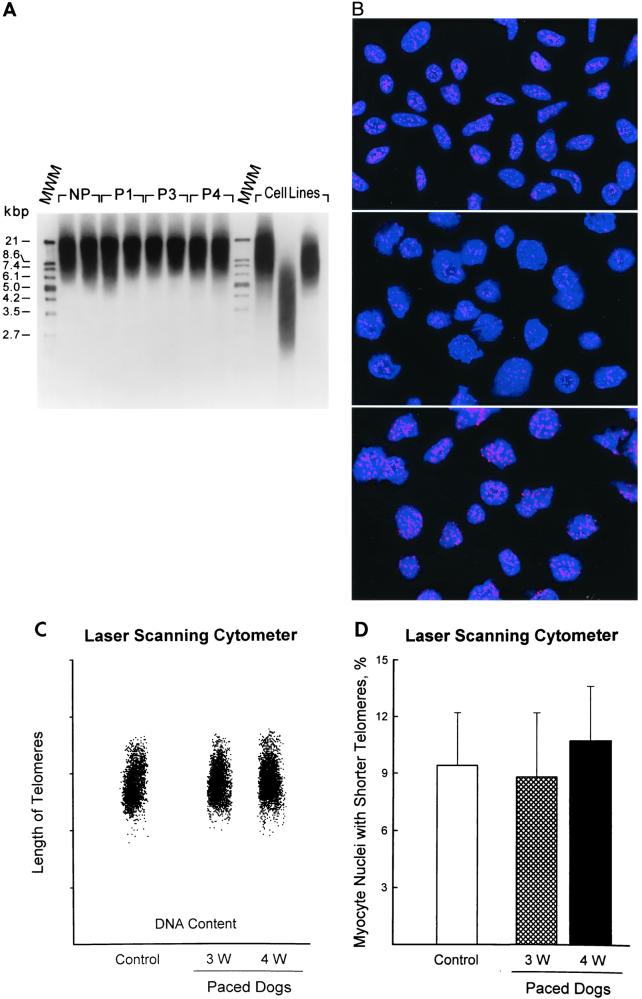
Digestion with restriction enzymes generated TRFs that were detected by Southern blot and a telomere-specific probe. Mean TRF length did not change with pacing (Fig. 4A). OD values were as follows: controls = 12.9 ± 3 (n = 8); pacing for 1 week = 12.6 ± 1.9 (n = 6), 3 weeks = 13.6 ± 2.2 (n = 6), and 4 weeks = 13.1 ± 2.9 (n = 8). The size distribution of TRFs was also comparable in the four groups of dogs: the smear produced by TRFs varied from 6 to 20 kbp in all conditions. A substantial portion of TRF is composed of subtelomeric DNA (non-TAAGGG) (27): the distance between restriction enzyme sites in subtelomeric DNA and the actual telomeric repeats averages 3 kbp (27, 28). However, mean TRF size is proportional to telomeric length evaluated by quantitative fluorescence in situ hybridization, with a high correlation coefficient (28). Therefore, myocyte nuclei were stained by in situ hybridization with a fluorescent PNA probe (Fig. 4B). Total nuclear DNA was assessed by PI and distinguished from the amount of fluorescence emitted by the PNA probe (24). The latter corresponded to the total telomeric length present in each nucleus (25). These parameters were used to obtain a bivariate distribution of telomeric length versus DNA in each nucleus (24). The length of telomeres was rather uniform in myocyte nuclei from control and paced hearts for 3 and 4 weeks (Fig. 4C). Telomeric length frequency histograms were analyzed and the fraction of nuclei with shorter telomeres was calculated (Fig. 4D). The percentage of myocytes with shorter telomeres did not change with pacing. The intensity of the fluorescent signal obtained in myocytes was compared with that of lymphoma cells (Fig. 4B) of known telomeric length, 7 and 48 kbp (20). Thus, telomere lengths in myocytes were 13.6 ± 1.1 in nonpaced and 13.2 ± 1.5, and 13.8 ± 1.2 kbp in paced dogs at 3 and 4 weeks, respectively.
Figure 4.
