Abstract
For more than a century immunologists and vaccinologists have existed in parallel universes. Immunologists have for long reveled in using “model antigens,” such as chicken egg ovalbumin or nitrophenyl haptens to study immune responses in model organisms such as mice. Such studies have yielded many seminal insights about the mechanisms of immune regulation, but their relevance to humans has been questioned. In another universe, vaccinologists have relied on human clinical trials to assess vaccine efficacy, but have done little to take advantage of such trials for studying the nature of immune responses to vaccination. The human model provides a nexus between these two universes, and recent studies have begun to use this model to study the molecular profile of innate and adaptive responses to vaccination. Such “systems vaccinology” studies are beginning to provide mechanistic insights about innate and adaptive immunity in humans. Here we present an overview of such studies, with particular examples from studies with the yellow fever and the seasonal influenza vaccines. Vaccination with the yellow fever vaccine causes a systemic acute viral infection and thus provides an attractive model to study innate and adaptive responses to a primary viral challenge. Vaccination with the live attenuated influenza vaccine causes a localized acute viral infection in mucosal tissues, and induces a recall response since most vaccinees have had prior exposure to influenza, and thus provides a unique opportunity to study innate and antigen-specific memory responses in mucosal tissues and in the blood. Vaccination with the inactivated influenza vaccine offers a model to study immune responses to an inactivated immunogen. Studies with these and other vaccines are beginning to reunite the estranged fields of immunology and vaccinology, and yielding unexpected insights about mechanisms of viral immunity. Therefore, vaccines that have been proven to be of immense benefit in saving lives offer us a new fringe benefit: lessons in viral immunology.
Introduction
Vaccines offer a practical and ethical way to perturb the immune system in humans. Infections, autoimmunity and cancers also perturb the immune system, but vaccination alone allows an exquisitely synchronized perturbation, in which the precise moment of immune stimulation can be known. This, together with the ability to obtain frequent blood samples from several minutes to several decades after vaccination, permits a detailed study of the kinetics of innate and adaptive responses. Since vaccines are administered to billions of people globally, and represent a wide spectrum of immunological stimuli ranging from live viruses and bacteria to recombinant proteins, they offer unique opportunities to probe the immune system with diverse stimuli. In addition to their enormous public health impact, many successful vaccines stimulate very robust antigen-specific T cells, and antibody responses that can persist a lifetime; however the mechanisms by which they do so remain largely unknown. Learning how to stimulate such robust and persistent immune responses in the context of vaccines against global pandemics such as HIV, malaria or TB represents a critical challenge in vaccinology. Yet, for over a century, we immunologists have done little to seize these opportunities.
Recent advances have begun to use vaccines as tools to probe the molecular and cellular networks orchestrating the immune response in humans, and such studies are beginning to yield novel insights about the immune system. These advances have relied largely on reductionistic approaches that aim to delineate a specific axis of interactions and study it in isolation from the confounding factors presented by the micro- (site, tissue) and macro- (organism and it’s history) environment. Such reductionist approaches have been spectacularly successful in guiding biological sciences for more than a century. In the past five years however, immunologists have begun to apply high throughput technologies such as transcriptomics and metabolomics to make systems wide measurements of immune responses, and use computational approaches to identify molecular signatures induced within a few days of vaccination, that correlate with and predict subsequent adaptive immune responses [1-3] (Figure 1). Several years ago, we [1], and Sekaly and colleagues [2], applied such a systems approaches to studying the innate and adaptive responses to vaccination with the live attenuated yellow fever vaccine (YF-17D), one of the most effective vaccines ever made. In order to identify signatures induced early after vaccination, which could be used to predict the magnitude of the later antigen-specific CD8+ T cell and neutralizing antibody responses to the vaccine. This study provides proof of concept evidence that systems approaches could indeed be used to identify early “signatures” that could predict the later immunogenicity of the vaccine. Subsequently, we extended this approach to identifying predictive signatures for the seasonal influenza vaccine [3], and, this approach is now being used to study immune responses to vaccines, by several groups, including our group [4-8].
Figure 1.
Systems biological approach to identifying molecular signatures of vaccination in humans. High throughput techniques such as transcriptomics (RNA-seq; microarrays), proteomics, CyTOF, luminex and metabolomics can be used to detect molecular signatures induced early after vaccination. Bioinformatics analyses of the data generated can be used to determine signatures that correlate with the ensuing adaptive immune response or protective immunity. The robustness of such signatures, and their ability to predict the immunogenicity and protective immunity to vaccination, can be tested in an independent “test trial,” (Trial 2).
Such studies involve collection of data on the magnitude and kinetics in global gene expression profiles, microRNA profiles and metabolomics profiles in peripheral blood mononuclear cells, shifts in compositions of cellular subtypes (FACS), data on cytokine profiles in peripheral blood, as well as antigen-specific T cell response data, and antigen-specific antibody titers (Figure 1). Together, these data provide an unbiased and detailed assessment of the state of immune system at various time points after vaccination. A key feature of such studies is the ability to discern the early molecular signatures that act as early markers of the downstream events. A variety of computational methods have been developed to identify early transcriptional signatures that correlate with, and predict, downstream factors that are known to serve as correlates of immune protection (Figure 1). Such studies have begun to yield novel insights about the mechanisms that control immune responses to vaccines. This interdisciplinary approach to the host’s response to the vaccine has been coined the systems vaccinology approach [9-12]. In this review, we highlight some key immunological lessons that are emerging from such studies.
What we are learning about viral immunity from studying human vaccines
What is now becoming increasingly clear is that many human vaccines induce robust adaptive immunity by triggering the receptors and cells of the innate immune system. The Nobel Prize in Physiology or Medicine 2011 was divided, one half jointly to Bruce A. Beutler and Jules A. Hoffmann “for their discoveries concerning the activation of innate immunity” and the other half to Ralph M. Steinman “for his discovery of the dendritic cell and its role in adaptive immunity (Nobelprize.org - http://www.nobelprize.org/nobel_prizes/medicine/laureates/2011/). The discovery of Toll-like receptors (TLRs) and dendritic cells (DCs), and the vital roles they play in sensing pathogens and tuning immune responses has provided a paradigm for how the innate immune system first recognizes pathogens [13-18]. In this paradigm, the innate immune system has the ability to detect components of viruses, bacteria and parasites through evolutionarily conserved receptors known as pattern-recognition receptors (PRRs). Such receptors include the TLRs, (of which 13 have been described in mammals), which are expressed on the surface and within the endosomal compartments of cells such as DCs. TLRs represent only one such family of receptors - other PRR families include the cytosolic RIG-I like receptors, which sense viral nucleic acids, and the C-type lectin like receptors and NOD-like receptors which sense diverse stimuli from viruses and microbes. Such PRRs are expressed by several different subsets of cells, including DCs and immune response triggered by a virus or microbe is an integrated function of all the PRRs that it triggers, as well as the types of DCs activated [13-18]. These issues have been reviewed extensively and are only mentioned here. Despite the plethora of knowledge, the question of the extent to which any of the licensed human vaccines activate these receptors and cell types, and if so whether such activation was essential for the induction of adaptive immunity remained unanswered. A few years ago, we began to address this, using the live attenuated yellow fever vaccine 17D (YF-17D) as a model [19-21].
