Abstract
The Pseudomonas aeruginosa heme utilization (Phu) system encodes several proteins involved in the acquisition of heme as an iron source. Once internalized heme is degraded by the iron-regulated heme oxygenase, HemO to biliverdin (BV) IXδ and β. In vitro studies have shown holo-PhuS transfers heme to the iron-regulated HemO. This protein-protein interaction is specific for HemO as PhuS does not interact with the α-regioselective heme oxygenase, BphO. Bacterial genetics and isotopic labeling (13C-heme) studies confirmed extracellular heme is converted to 13C-BVIX δ and β through the catalytic action of HemO. In an effort to further understand the role of PhuS similar studies were performed on the P. aeruginosa PAO1 ΔphuS and ΔphuS/ΔhemO strains. In contrast to wild type strain the absence of PhuS results in extracellular heme uptake and degradation via the catalytic action of HemO and BphO. At low heme concentrations loss of PhuS leads to inefficient extracellular heme uptake supported by the fact the mRNA levels of PhuR, HemO and BphO remain elevated when compared to the wild type PAO1. On increasing extracellular heme concentrations the elevated levels of PhuR, HemO and BphO allow “leaky uptake” and degradation of heme via HemO and BphO. Similarly, in the ΔphuS/ΔhemO strain the higher heme concentrations combined with elevated levels of PhuR and BphO leads to non-specific heme uptake and degradation by BphO. Thus we propose heme flux into the cell is driven by the catalytic action of HemO with PhuS acting as a “control-valve” to regulate extracellular heme flux.
INTRODUCTION
The ability of bacterial pathogens to acquire iron is essential for their survival and virulence. In addition to iron-siderophore scavenging mechanisms many bacteria encode systems for the utilization of heme as a source of iron (1–3). The opportunistic gram-negative pathogen Pseudomonas aeruginosa (PAO1) encodes two inter-dependent heme uptake systems, the Pseudomonas heme utilization (phu) system and the heme assimilation system (has) (4). The phu system encodes the outer-membrane receptor, PhuR, and the periplasmic transport system comprising a soluble receptor (PhuT) and the ATP-dependent permease (ABC-transporter), PhuUV. In contrast, the has system encodes a soluble hemophore (HasA) which is secreted to the extracellular media, extracts heme from hemoglobin and returns it to a TonB- dependent outer-membrane receptor (HasR). However, the has operon lacks a periplasmic uptake system and is presumed to utilize the phu encoded ABC-transporter (5–7).
Once internalized heme is then sequestered by the cytoplasmic heme binding protein PhuS. In a series of in vitro studies we have shown that the cytoplasmic heme binding protein PhuS forms a specific protein complex with the iron-regulated heme oxygenase, HemO (8–10). Heme is transferred to HemO for further degradation with the release of iron, CO, BVIXδ and BVIXβ (11, 12). P. aeruginosa encodes a second non-iron regulated heme oxygenase, BphO, which is directly upstream of the phytochrome two-component sensor kinase, BphP (13, 14). BphO catalyzes the degradation of heme to BVIXα that then acts as a chromophore for the sensor kinase BphP. Furthermore, in contrast to HemO the cytoplasmic heme binding protein PhuS does not interact with the non-iron regulated BphO (8). In keeping with the specific interaction of PhuS with HemO 13C-heme isotopic labeling studies confirmed extracellular heme is almost exclusively metabolized to 13C-BVIXδ and 13C-BVIX β (15). Furthermore, on deletion of the hemO gene no BVIX is detected confirming that in the presence of PhuS the α-selective BphO cannot compensate for the loss of HemO.
In the current study we have investigated the role of the cytoplasmic heme binding protein, PhuS in extracellular heme metabolism. Through a combination of bacterial genetics and 13C-heme isotopic labeling experiments we show that deletion of phuS leads to an uncoupling in the regulation and specificity of heme utilization. Based on these findings we propose a model whereby PhuS acts as a heme-dependent regulator of extracellular heme uptake whose regulatory function is coupled to the catalytic action of HemO.