(A) TRF length in myocytes from nonpaced (NP), P1, P3, and P4 dogs. Immortal cell lines used for comparison have, from left to right, a mean TRF of 10.2, 3.9, and 7 kbp, respectively. MWM, molecular weight marker. (B) Myocyte nuclei (Top) and lymphoma cells with short (Middle) and long (Bottom) telomeres were stained by in situ hybridization with a PNA probe specific for telomeric sequence. Nuclei are illustrated by the blue fluorescence of PI, and the red fluorescent dots correspond to individual telomeres. Confocal microscopy, ×1,000. (C) Bivariate distribution of DNA content and telomere length in myocyte nuclei from the left ventricle of nonpaced and paced dog hearts. (D) Percentages of myocyte nuclei with shorter telomeres. Results are mean ± SD; n = 6 in each group of dogs.
Discussion
The current findings indicate that ventricular dysfunction and failure were coupled with activation of myocyte proliferation and preservation of telomeric length. Increases in telomerase level and activity positively interfered with telomeric shortening during myocyte division. The transition from moderate to severe cardiac decompensation was characterized by a progressive increase in the number of replicating myocytes, raising the possibility that myocyte regeneration and telomerase activity played an important role in repairing dying myocardium (6, 7). A subpopulation of myocytes expressed telomerase, suggesting that they had the potential of reentering the cell cycle and undergoing multiple mitotic divisions. This contention is supported by the large fraction of Ki67-positive cycling myocytes, which were telomerase competent. The nuclear expression of Ki67 was indicative of cell division instead of nuclear division. Cycling myocytes in the paced heart do not result in an increase in the number of nuclei per cell. The proportion of mononucleated and binucleated myocytes is not altered (6, 8); when binucleated myocytes replicate both nuclei divide, giving rise to two new cells (8). Additionally, cyclins and cyclin-dependent kinases, BrdUrd labeling, and mitosis have been demonstrated in this animal model (8). Mitotic indices have been measured in tissue sections and isolated myocytes. Telomerase is a property of dividing cells. In germ cells, this protein ensures telomere length and unlimited cell multiplication (9). Hemopoietic, epithelial, and basal stem cells are telomerase competent (9, 23). They are normally quiescent, dividing upon stimulation; telomerase activity is present in the various phases of the cell cycle, but is repressed in G0 (29). Importantly, progenitor and rapidly replicating amplifying cells have high levels of telomerase activity (29). Recently, telomerase has been found in neurons and myocytes that belong to organs with low turnover rates (17, 30). The identification of telomerase in more than one-fifth of myocytes of the canine myocardium, combined with their ability to multiply and express enzyme activity, suggests that proliferation of a subgroup of myocytes contributed significantly to cardiac growth. Myocytes behave as neuronal cells in the central nervous system (3). The biology of the different organs in the organism may follow a common pattern of growth. The preservation of telomeric length found here in young dogs with rapidly developing cardiac failure differs from myocardial aging. Regeneration of myocytes with age in rodents leads to progressive telomeric shortening (24).
The cell cycle regulation of telomerase is only partially understood. The unexpected behavior of telomerase in cells exposed to DNA damaging agents, cell cycle blockers, and protein kinase inhibitors has complicated the recognition of the role of this enzyme in replicating cells (31–33). Chemical interference with tubulin assembly and S-phase progression does not alter telomerase activity of cancer cells in vitro (31, 33). Similarly, proliferating T lymphocytes possess high levels of telomerase that are not affected by hydroxyurea (32). However, these findings are restricted to cells subjected to multiple interventions that cannot be compared with unmanipulated in vitro conditions or in vivo states. In the absence of telomerase, normal cells in culture have a finite lifespan (22). Conversely, forced expression of telomerase restores cell growth beyond the Hayflick limit, increasing maximum lifespan (34). The inactivation of cell cycle modulators, such as Rb and p16, is required to achieve cell immortalization (34). Together this information strongly suggests the difficulty of extrapolating in vitro results to in vivo systems.