The yellow fever and the seasonal influenza vaccines
The live attenuated yellow fever vaccine 17D (YF-17D) is one of the most successful vaccines ever developed, having an efficacy of greater than 90%, and having being administered to more than 600 million people globally [9, 22, 23]. The vaccine consists of a positive single stranded RNA virus, which was developed by Max Theiler at Rockefeller. The Asibi strain of the yellow fever virus, which was isolated from an infected individual (Mr. Asibi) in West Africa in 1927 and subsequently passaged in rhesus monkeys, served as the starting material for the vaccine development effort [24]. This Asibi strain was attenuated by passing it more than 200 times, first in mouse brain and mouse embryo tissue culture to reduce its viscerotropism, (which can lead to hepatic and renal failure and hemorrhage), and next in chicken embryo and chicken embryo tissue culture with the brain and spinal cord removed to attenuate its neurotropism [9, 22-24]. This work culminated in the 1937 Journal of Experimental Medicine paper in which YF-17D was first used in human volunteers, including the authors, and shown to induce neutralizing antibodies [25, 26]. In 1945 the World Health Organization organized the establishment of two sub-strain seed lots of this vaccine: 17DD, which is used in South America, and 17D-204, which is used in the majority of the rest of the world. The Nobel Prize in Physiology or Medicine 1951 was awarded to MaxTheiler “for his discoveries concerning yellow fever and how to combat it” (http://www.nobelprize.org/nobel_prizes/medicine/laureates/1951/). Remarkably, a single subcutaneous injection of the vaccine results in very robust antigen-specific CD8+ T cells [9, 19, 22, 23, 27], and a balanced Th1/Th2 responses and neutralizing antibody responses that can last several decades [9, 22, 23] (Figure 2). Vaccination with the yellow fever vaccine causes a systemic acute viral infection, and the fact that most individuals in developed countries have not been exposed to yellow fever provides an attractive model to study innate and adaptive responses to a primary viral challenge.
Figure 2. Innate and adaptive responses to the live attenuated yellow fever vaccine YF-17D.
Vaccination with YF-17D results in an acute viral infection during which viral replication peaks between days 5 and 7 becomes undetectable by 14 days post vaccination. Vaccination rapidly induces IgM neutralizing antibody titers which peak at 2 and then decline, but can be detected for as long as 18 months post vaccination. Virus-specific IgG neutralizing antibody titers develop more slowly and can persist for up to 40 years. Vaccination induces virus-specific CD8+ T cell responses, which develop rapidly after immunization, peaking at day 15 (with roughly 2-13% of CD8+ T cells being activated at day 15) but reaching near baseline levels by day 30. Vaccination also induces a mixed Th1 and Th2 CD4+ T cell response. A robust innate response develops within hours and seems to persist for more than 15 days, likely caused by persisting viral replication in the blood, during the first 7 days or so. YF-17D activates multiple subsets of DCs via several different PRRs. This results in a mixed Th1/Th2 response, and may also impact the persistence of the antibody response. YF-17D also activates mammalian target of rapamycin (mTOR) in plasmacytoid DCs, via a mechanism dependent on TLR7, and this leads to phosphorylation of interferon regulatory factor 7 (IRF7). This results in induction of IFNα which activate CD8+ T cells.
Two types of vaccines for seasonal influenza are licensed for use in the USA: trivalent inactivated influenza vaccine (TIV), given by intramuscular injection; and live attenuated influenza vaccine (LAIV), administered intranasally [28]. These vaccines are derived from three strains of influenza viruses, which are negative single stranded RNA viruses. TIV is made from influenza viruses grown in the allantoic cavities of embryonated chicken eggs, and subsequently inactivated by formalin, and purified to remove egg-derived contaminants [29, 30]. The monovalent vaccines are combined to formulate the final trivalent bulk vaccines. Although the resulting whole-virus vaccines are still used in some countries, most vaccines manufactured since the 1970s have been the so called “split” or sub-virion vaccines. Split vaccines are less reactogenic than the whole virus vaccines, and are prepared by disrupting the viral lipid envelope using a detergent. The LAIV, (also known as “cold-adapted” influenza vaccine) were produced by first isolating the two “backbone” viruses (ca A/Ann Arbor/6/60 (H2N2) (A/AA ca) and ca B/Ann Arbor/1/66 (B/AA ca)) from infected individuals, and then growing these viruses at successively lower temperatures in primary chick kidney cells to obtain viruses that grew efficiently at 25° C as they did at 33° C [31]. Clones of these two viruses was isolated by serial plaque-to-plaque purifications. This viruses grew well at 25° C, and its replication was reduced by at least 100-fold of the median tissue culture infective dose (TCID50) at 39° C, and was attenuated behavior ferrets. The vaccine viruses are then generated by classical reassortment, by coinfection with vaccine donor viruses and wild type influenza viruses. Alternatively reverse genetics techniques can be used. The resulting recombinant virus contains six internal gene segments of the vaccine strain combined with the two antigen-encoding gene segments, HA and NA.
The widespread use of TIV and LAIV during every influenza season provides ample opportunities to perform clinical studies. However there are several immunological challenges with the influenza vaccines. First, unlike the case with YF-17D, adults have been pre-exposed to influenza viruses or vaccines, and thus have pre-existing immunity to influenza. So vaccination results in a recall response, and thus the extent to which preexisting antibody derived from prior infection or vaccination, influence immunity to a subsequent vaccination, and contribute to protective immunity remains poorly understood. In this context, a recent study demonstrates that pre-existing influenza specific CD4+ T cells correlate with protection against influenza challenge in humans [32]. Second, the viral strain is usually changed annually on the basis of the results of global influenza surveillance data [33]. The efficacy of a vaccine against influenza, therefore, depends on the match of antigenicity between the vaccine and circulating influenza strains [34]. Furthermore, the extent to which the so-called “antigenic drift” (caused by accumulation of mutations within the genes that code for antibody binding sites, resulting in a new strain of virus which cannot be inhibited as effectively by the antibodies that were originally targeted against previous strains [35], versus “antigenic shift” (caused by two or more different strains of the virus combining to form a new subtype having a mixture of the antigens of the two or more original strains [35]), impacts on the innate signatures induced by vaccination remains unknown. Additionally, other factors such as the age and immunocompetence of vaccinees contribute to mechanisms that mediate the efficacy of vaccines against influenza [36].