RESULTS AND DISCUSSION
Heme repression of the wild type PAO1 Phu system
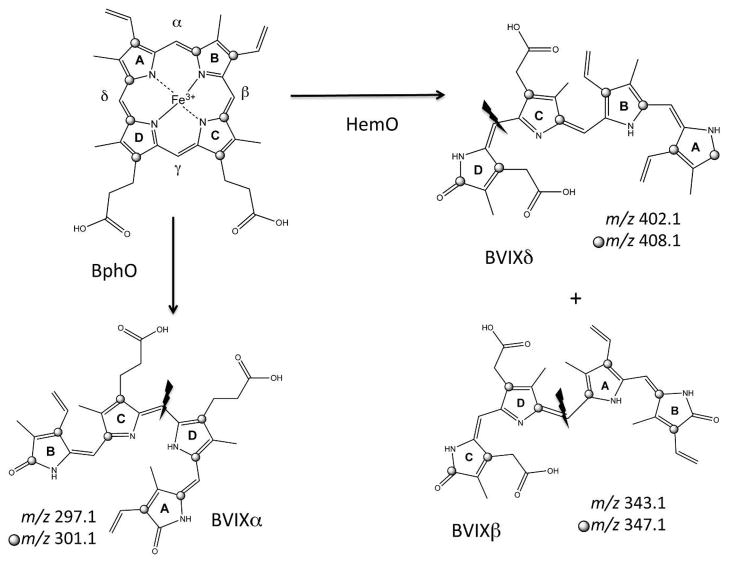
We have previously shown by isotopic 13C-heme labeling and LC-MS that we can distinguish BVIX derived from the degradation of intracellular biosynthesized heme (12C-heme) from that of extracellular heme uptake (13C-heme) (Scheme 1) (15). Furthermore, by utilizing tandem ESI-MS/MS the dominant fragment ions can distinguish BVIX isomers derived from oxidative cleavage by HemO (BVIXβ and BVIXδ) and that of BphO (BVIXα) (Scheme 1). Therefore by monitoring the 13C-heme metabolite profile along with the mRNA expression levels of the heme uptake proteins we have further characterized the role of PhuS in extracellular heme uptake.
Scheme 1. MS/MS Fragmentation patterns of the 13C-labeled and unlabeled BVIX isomers.
13C-labeling pattern marked by grey circles.
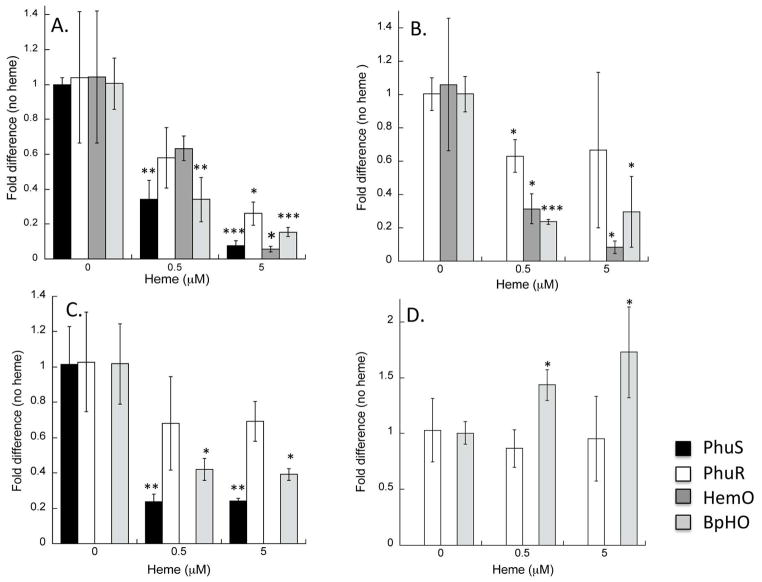
Samples for 13C-heme metabolite analysis were taken at 8 hours (mid-log phase) based on the growth curves of wild type PAO1 and mutant strains in the presence of 0, 0.5 or 5 μM heme (Supplementary Figure S1). In order to assess the regulatory effect of heme uptake and utilization on the mRNA levels of the outer-membrane receptor PhuR, PhuS and HemO qRT-PCR analysis at mid-log phase was performed. In addition mRNA levels of the non-iron regulated heme oxygenase, BphO were also monitored as a function of heme concentration. Addition of 0.5 μM heme to the media repressed the steady-state mRNA levels of phuR, phuS and hemO by 2 to 3-fold when normalized to the constitutively expressed omlA gene, whereas at 5 μM heme a 10-fold suppression of phuS and hemO was observed (Fig 1A). The heme-dependent decrease in the mRNA levels is most likely a direct consequence of iron released on heme cleavage leading to Fur-dependent repression of phuR, phuS and hemO. However, somewhat surprisingly a similar 5-fold decrease in mRNA levels was observed for the non-Fur regulated BphO suggesting the enzyme while not iron-regulated is heme dependent (Figure 1A).