Telomerase activity increased during the onset and progression of ventricular dysfunction. Similarly, the extent of myocyte proliferation paralleled the duration and severity of the overload. Myocyte apoptosis and necrosis are both involved in pacing-induced dilated cardiomyopathy (6, 7). Late in the disease, myocyte death exceeds regeneration resulting in a reduction in the total number of left ventricular myocytes (6). The imbalance between cell death and cell multiplication may be critical for the development of terminal failure.
By using immunocytochemistry and confocal microscopy, we documented that the nuclear expression of telomerase was restricted to myocytes. These findings are consistent with previous observations from our laboratory (17) but are in contrast with data in the developing heart (35). In the latter report, ventricular tissue was used and the postnatal disappearance of active telomerase was considered the hallmark of myocytes permanently withdrawing from the cell cycle. However, fibroblasts do not possess telomerase and the same apply to smooth muscle cells and endothelial cells (36–39). The analysis performed here confirmed the absence of telomerase in all cardiac nonmyocyte populations. Because nearly 70% of nuclei in the heart belong to interstitial cells, contamination from these cells lacking telomerase is the most likely cause of the results in the whole myocardium (35).
In conclusion, our observations strongly suggest that the transition from ventricular dysfunction to severe cardiac decompensation is characterized by enhanced expression of Ki67 and telomerase activity in myocytes. These findings are consistent with the notion that cell proliferation expand the functioning myocardium in an attempt to delay the onset of terminal failure. Cell regeneration counteracts, at least in part, myocyte loss, interfering with the effects of cell death mechanisms on the remodeling process of the pathologic heart.
Acknowledgments
This work was supported by National Institutes of Health Grants HL-38132, HL-39902, HL-43023, AG-15756, HL-66923, AG-17042, and HL-65577.
Abbreviations
- TERT
telomerase reverse transcriptase
- TRF
telomeric restriction fragment
- PNA
peptide nucleic acid
- PI
propidium iodide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Anversa P, Kajstura J. Circ Res. 1998;83:1–14. doi: 10.1161/01.res.83.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Soonpaa M H, Field L J. Circ Res. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Horner P J, Gage F H. Nature (London) 2000;407:963–969. doi: 10.1038/35039559. [DOI] [PubMed] [Google Scholar]
- 4.Kajstura J, Leri A, Finato N, Di Loreto C, Beltrami C A, Anversa P. Proc Natl Acad Sci USA. 1998;95:8801–8805. doi: 10.1073/pnas.95.15.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltrami A P, Urbanek K, Kajstura J, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami C A, Anversa P. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 6.Kajstura J, Zhang X, Liu Y, Szoke E, Cheng W, Olivetti G, Hintze T H, Anversa P. Circulation. 1995;92:2306–2317. doi: 10.1161/01.cir.92.8.2306. [DOI] [PubMed] [Google Scholar]
- 7.Leri A, Liu Y, Malhotra A, Li Q, Stiegler P, Claudio P P, Giordano A, Kajstura J, Anversa P. Circulation. 1998;97:194–203. doi: 10.1161/01.cir.97.2.194. [DOI] [PubMed] [Google Scholar]
- 8.Setoguchi M, Leri A, Wang S, Liu Y, De Luca A, Giordano A, Hintze T H, Kajstura J, Anversa P. Lab Invest. 1999;79:1545–1558. [PubMed] [Google Scholar]
- 9.Greider C W. Proc Natl Acad Sci USA. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Ijpma A, Greider C W. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Rivera L, Herrera E, Albar J P, Blasco M A. Proc Natl Acad Sci USA. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda A R, Vernos I. Cell. 2000;102:425–435. doi: 10.1016/s0092-8674(00)00048-9. [DOI] [PubMed] [Google Scholar]