Insights into innate responses to vaccination
Several years ago we began to study YF-17D, with a view to learning new insights about the mechanisms by which it stimulates such effective immunity. The model system provided by this “gold standard” vaccine proved to be instructive. To our surprise we observed that this vaccine was able to trigger activation of both myeloid dendritic cells (DCs) and plasmacytoid DCs, via several different Toll-like receptors (TLRs), including TLRs 2, 7, 8 and 9 [19] and TLR3 (unpublished), as well as the cytosolic nucleic acid sensing receptors RIG-I and MDA 5 [1] (Figure 2). Interestingly, mice deficient in MyD88, an adaptor protein critical for signaling via all TLRs except TLR3, were severely impaired in antigen-specific CD8+ and CD4+ T cells that produced interferon-gamma (IFNγ) in response to vaccination with YF-17D, whereas mice deficient in TLR2 displayed strikingly enhanced responses. This demonstrated that YF-17D mediated TLR2 activation resulted in a regulation of Th1 and Tc1 responses, consistent with several previous reports demonstrating a regulatory role for TLR2 in several different settings [37] (Figure 2). In addition to the CD4 stimulatory cascade, a parallel chain of signal transduction occurs in plasmacytoid DCs, mediated by the TLR7/9 – mTOR axis (Figure 2). This signaling cascade culminates in the secretion of type I interferons, which stimulate the unfolding of the cytotoxic T cell response [38]. Consistent with this, the transcriptional signatures induced by vaccination of humans with YF-17D reveals a very robust signature comprising of a network of genes orchestrating type I IFN signaling and anti viral immunity [1, 2]. Therefore, it appears that it is this broad spectrum of the channels through which the sensory signals are amplified within the antigen-sensing cells, that results in the broad spectrum of stimulatory messages that are been sent to the naïve T cells, resulting in potent stimulation of humoral and cytotoxic responses.
There is relatively little data on how the seasonal influenza vaccines trigger PRRs and DC subsets. Vaccination with the inactivated whole virus requires TLR7-mediated production of type I interferons by plasmacytoid DCs for its immunogenicity [39]. Furthermore, the immunogenicity of the whole inactivated vaccine against H5N1 influenza is dependent on MyD88-dependent TLR7 signaling [40]. Consistent with this, vaccination of humans with LAIV induces a robust type I IFN anti viral transcriptomic signature in the blood [3], and vaccination with TIV also induces some type I IFN genes, as well as genes encoding pro inflammatory mediators [3, 8]. Pathogenic influenza viruses activates plasmacytoid DCs via TLR7 and myeloid DCs through the adaptor IPS-1, which signals downstream of RIG-I [41]. Infection of mice deficient in MyD88 and IPS-1, results in reduced innate responses. Antigen-specific antibody responses and CD4+ T cell responses are MyD88 dependent but are not dependent on IPS-1. In contrast, induction of antigen-specific CD8+ T cell responses is normal in MyD88- or IPS-1-deficient mice. It is not clear whether LAIV or TIV trigger the same PRRs.
Insights into the mechanisms controlling antibody responses
Vaccination with YF-17D induces robust neutralizing antibodies that can last for 30-40 years (Figure 2) [42]. Similarly, in people who received the smallpox vaccine, vaccinia-specific serum antibody titers, measured by ELISA, were very stable up to 75 years after vaccination, and based on a correlative analysis between the ELISA binding titers and neutralizing antibody titers, at least 50% of the vaccinees had neutralizing titers that were greater than 1:32 [43], which is estimated to be the half maximal titer necessary for protective immunity [44]. Consistent with this, smallpox-specific memory B cells lasted for >50 years [45].
Despite the enormous public health success of such vaccines, the mechanisms by which they stimulate such robust and long-lived immune responses remain unknown. In the case of YF-17D, a key question was whether the activation of several different TLRs [19] impacted the antibody responses induced by YF-17D in any way. Our results indicate that induction of antigen-specific antibody responses to YF-17D was severely impaired in mice deficient in MyD88 (unpublished observations). In order to assess the impact of triggering multiple TLRs on the adaptive immune response we designed a nanoparticle based vaccine, similar in size and composition to a virus. We synthesized nanoparticles roughly 300nM in diameter, using poly(D,L-lactic-co-glycolic acid) – a biodegradable synthetic polymer. These particles contained the TLR ligands MPL (TLR4), R837 (TLR7), or both ligands, together with an antigen, such as recombinant hemagglutinin from the H5N1 avian influenza strain. Immunization of mice with nanoparticles containing both TLR ligands induced synergistic increases in antigen-specific, neutralizing antibodies compared to immunization with a single TLR ligand. Strikingly there was enhanced persistence of germinal centers (GCs), similar to what has been previously reported with acute viral infections [46], and unlike the relatively short lived response observed with immunization with protein antigens plus adjuvants such as alum [47]. Consistent with the long-lived germinal center responses, and given the critical importance of germinal centers in the differentiation of memory B cells and long-lived plasma cells, we observed persistent plasma cell responses, which persisted in the lymph nodes for >1.5 years. Surprisingly, there was no enhancement of the early short-lived plasma cell response, relative to that observed with single TLR ligands. Importantly, antibody responses were dependent on direct triggering of both TLRs on B cells and dendritic cells (DCs), as well as on T-cell help. Thus, activation of specific combinations of TLRs results in long-lived germinal centers and plasma cells responses. A key question here is whether TLR signaling modulates the function of B cells and other cells within germinal centers. Given that TLRs are expressed in germinal center B cells and on follicular dendritic cells, a key question is how TLR signaling in these cells modulates the germinal center reaction and the generation of memory B cells and long-lived plasma cells. Since live viral vaccines persist at least for a few days after vaccination, it is conceivable that TLR ligands derived from such vaccines activate B cells or FDCs in nascent germinal centers to promote memory B cell formation. The extent to which such a mechanism involving the combinatorial activation of TLRs on both DCs and B cells, contributes to the longevity of the B cell response to YF-17D and other viral vaccines, such as the smallpox vaccine, remains to be determined.
In the case of the seasonal influenza vaccines, the extent to which antibody responses persist is less well studied. Possible exposure of vaccinees to influenza virus during the influenza season, as well as pre-existing antibodies from previous exposures renders it difficult to evaluate the persistence of antigen-specific antibody responses to vaccination. However, analysis of long lived plasma cells in the bone marrow that produce influenza specific antibodies in subjects that received the seasonal vaccine is currently underway (Rafi Ahmed - personal communication). A recent study of subjects vaccinated with TIV indicates that the number of influenza-specific T follicular cells (TFH) expressing the costimulatory molecule ICOS, as well as the chemokine receptor CXCR3 and CXCR5 in the blood, correlates with antibody responses [48]. These cells could induce memory B cells to differentiate into plasma cells [48].
Insights into antigen-specific CD8+ T cell responses
Our understanding of CD8+ T cell responses to viral infections is based largely on experiments in mice. For example, infection of mice with the Armstrong strain of lymphocytic choriomeningitis virus (LCMV) or vaccinia virus results in acute viral infections, during which there is a massive activation and expansion of CD8+ T cells, with as much as 30-80% of the splenic CD8+ T cells being virus-specific, 8 days after infection [49]. At this time point, which is the peak of the response, the effector CD8+ T cells produce cytokines, proliferate rapidly and die rapidly through apoptosis. This causes a contraction of the virus specific CD8+ T cell population, in which the few surviving cells develop into long lived memory cells. In humans, CD8+ T cell responses have been studied extensively in chronic viral infections such as human immunodeficiency virus (HIV), cytomegalovirus (CMV), hepatitis C virus (HCV) and Epstein-Barr virus (EBV) [50-59]. The responses in such infections can be very robust, but the phenotype of the CD8+ T cells varies depending on the virus.