Figure 1. Effect of heme on the expression of the heme utilization proteins.
RNA isolated from the indicated strains following 8 hours growth in media supplemented with heme was used as described in the Methods. mRNAs levels for PhuR (white bars), PhuS (black bars), HemO (dark grey bars), BphO (light grey bars) represent the standard deviation from at least three independent experiments in triplicate. (A) PAO1; (B) PAO1 ΔphuS; (C) PAO1 ΔhemO; and (D) PAO1 ΔphuS/ΔhemO. The indicated p-values were normalized to mRNA levels of the respective genes in the absence of heme where *p< 0.05, ** p< 0.005 or ***p<0.001.
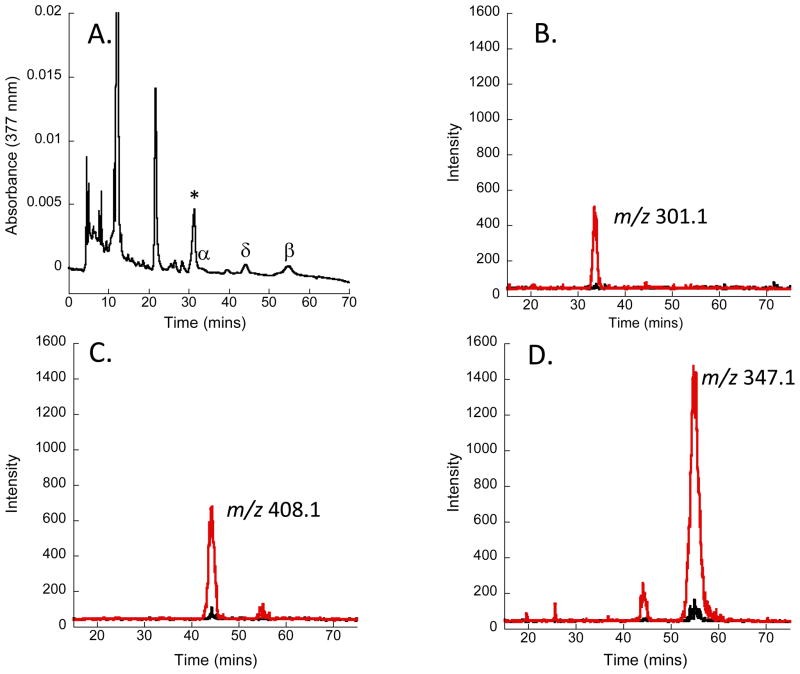
The heme dependent repression of proteins critical for heme uptake (PhuR) and utilization (PhuS and HemO) is consistent with the metabolite profile. Separation of the BVIX isomers by UPLC followed by MS/MS fragmentation of the parent ion and multiple reaction monitoring (MRM) allows for the identification of the major fragments for BVIXα, BVIXβ and BVIXδ derived from both extracellular (13C-heme) and intracellular (12C-heme) heme as a function of time. As shown in Figure 2 growth of the cells in 0.5 μM heme shows that the major fragments for each of the BVIX isomers is derived solely from 13C-heme. Furthermore, based on fragmentation patterns typical of BVIXδ (m/z 408.1) and BVIXβ (m/z 347.1) the major fraction arises from extracellular 13C-heme degraded by the catalytic action of HemO (Figure 2C and D) with a smaller fraction being degraded through BphO (Figure 2A). It was perhaps surprising to observe BVIXα derived from extracellular heme given the previous observation that holo-PhuS interacts with HemO but not BphO (8). However, as will be described below in the absence of HemO no 13C-heme is detected, confirming that extracellular heme uptake is driven by the catalytic action of HemO. Therefore, we believe the conversion to BVIXα is a result of heme “leakage” independent of the PhuS-HemO coupled reaction.