- 13.Scholzen T, Gerdes J. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.MacCallum D E, Hall P A. J Pathol. 2000;190:537–544. doi: 10.1002/(SICI)1096-9896(200004)190:5<537::AID-PATH577>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 15.Hintze T H, Wang J, Seyedi N, Wolin M S. In: Flow-Dependent Regulation in Cardiovascular Function. Bevan J A, Kaley G, Rubanyi G M, editors. New York: Oxford Univ. Press; 1995. pp. 320–337. [Google Scholar]
- 16.Barlucchi L, Leri A, Dostal D E, Fiordaliso F, Tada H, Hintze T H, Kajstura J, Nadal-Ginard B, Anversa P. Circ Res. 2001;88:298–304. doi: 10.1161/01.res.88.3.298. [DOI] [PubMed] [Google Scholar]
- 17.Leri A, Malhotra A, Liew C-C, Kajstura J, Anversa P. J Mol Cell Cardiol. 2000;32:385–390. doi: 10.1006/jmcc.1999.1084. [DOI] [PubMed] [Google Scholar]
- 18.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson S M, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine D M, et al. Nature (London) 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 19.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 20.McIlrath J, Bouffler S D, Samper E, Cuthbert A, Wojcik A, Szumiel I, Bryant P E, Riches A C, Thompson A, Blasco M A, et al. Cancer Res. 2001;61:912–915. [PubMed] [Google Scholar]
- 21.Reimann N, Rogalla P, Kazmierczak B, Bonk U, Nolte I, Grzonka T, Barnitzke S, Bullerdiek J. Cytogenet Cell Genet. 1994;67:81–85. doi: 10.1159/000133804. [DOI] [PubMed] [Google Scholar]
- 22.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaziri H, Dragowska W, Allsopp R C, Thomas T E, Harley C B, Landsorp P M. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajstura J, Pertoldi B, Leri A, Beltrami C A, Deptala A, Darzynkiewicz Z, Anversa P. Am J Pathol. 2000;156:813–819. doi: 10.1016/S0002-9440(10)64949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePauw E S D, Verwoerd N P, Duinkerken N, Willemze R, Raap A K, Fibbe W E, Tanke H J. Cytometry. 1998;32:163–169. doi: 10.1002/(sici)1097-0320(19980701)32:3<163::aid-cyto1>3.3.co;2-v. [DOI] [PubMed] [Google Scholar]
- 26.Poon S S, Martens U M, Ward R K, Lansdorp P M. Cytometry. 1999;36:267–278. doi: 10.1002/(sici)1097-0320(19990801)36:4<267::aid-cyto1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 27.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp P M. Nat Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 28.Hultdin M, Gronlund E, Norrback K-F, Eriksson-Lindstrom E, Just T, Roos G. Nucleic Acids Res. 1998;26:3651–3656. doi: 10.1093/nar/26.16.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yui J, Chiu C-P, Lansdorp P M. Blood. 1998;91:3255–3262. [PubMed] [Google Scholar]
- 30.Mattson M, Klapper W. J Neurosci Res. 2001;63:1–9. doi: 10.1002/1097-4547(20010101)63:1<1::AID-JNR1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Kumar R, Mandal M, Sharma N, Sharma H W, Dhingra U, Sokoloski J A, Hsiao R, Narayanan R. Proc Natl Acad Sci USA. 1996;93:6091–6095. doi: 10.1073/pnas.93.12.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchkovich K J, Greider C W. Mol Biol Cell. 1996;7:1443–1454. doi: 10.1091/mbc.7.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ku W-C, Cheng A-J, Wang T-C. Biochem Biophys Res Commun. 1997;241:730–736. doi: 10.1006/bbrc.1997.7874. [DOI] [PubMed] [Google Scholar]
- 34.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 35.Borges A, Liew C-C. J Mol Cell Cardiol. 1997;29:2717–2724. doi: 10.1006/jmcc.1997.0503. [DOI] [PubMed] [Google Scholar]
- 36.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 37.Hsiao R, Sharma H W, Ramakrishnan S, Keith E, Narayanan R. Anticancer Res. 1997;17:827–832. [PubMed] [Google Scholar]
- 38.Egan C A, Savre-Train I, Shay J W, Wilson S E, Bourne W M. Invest Opthalmol Vis Sci. 1998;39:648–653. [PubMed] [Google Scholar]
- 39.Yang J, Chang E, Cherry A M, Bangs C D, Oei Y, Bodnar A, Bronstein A, Chiu C P, Herron G S. J Biol Chem. 1999;274:26141–26148. doi: 10.1074/jbc.274.37.26141. [DOI] [PubMed] [Google Scholar]