Recently Ahmed and colleagues have studied the CD8+ T cell responses to yellow fever and smallpox vaccination in humans [60]. Both vaccines cause acute viral infections in humans. Both vaccines induced a rapid expression of Ki-67+ CD8+ T cells which downregulated bcl-2 resulting in enhanced susceptibility to apoptosis [61] The activated human T cells also upregulated HLA-DR and CD38, so the rapidly proliferating CD8+ T cells expressed the phenotype Ki-67+ Bcl-2lo HLA-DR+ CD38+. Two weeks after vaccination with YF-17D, at the peak of the response, such cells constituted 2-13% of all CD8 T cells. In the case of Dryvax vaccination, the frequency of such cells was 10-40% of all CD8 T cells, 2 weeks after vaccination [61]. Subsequently over the next 2 weeks the CD8+ T cell population contracted more than 90-99% and gradually differentiated into long-lived memory CD8+ T cells [62]. There was minimal bystander expansion, and the breadth of the virus specific response induced by both vaccines was high, as judged by epitope mapping studies. Furthermore, tetramer-based analysis showed that vaccination with either vaccine resulted in the typical expansion, contraction and memory phase observed with experimental infections in mice. Despite a major contraction, tetramer-specific CD8+ T cells could be detected 1 year after vaccination and these cells were polyfunctional with respect to cytokine production [62].
Insights into the potential mechanisms underlying severe adverse reactions
YF-17D has an excellent safety record, but in very rare cases (1 in 250,000 to 1 in 500,000), vaccinees develop serious, often fatal, adverse events [9, 22, 23]. One type of serious adverse event, the so-called neurotropic disease is caused by neuroinvasion of YF-17D and typically includes symptoms such as encephalitis, Guillain–Barré syndrome and autoimmune disease in the central or peripheral nervous system [9, 22, 23]. The other type of serious adverse event is viscerotropic disease, which causes a fulminant yellow fever infection of the liver and other visceral organs, is typified by multiple organ systems failure, similar to what has been observed during infection with the pathogenic virus [9, 22, 23]. High levels of yellow fever virus antigen can be found in the liver, heart and other organs, primarily in tissue-associated macrophages [9][22]. The fatality rate for patients with viscerotropic disease is greater than 60%. Vaccinees with viscerotropic disease develop a high fever, malaise and myalgia, within 2 to 5 days post vaccination. This is followed by jaundice, oliguria, cardiovascular instability, haemorrhage, and renal and respiratory failure. The immunological mechanisms that result in serious adverse reactions remain unknown. It is likely that such events are not caused by gene mutations in YF-17D, as virus isolated from patients with serious adverse events had the same nucleotide sequence as the vaccine strain Host genetics may play a role. Given the critical role type I IFN signaling in controlling viral infections, it is possible that mutations in genes involved in this network render the host susceptible. Thus, vaccination of mice deficient in the type I IFN receptor developed infection and disease, displaying many of the features of viscerotropic disease, after vaccination with YF-17D [63]. Furthermore, a mutation in an allele of the gene encoding (Oas1b) encoding 2′,5′-oligoadenylate synthetase (OAS), an enzyme important in type 1 interferon-mediated innate immunity has been mapped to flavivirus susceptibility in mice. Consistent with this, in a collaborative study with Belsher et al [64] we found polymorphisms in Oas1 and Oas2 genes, in a 22 year old female who developed fatal viscerotropic disease. However, in a different case of viscerotropic disease in a 65 year old male, we found no such polymorphisms [65], but rather polymorphisms in genetic polymorphisms in CC-chemokine receptor 5 and its ligand, CCL5 [65]. In this individual there were no polymorphisms in the genes encoding TLR3 or DC-SIGN, other genes known to be involved in sensing viruses. Interestingly, there were no polymorphisms in the genes encoding CCR5 or RANTES in the 22 year old female [64]. These results suggest that either: 1) that none of these polymorphisms are relevant to the disease, or 2) there are many different genetic polymorphisms that could result in viscerotropic disease. Evaluation of such polymorphisms in other viscerotropic cases, coupled with mechanistic studies using humanized mice that are reconstituted with hematopoietic cells from such patients may provide greater insights into the role of such genes in regulating viscerotropic disease.
The rarity of these cases, and the practical difficulties of obtaining blood samples at frequent intervals after vaccination and after the development of adverse symptoms poses challenges to performing studies. As such, only very few studies have begun to examine host responses during adverse reactions. In the aforementioned study involving the 64 year old male [65], we could detect viral RNA in the blood 33 days after vaccination, in contrast to the rapid disappearance of virus by around 7 days post vaccination in healthy subjects. One hypothesis that could have explained the persistent viral loads was that there was suboptimal antibody responses and CD8+ T cell responses which lead to impaired viral clearance. However to our surprise, we observed robust and sustained antigen-specific CD8+ T and B cell responses, which suggested that persistent virus was not due to adaptive immunity of suboptimal magnitude. The genes encoding OAS1, OAS2, TLR3, and DC-SIGN, which mediate antiviral innate immunity, did not carry any SNPs. However, as stated above there were heterozygous genetic polymorphisms in chemokine receptor CCR5, and CCL5, which influence the migration of effector T cells and CD14+CD16bright monocytes to tissues [65]. Consistent with this, there was a 200-fold increase in the number of CD14+CD16bright monocytes in the blood during viremia and a 20-fold increase in these cells, even several months after virus clearance. Such cells are known to produce inflammatory mediators, and consistent with this, there were elevated levels of interleukin-1α (IL-1α), IL-6, CXC-chemokine ligand 10, CC-chemokine ligand 2 (CCL2) and CCL5 [65]. These results indicate that in this patient, viscerotropic disease was not due to the impaired magnitude of adaptive immunity but rather due to potential anomalies in the innate immune system and a possible disruption of the CCR5-RANTES axis.
Novel biological hypotheses enabled by the systems vaccinology approach
The systems biological approaches to study vaccine responses have yielded a veritable treasure trove of data, and have enabled the creation of many exciting and novel hypotheses about the molecular mechanisms that regulate vaccine immunity. The experimental testing of several of these hypotheses in animal models is well underway, and are beginning to yield many novel and unexpected insights about mechanisms of immunity. Here we discuss five of these hypotheses.
Hypothesis 1: Viral induced integrated stress response in dendritic cells stimulate antigen-specific CD8+ T cells
In our systems biology studies to predict the immunogenicity of the yellow fever vaccine in humans [1] we identified transcriptional signatures in the blood, induced within a few days after vaccination that correlated with, and predicted the magnitude of the later CD8+ T cell response to the vaccine [1]. The gene signatures included solute carrier family 2, member 6 (SLC2A6), complement protein C1qB and eukaryotic translation initiation factor 2 alpha kinase 4—an orchestrator of the integrated stress response (Figure 3) [1]. EIF2AK4/GCN2 plays a critical role in the so-called integrated stress response [66-69] and regulates protein synthesis in response to amino acid starvation, by phosphorylating elongation initiation factor 2α (EIF2α)(Figure 2) [66-69]. The translation of house keeping mRNA is then stopped, and the untranslated mRNA are redirected from polysomes to small cytoplasmic compartments known as stress granules, for transient storage [66-69]. Consistent with this, YF-17D induced the phosphorylation of EIF2α and the formation of stress granules (Fig. 2AC and [1]).