Figure 2. LC-MS/MS BVIX isomer fragmentation patterns for PAO1 wild type supplemented with 0.5 μM 13C-heme.
(A) HPLC analysis of BVIX isomers following extraction from the extracellular media. BVIX isomer peaks as marked. *Indicates a non-BVIX contaminant; (B) MS/MS fragmentation of 13C-BVIXα (red line) and 12C-BVIXα (black); (C) As in (B) for BVIXδ; (D) As in (B) for BVIXβ. LC-MS/MS was performed as described in the Methods with multiple reaction monitoring.
At the higher heme concentration the primary metabolite observed is BVIXα that on fragmentation has an m/z of 297.1 consistent with metabolism of intracellular 12C-heme by BphO (Figure 3). Therefore, at higher heme concentrations the increased heme uptake and subsequent iron release down regulates the expression levels of the heme uptake proteins. This down regulation of the heme uptake system allows for the redistribution of intracellular heme iron through the catalytic action of BphO. The current data is consistent with HemO being the major contributor to the metabolism of heme acquired from the extracellular environment.
Figure 3. LC-MS/MS BVIX isomer fragmentation patterns for PAO1 wild type supplemented with 5 μM 13C-heme.

(A) MS/MS fragmentation of 13C-BVIXα (red line) and 12C-BVIXα (black); (B) As in (A) for BVIXδ; (C) As in (A) for BVIXβ. LC-MS/MS was performed as described in the Methods with multiple reaction monitoring.
PhuS is an extracellular heme chaperone to the iron-regulated HemO
Interestingly, in the ΔphuS strain the addition of 0.5 μM heme to the media leads to the suppression of mRNA levels as observed in wild type PAO1 (Figure 1B). However, when compared to wild type PAO1 the ΔphuS strain shows increased mRNA levels for PhuR, HemO and BphO when grown with or without heme (Supplementary Figure S2). The increased mRNA levels may arise either as a consequence of increased transcription or alternatively post-transcriptional stabilization of the mRNAs, perhaps via the heme regulated PrrH (Pseudomonas RNA regulated by Heme) sRNA network (16, 17).
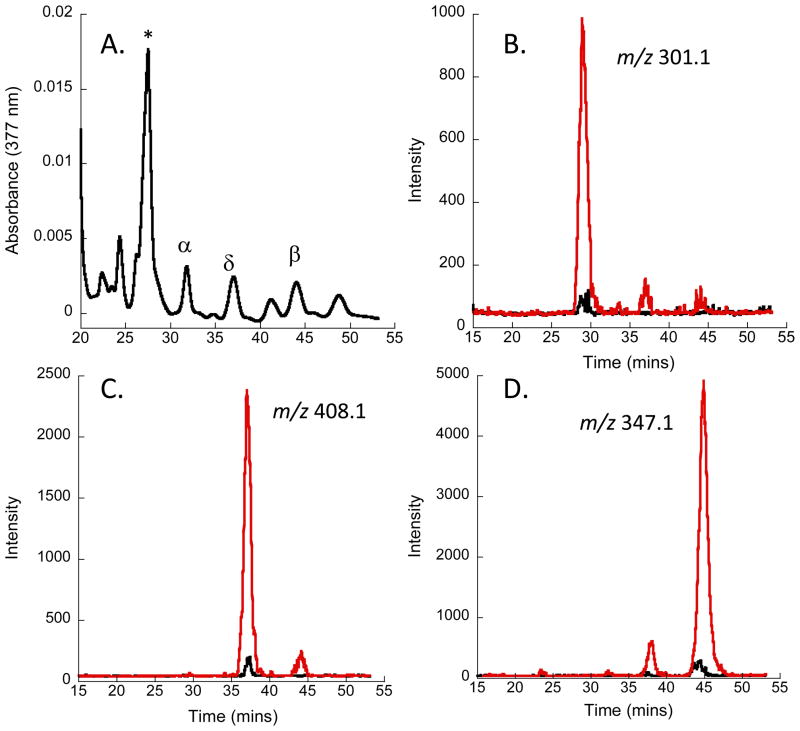
Perhaps surprisingly the increased mRNA levels for PhuR, HemO and BphO in the ΔphuS strain when compared to the wild type PAO1 at the lower heme concentration (0.5 μM heme) is not reflected in the metabolite profile where there is no evidence of BVIX derived from either extracellular or intracellular heme (Supplementary Figure S3). In contrast to the wild type PAO1 at high heme concentration where extracellular heme uptake is shut down (Figure 3), the ΔphuS strain yielded BVIX metabolites derived from extracellular heme as judged by the fragment ions at m/z 408.1, 347.1 and 301.1, for 13C-BVIXδ, 13C-BVIXβ and 13C-BVIXα, respectively (Figure 4). Interestingly, in the absence of PhuS heme is shunted through HemO and BphO consistent with previous in vitro studies suggesting PhuS acts as a specific chaperone to the iron-regulated HemO (8). Therefore, we propose PhuS is required for the efficient uptake of heme and coupled to the catalytic action of HemO, regulates the flux of heme into the cell. This data is supported by previous genetic studies of deletions of phuS homologs in Yersinia entercolitica and Shigella dysenteriae where the strains were unable to efficiently utilize heme at low concentrations and showed a heme toxicity phenotype at high heme concentrations (18, 19).