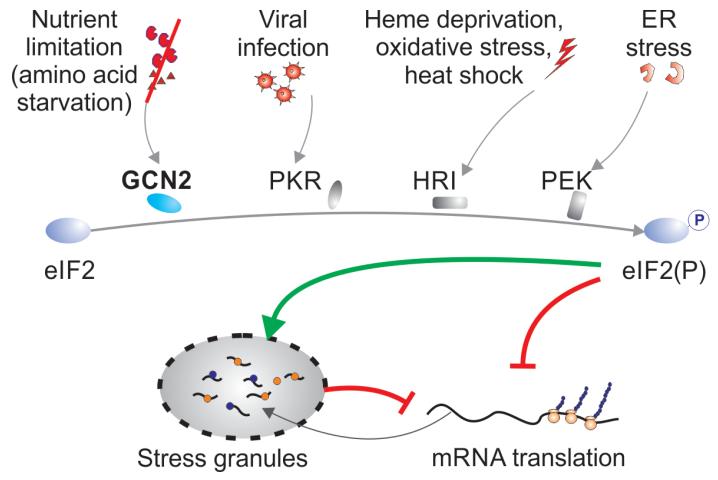
Figure 3. The integrated stress response.
Mammalian cells have evolved at least four different sensors that can detect environmental stresses of various kinds. GCN2 (EIF2AK4) senses changes in amino acid concentrations, PKR senses viral infections, HRI senses heme deprivation oxidative stress and heat shock and PEK or PERK senses endoplasmic reticulum stress. Activation of any one of these sensors results in the phosphorylation of eIF2α, which leads to the shut down of housekeeping mRNA, and their compartmentalization in stress granules in the cytosol.
EIF2AK4/GCN2 kinase is but one of several related sensors that mediate cellular responses to stress. Other members of this family include EIF2AK2/PKR, EIF2AK1/HRI and EIF2AK3/PERK (Figure 2). These sentinel molecules are activated in response to dsRNA [70], depletion of heme [71-73], and the overabundance of misfolded/malfolded proteins in the endoplasmic reticulum [74]. All four of these kinases have a common target, and phosphorylate eIF2alpa on Ser51 (Figure 3). Due to the integration of the signals from diverse physiological stimuli through the same factor, the four pathways are collectively referred to as integrated stress response [75-77]. Activation of this pathway impacts diverse physiological processes, including regulation of cell lineage-specific differentiation [78], metabolic adaptation [75, 79, 80], cell death [81] and organ homeostasis [82]. While the heme-sensing pathway is of particular relevance in maturing erythroid cells [78], sensing of dsRNA and ER stress have an immediate effect on the regulation of inflammatory response [83] and dictate the decision between metabolic adaptation and apoptosis in the affected cells [84].
Recent work has shown an antiviral effect of EIF2AK4 against RNA viruses [66-69] but the effect of this factor on adaptive immune responses is not known. This prompted us to further investigate the role of EIF2AK4 and the integrated stress response in the immune reaction to YF-17D. Our experimental studies suggest a critical role for GCN2 in programming DCs to stimulate antigen-specific CD8+ T cell response to YF-17D vaccination (Nair et al - manuscript in preparation). This work reveals a previously unknown function of GCN2 pathway to vaccine-induced immunity.
Hypothesis 2: Intestinal microbiota regulates vaccine immunity
One of the intriguing molecular signatures uncovered through the use of systems biology in human vaccine studies with the seasonal TIV is a cluster of genes associated with and including TLR5 [3]. Our study found that the expression of TLR5 within a few days after vaccination was strongly correlated to the magnitude of the HAI titers 4 weeks after vaccination. Given that TLR5 is known to be a sensor of bacterial flagellin and is not thought to be involved in sensing viruses, this was a surprising observation. The influenza vaccine itself does not appear to trigger TLR5 signaling directly (unpublished data). Thus, it raises the possibility that other ligands for TLR5, such as those derived from the commensal flora, may be contributing to vaccine responses in humans. Studies focusing on this and other possibilities are currently underway and already suggest a critical role for the intestinal microbiota in influencing adaptive immunity to vaccination (Oh et al – manuscript in preparation).
The human commensal flora is composed of diverse and complex communities of microorganisms. They inhabit barrier surfaces of major organs (e.g. skin) and tissues (e.g. gastrointestinal tract) and have evolved beneficial properties for the host that permits a symbiotic relationship to be maintained with the host (reviewed in [85]). Recently, the notion that the host microbiota, in particular those residing in the gastrointestinal tract, can shape immune responses at other sites has begun to emerge. In the context of influenza A virus infection, Ichinhoe et al. have shown that neomycin-sensitive bacteria in the gut impacts the virulence and the capacity to which the host immune system mounts immune responses in the respiratory tract [86]. Similarly, others have shown that immune function can be compromised not only in local mucosal tissues [87-89], but also systemically as was demonstrated in a model of LCMV infection [90]. Thus, the absence or dysbiosis of microbiota can invoke critical changes to the host immune system. It is also clear, however, that microbial signals from the gut are not the only factors contributing to proper immune function. Naik et al. recently demonstrated that skin-resident microorganisms play crucial roles in inflammation and T cell responses of the skin [91]. Thus, the growing body of literature indicates that the microbiota affects immune function in multiple tissues and organs. Importantly, these emerging evidence support the possibility that microbiota can serve critical roles during vaccine-induced immune responses. The data generated from our clinical studies suggest that sensing of flagellated bacteria is one key component of the microbiota’s capacity to provide crucial signals necessary for efficient priming of adaptive immunity.
The possibility that the microbiota plays a role in immune responses to vaccines certainly bears major implications for global public health. Our analyses predict that the status of the host microbiota may be a critical determinant of vaccine efficacy. Currently, there is no information in both humans and animal models whether the microbiota impacts the magnitude and/or quality of vaccine-induced immune response. Examining the relationship between microbiota and vaccine efficacy is a novel concept, and understanding how such a phenomenon operates is a critical field to study. Specifically, how the microbiota may be regulating immune responses to parenterally administered vaccines remains unclear, and current studies in our lab are addressing the potential mechanism underlying this phenomenon.
Hypothesis 3: Calcium/calmodulin-dependent protein kinase IV (CaMKIV) is a regulator of antibody responses to vaccines
The clinical study with the seasonal TIV has revealed an intriguing molecular signature associated with one of the most fundamental element of cellular processes, calcium signaling [3]. In the study, we observed an inverse correlation between the early expression of the gene encoding calcium/calmodulin-dependent protein kinase IV (CaMKIV), and the later hemagglutinin titers to influenza vaccination. Specifically, the change in expression of the CaMKIV gene on day 3 post vaccination was negatively correlated with the magnitude of antibody response on day 28 as well as with an expanding population of IgG-secreting plasmablasts on day 7 post vaccination. This observation led us to hypothesize that CaMKIV may function as a negative regulator of antibody response to influenza vaccination. Results obtained from a comparison of vaccine responses in wild type and Camk4−/− mice indeed support this hypothesis as the antibody responses were significantly higher in the absence of CaMKIV expression [3]. CaMKIV is a serine/threonine kinase, which belongs to a family of calcium/calmodulin-dependent kinases. Although other members in this family of kinases, such as CaMKI and CaMKII, are ubiquitously expressed, CaMKIV is predominantly expressed in cells of the nervous system and the immune system [92, 93]. Specialized function of CaMKIV has previously been ascribed to neural activity and long-term memory - neural memory that is, and not immune memory. Thus, the role of CaMKIV in regulating immunity is poorly understood. Previous studies suggest that CaMKIV is involved in several processes across multiple cell-types, such as regulating survival of hematopoietic progenitor cells [94], thymic selection [95, 96], and inflammatory responses [97]. Some reports have indicated a possible role of CaMKIV in mediating activation of CD4 T cells and survival of DCs [95, 97]. Consistent with these observations, downstream targets of CaMKIV such as CREM and CREB have been observed at elevated levels in SLE patients [98]. Thus, existing evidence in literature points to a positive role of CaMKIV in immune cell activation. Our studies suggest that CaMKIV may exert a suppressive effect on the immune response. Our current studies are examining whether function(s) of CaMKIV is cell-type specific and/or influenced by the properties of the antigen or the context in which it is encountered (ie. adjuvant, PAMPS, etc).
Hypotheses 4: Role of TNF Super Family and TNF-R Super Family in Programming of B Cell Fates
In our previous study with the yellow fever vaccine, a striking correlation was found between the expression of several genes belonging to the TNF superfamily and antibody titers following vaccination against the yellow fever vaccine, YF17D [1]. We have since found correlations also evident following vaccination against the seasonal influenza [3]. Specifically, our data show that expression levels of TNFSF13 (APRIL), TNFSF13B (BAFF), and their receptors TNFRSF17 (BCMA) and TNFRSF13B (TACI) all strongly correlate with the magnitude of the antibody response. These molecules are known for their roles in not only the development of B cells, but maturation and differentiation affecting multiple aspects of the humoral immune response. BAFF-deficient mice lack follicular B cells [99], and conversely exogenous BAFF has been demonstrated to induce expansion of B2 and marginal zone B cells in vivo [100]. Subsequent studies have since identified more specific roles of BAFF and other members of this superfamily, which include germinal center reactions [101-103], class switch recombination [104], and plasma cell differentiation [105]. However exactly if and how these molecules regulate the generation of short lived plasma cells, (which largely contribute to influenza specific antibody titers measured 28 days after vaccination), versus long lived plasma cells (which likely contribute to yellow fever specific neutralizing antibody responses measured months to a year later), and B cell memory is unknown. It is possible that these molecules differentially regulate short-lived versus long lived antibody responses. Experiments are currently underway to test these hypotheses.
Hypothesis 5: Role of ‘unfolded protein response’ in vaccine immunity
In our analyses of the transcriptional signatures induced by TIV, we found a striking correlation of adaptive immune responses [3] to the early expression of genes encoding PKR-like ER kinase (EIF2AK3/PERK) [19]. In addition, meta-analysis of the collection of genes whose expression changes in response to the TIV vaccine revealed a significant group of coordinately regulated genes involved in the unfolded protein response [3]. Taken together, these data prompted us to investigate the role of PERK, in the shaping of the innate and adaptive immune responses.
PERK is a sensor of endoplastic reticulum (ER) stress most notably invoked during periods of rapid and high levels of protein synthesis [106]. If the rate of protein synthesis exceeds the rate of proper folding and post-translational modifications, proteins remain in unfolded form and may result into accumulation inside the cell. Under normal condition these unfolded proteins are guided back to the cytoplasm for degradation through ER-associated ubiquitin/proteasome degradation (ERAD) pathway [107]. However, under certain conditions as well as certain cells types with secretory functions, misfolded proteins often accumulate inside the cells due to higher rate of protein translation causing ER stress and subsequent activation of unfolded protein response (UPR) [108, 109]. PERK is one of three critical mediators of the UPR pathway (others being ATF6p90 and IRE1) [110, 111]. These conditions are likely to manifest during periods rapid cytokine/chemokine production such as those occurring in dendritic cells as well antibody production in activated B cell subsets. Thus, understanding how the gene products of these early molecular signatures contribute to the adaptive immune response will be critical to understanding determinants of vaccine efficacy.
Perspectives
Data >>> Knowledge >>> Understanding
Studying immune responses to vaccination in humans is beginning to yield many important insights into viral immunity. Several major challenges remain [112]. A systems vaccinology study generates a multidimensional dataset. Data on changes in expression levels of thousands of genes are collected, and genes are sorted and binned according to the correlations between changes in their expression level and cellular or humoral responses (Figure 1). Over the coming years, several such studies will be done, and will generate mountains of data. The single most important challenge is how to go from data to knowledge to understanding. Several issues need to be considered. First, how can we validate the biological role of identified genes and reconstruct the chain of events leading to the altered transcriptional activity of genes of interest? Second, apparent transcriptional changes can be caused by either changed transcriptional activity of a gene, or changes in frequency of the cell subtype in which the gene is expressed; we need to have a tool at our disposal that would enable us to trace the transcript frequency to the cell type. Third, how can we integrate the knowledge obtained in previous transcriptomic studies of the immune system in order to functionally validate the regulated genes? The first problem concerns hypothesis validation. The latter two problems are united by the common theme of hypothesis generation. The twin aspects of hypotheses generation and hypotheses validation represent the key issues in systems approaches to biological sciences in general.
Several examples of hypothesis building and testing have been illustrated in this review. The hypotheses that led to the unfolding of these stories were created by an iterative process involving: a) observed correlations between gene expression and immune outcome; b) applying biological intuition and literature survey to the observed correlations in order to generate plausible and interesting hypotheses; c) initial experimental verification of the first tenets of hypotheses; d) and if the initial experiments yielded interesting leads, then deeper experimental verification of the hypotheses. While these approaches proved to be effective tools in hypothesis building and validation, they do not exhaust the potential for data mining contained within the systems biology data. How can we accelerate this process? One approach is through the integration of multiple lines of evidence obtained through independent technologies (reviewed in [113]). For example, Amit at al [114] have identified a transcription regulation network mediating TLR signaling in mouse dendritic cells. Starting off with the genomewide transcriptional profiling of cells treated with TLR ligands, the authors identified 144 candidate regulatory factors whose expression tracked with that of blocks of co-expressed genes. Knocking down the expression of these putative regulators by siRNA, the authors identified key transcriptional regulatory cascades responsible for antiviral and inflammatory reactions precipitated by TLR ligation. In this example, high throughput studies were supplemented by a gene-by-gene functional validation assays using the RNA interference technology. This technology provides an ultimate functional validation tool as it allows to rapidly and specifically decrease the mRNA level of target genes and monitor the resulting phenotype. An important step that brings this methodology into the field of –omics studies is the development of large scale shRNA libraries [115]. Several commercial and not-for-profit (The RNAi consortium at Broad Institute, http://www.broadinstitute.org/rnai/trc ) institutions have developed such libraries. These libraries can be readily utilized to validate the role and function of nearly each one of the genes that are identified in the clinical studies. Such approach holds promise for the development of systems vaccinology. Following the systems vaccinology study in human cohort, specific roles of candidate genes and interactions can be investigated in a cell culture model of relevant cell types. Complementing the high-throughput tanscriptomics data with cell-based assays such as M2H, or other –omics data, such as phosphoproteomics, will guide the selection of the selection of targets for siRNA knockdown [116].
Even though the field of systems vaccinology is still in it’s formative stage, it has already provided insights into the sequence of molecular events leading to successful vaccination, allowed to develop early predictive markers of vaccine efficacy, and generated leads into several novel projects increasing our understanding of the vaccine perception by a human organism. Further expansion of the arsenal of available tools and integration of expertise of multiple research groups paves the way the development of novel principles guiding rational vaccine development.
Acknowledgements
Research in the Pulendran lab is supported by the US National Institutes of Health (U19AI090023, U54AI057157, R37AI48638, R37DK057665, U19AI057266, PO1A1096187, Scripps CHAVI-ID Award (UM1AI100663) and the Bill and Melinda Gates Foundation.
Footnotes
Conflicts of interest B.P, J.Z.O, H.I.N, R.R, D.A.Z are not aware of any conflicts of interest.
References
- 1.Querec TD, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10(1):116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaucher D, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205(13):3119–31. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucasas KL, et al. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. Journal of Infectious Diseases. 2011;203(7):921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9(1) doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zak DE, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8+ T-cell responses but is attenuated by preexisting Ad5 immunity. Proceedings of the National Academy of Sciences. 2012;109(50):E3503–E3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vahey MT, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS, S malaria vaccine. Journal of Infectious Diseases. 2010;201(4):580–589. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- 8.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38(4):831–44. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9(10):741–7. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 10.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33(4):516–29. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2012;4(2):193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, et al. Systems biological approaches to measure and understand vaccine immunity in humans Seminars in Immunology. 2013 doi: 10.1016/j.smim.2013.05.003. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinman RM. Dendritic cells in vivo: a key target for a new vaccine science. Immunity. 2008;29(3):319–24. doi: 10.1016/j.immuni.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327(5963):291–5. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 16.Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22(1):41–7. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9(7):465–79. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327(5963):286–90. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Querec T, et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203(2):413–24. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Querec TD, Pulendran B. Understanding the role of innate immunity in the mechanism of action of the live attenuated Yellow Fever Vaccine 17D. Adv Exp Med Biol. 2007;590:43–53. doi: 10.1007/978-0-387-34814-8_3. [DOI] [PubMed] [Google Scholar]
- 21.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;12(6):509–17. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett AD, Monath TP. Epidemiology and ecology of yellow fever virus. Adv Virus Res. 2003;61:291–315. doi: 10.1016/s0065-3527(03)61007-9. [DOI] [PubMed] [Google Scholar]
- 23.Monath TP. In: Milestones in the Conquest of Yellow Fever, in Microbe hunters - then and now. Koprowski H, Oldstone MBA, editors. Medi-Ed Press; Bloomington, Ill.: 1996. p. 456. [Google Scholar]
- 24.Theiler M, Smith HH. The effect of prolonged cultivation in vitro upon the pathogenicity of yellow fever virus. J Exp Med. 1937;65(6):767–786. doi: 10.1084/jem.65.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theiler M, Smith HH. The use of yellow fever virus modified by in vitro cultivation for human immunization. J Exp Med. 1937;65(6):787–800. doi: 10.1084/jem.65.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theiler M, Smith HH. The use of yellow fever virus modified by in vitro cultivation for human immunization. J. Exp. Med. 65, 787-800 (1937). Rev Med Virol. 2000;10(1):6–16. discussion 3-5. [PubMed] [Google Scholar]
- 27.Santos AP, et al. Detection of Th1/Th2 cytokine signatures in yellow fever 17DD first-time vaccinees through ELISpot assay. Cytokine. 2008;42(2):152–5. doi: 10.1016/j.cyto.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki S, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol. 2007;81(1):215–28. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerin JL, Anderson NG. Purification of influenza virus in the K-II zonal centrifuge. 1969 doi: 10.1038/2211255a0. [DOI] [PubMed] [Google Scholar]
- 30.Reimer C, et al. Purification of large quantities of influenza virus by density gradient centrifugation. J Virol. 1967;1(6):1207–1216. doi: 10.1128/jvi.1.6.1207-1216.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luke CJ, Lakdawala SS, Subbarao K. In: Influenza vaccine-live in Vaccines. Plotkin SA, Orenstein W, Offit PA, editors. Saunders; 2012. [Google Scholar]
- 32.Wilkinson TM, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012;18(2):274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 33.Fiore AE, et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR. Recommendations and Reports: Morbidity and mortality weekly report. Recommendations and reports/Centers for Disease Control. 2010;59(RR-8):1. [PubMed] [Google Scholar]
- 34.Sasaki S, et al. Influence of prior influenza vaccination on antibody and B-cell responses. PLoS One. 2008;3(8):e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 36.Zeman AM, et al. Humoral and cellular immune responses in children given annual immunization with trivalent inactivated influenza vaccine. Pediatr Infect Dis J. 2007;26(2):107–115. doi: 10.1097/01.inf.0000253251.03785.9b. [DOI] [PubMed] [Google Scholar]
- 37.Pulendran B, Tang H, Manicassamy S. Programming dendritic cells to induce T(H)2 and tolerogenic responses. Nat Immunol. 2010;11(8):647–55. doi: 10.1038/ni.1894. [DOI] [PubMed] [Google Scholar]
- 38.Cao W, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6Kpathway. Nat Immunol. 2008;9(10):1157–64. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koyama S, et al. Plasmacytoid dendritic cells delineate immunogenicity of influenza vaccine subtypes. Sci Transl Med. 2010;2(25):25ra24. doi: 10.1126/scitranslmed.3000759. [DOI] [PubMed] [Google Scholar]
- 40.Geeraedts F, et al. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 2008;4(8):e1000138. doi: 10.1371/journal.ppat.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koyama S, et al. Differential role of TLR-and RLR-signaling in the immune responses to influenza A virus infection and vaccination. The Journal of Immunology. 2007;179(7):4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 42.Poland J, et al. Persistence of neutralizing antibody 30-35 years after immunization with 17D yellow fever vaccine. Bulletin of the World Health Organization. 1981;59(6):895. [PMC free article] [PubMed] [Google Scholar]
- 43.Hammarlund E, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9(9):1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 44.Mack TM, Noble J, Jr., Thomas DB. A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg. 1972;21(2):214–8. doi: 10.4269/ajtmh.1972.21.214. [DOI] [PubMed] [Google Scholar]
- 45.Crotty S, et al. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171(10):4969–73. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 46.Bachmann MF, et al. Induction of long-lived germinal centers associated with persisting antigen after viral infection. J Exp Med. 1996;183(5):2259–69. doi: 10.1084/jem.183.5.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McHeyzer-Williams M, et al. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12(1):24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bentebibel SE, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5(176):176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sallusto F, et al. From vaccines to memory and back. Immunity. 2010;33(4):451–63. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barouch DH, Letvin NL. CD8< sup>+</sup> cytotoxic T lymphocyte responses to lentiviruses and herpesviruses. Curr Opin Immunol. 2001;13(4):479–482. doi: 10.1016/s0952-7915(00)00244-2. [DOI] [PubMed] [Google Scholar]
- 51.Callan M, et al. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187(9):1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamann D, et al. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med. 1997;186(9):1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lauer GM, et al. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol. 2002;76(12):6104–6113. doi: 10.1128/JVI.76.12.6104-6113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lechner F, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191(9):1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410(6831):980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 56.Roos MTL, et al. Changes in the composition of circulating CD8+ T cell subsets during acute Epstein-Barr and human immunodeficiency virus infections in humans. Journal of Infectious Diseases. 2000;182(2):451–458. doi: 10.1086/315737. [DOI] [PubMed] [Google Scholar]
- 57.Tan LC, et al. A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. The Journal of Immunology. 1999;162(3):1827–1835. [PubMed] [Google Scholar]
- 58.Urbani S, et al. Virus-specific CD8+ lymphocytes share the same effector-memory phenotype but exhibit functional differences in acute hepatitis B and C. J Virol. 2002;76(24):12423–12434. doi: 10.1128/JVI.76.24.12423-12434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Leeuwen EM, et al. Proliferation requirements of cytomegalovirus-specific, effector-type human CD8+ T cells. The Journal of Immunology. 2002;169(10):5838–5843. doi: 10.4049/jimmunol.169.10.5838. [DOI] [PubMed] [Google Scholar]
- 60.Ahmed R, Akondy RS. Insights into human CD8+ T-cell memory using the yellow fever and smallpox vaccines. Immunol Cell Biol. 2011;89(3):340–345. doi: 10.1038/icb.2010.155. [DOI] [PubMed] [Google Scholar]
- 61.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–22. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 62.Akondy RS, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183(12):7919–30. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meier KC, et al. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS Pathog. 2009;5(10):e1000614. doi: 10.1371/journal.ppat.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belsher JL, et al. Fatal multiorgan failure due to yellow fever vaccine-associated viscerotropic disease. Vaccine. 2007;25(50):8480–5. doi: 10.1016/j.vaccine.2007.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pulendran B, et al. Case of yellow fever vaccine--associated viscerotropic disease with prolonged viremia, robust adaptive immune responses, and polymorphisms in CCR5 and RANTES genes. J Infect Dis. 2008;198(4):500–7. doi: 10.1086/590187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proceedings of the National Academy of Sciences. 1984;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sood R, et al. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics. 2000;154(2):787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu J, et al. The interferon-induced double-stranded RNA-activated protein kinase PKR will phosphorylate serine, threonine, or tyrosine at residue 51 in eukaryotic initiation factor 2α. Journal of Biological Chemistry. 1999;274(45):32198–32203. doi: 10.1074/jbc.274.45.32198. [DOI] [PubMed] [Google Scholar]
- 69.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7(2):213–21. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Proud CG. PKR: a new name and new roles. Trends Biochem Sci. 1995;20(6):241–6. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 71.Chen JJ, London IM. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci. 1995;20(3):105–8. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 72.Chefalo PJ, et al. Inhibition of protein synthesis in insect cells by baculovirus-expressed heme-regulated eIF-2 alpha kinase. J Biol Chem. 1994;269(41):25788–94. [PubMed] [Google Scholar]
- 73.Chefalo PJ, et al. Heme-regulated eIF-2alpha kinase purifies as a hemoprotein. Eur J Biochem. 1998;258(2):820–30. doi: 10.1046/j.1432-1327.1998.2580820.x. [DOI] [PubMed] [Google Scholar]
- 74.Harding HP, et al. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 75.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 76.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108(4):545–56. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 77.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14(1):20–8. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Crosby JS, et al. Regulation of hemoglobin synthesis and proliferation of differentiating erythroid cells by heme-regulated eIF-2alpha kinase. Blood. 2000;96(9):3241–8. [PubMed] [Google Scholar]
- 79.Anthony TG, et al. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 2004;279(35):36553–61. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- 80.Zhang P, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22(19):6681–8. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11(4):372–80. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 82.Zhang P, et al. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–74. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488(7413):670–4. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niwa M, Walter P. Pausing to decide. Proc Natl Acad Sci U S A. 2000;97(23):12396–7. doi: 10.1073/pnas.250476097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108(13):5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific Tcell responses. Science. 2012;337(6101):1553–6. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 89.Mazmanian SK, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 90.Abt MC, et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity. 2012 doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ribar TJ, et al. Cerebellar defects in Ca2+/calmodulin kinase IV-deficient mice. J Neurosci. 2000;20(22):RC107. doi: 10.1523/JNEUROSCI.20-22-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wayman GA, et al. Analysis of CaM-kinase signaling in cells. Cell Calcium. 2011;50(1):1–8. doi: 10.1016/j.ceca.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kitsos CM, et al. Calmodulin-dependent protein kinase IV regulates hematopoietic stem cell maintenance. J Biol Chem. 2005;280(39):33101–8. doi: 10.1074/jbc.M505208200. [DOI] [PubMed] [Google Scholar]
- 95.Anderson KA, Means AR. Defective signaling in a subpopulation of CD4(+) T cells in the absence of Ca(2+)/calmodulin-dependent protein kinase IV. Mol Cell Biol. 2002;22(1):23–9. doi: 10.1128/MCB.22.1.23-29.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Illario M, et al. Calmodulin-dependent kinase IV links Toll-like receptor 4 signaling with survival pathway of activated dendritic cells. Blood. 2008;111(2):723–31. doi: 10.1182/blood-2007-05-091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Juang YT, et al. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J Clin Invest. 2005;115(4):996–1005. doi: 10.1172/JCI200522854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schiemann B, et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293(5537):2111–4. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- 100.Moore PA, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285(5425):260–3. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 101.Vora KA, et al. Cutting edge: germinal centers formed in the absence of B cell-activating factor belonging to the TNF family exhibit impaired maturation and function. J Immunol. 2003;171(2):547–51. doi: 10.4049/jimmunol.171.2.547. [DOI] [PubMed] [Google Scholar]
- 102.Yan M, et al. Identification of a receptor for BLyS demonstrates a crucial role in humoral immunity. Nat Immunol. 2000;1(1):37–41. doi: 10.1038/76889. [DOI] [PubMed] [Google Scholar]
- 103.Rahman ZS, et al. Normal induction but attenuated progression of germinal center responses in BAFF and BAFF-R signaling-deficient mice. J Exp Med. 2003;198(8):1157–69. doi: 10.1084/jem.20030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3(9):822–9. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Darce JR, et al. Divergent effects of BAFF on human memory B cell differentiation into Ig-secreting cells. J Immunol. 2007;178(9):5612–22. doi: 10.4049/jimmunol.178.9.5612. [DOI] [PubMed] [Google Scholar]
- 106.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8(9):663–74. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 107.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426(6968):891–4. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 108.Ron D. Translational control in the endoplasmic reticulum stress response. J Clin Invest. 2002;110(10):1383–8. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol. 2001;13(3):349–55. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- 110.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 111.Shi Y, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18(12):7499–509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nakaya HI, Pulendran B. Systems vaccinology: its promise and challenge for HIV vaccine development. Curr Opin HIV AIDS. 2012;7(1):24–31. doi: 10.1097/COH.0b013e32834dc37b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shapira SD, Hacohen N. Systems biology approaches to dissect mammalian innate immunity. Curr Opin Immunol. 2011;23(1):71–7. doi: 10.1016/j.coi.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Amit I, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326(5950):257–63. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Root DE, et al. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–9. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 116.Amit I, Regev A, Hacohen N. Strategies to discover regulatory circuits of the mammalian immune system. Nature Reviews Immunology. 2011;11(12):873–880. doi: 10.1038/nri3109. [DOI] [PMC free article] [PubMed] [Google Scholar]