Figure 4. LC-MS/MS BVIX isomer fragmentation patterns for PAO1 ΔphuS strain supplemented with 5 μM 13C-heme.
(A) HPLC analysis of BV isomers following extraction from the extracellular media. BVIX isomer peaks as marked. *Indicates a non-BVIX contaminant; (B) MS/MS fragmentation of 13C-BVIXα (red line) and 12C-BVIXα (black); (C) As in (B) for BVIXδ; (D) As in (B) for BVIXβ. LC-MS/MS was performed as described in the Methods with multiple reaction monitoring.
The current data identifies PhuS as a heme titratable regulator of extracellular heme flux into the cell through its interaction with HemO, and that in its absence the inability to control extracellular heme flux leads directly or indirectly to a disruption in regulation of the heme uptake system.
HemO drives the metabolic flux of extracellular heme into the cell
In a previous study we have shown deletion of the hemO gene effectively shuts down extracellular heme uptake (15). In the current study we further examined the loss of HemO in the context of the expression levels of the outer membrane receptor, PhuR and the heme chaperone, PhuS. In comparison to wild type PAO1 the expression of PhuR in the hemO deletion was not significantly suppressed on the addition of 5 μM extracellular 13C-heme to the media (Figure 1C). Furthermore, the HemO and BphO mRNA levels while suppressed when grown in 0.5 μM heme were not further suppressed at the higher heme concentration. As observed in previous studies at either 0.5 or 5 μM heme no BVIXδ or BVIXβ was detected due to the loss of HemO (Supplementary Figure S4 and Figure 5) (15). Furthermore, barely detectable BVIXα (m/z 297.1) levels were seen at either heme concentration (Supplementary Figure S4 and Figure 5). Therefore, consistent with our previous studies the catalytic activity of HemO is required for driving extracellular heme uptake (15).
Figure 5. LC-MS/MS BVIX isomer fragmentation patterns for PAO1 ΔhemO strain supplemented with 5 μM 13C-heme.
(A) HPLC analysis of BV isomers following extraction from the extracellular media. BVIX isomer peaks as marked. *Indicates a non-BVIX contaminant; (B) MS/MS fragmentation of 13C-BVIXα (red line) and 12C-BVIXα (black); (C) MS/MS fragmentation of 13C-BVIXδ (red line) and 12C-BVIXδ (black); MS/MS fragmentation of 13C-BVIXβ (red line) and 12C-BVIXβ (black). LC-MS/MS was performed as described in the Methods with multiple reaction monitoring.
The metabolite profiles of the ΔphuS and ΔhemO strains taken together suggest the metabolic flux of heme into the cell is driven by the catalytic action of HemO, with PhuS acting as the “control-valve” in regulating uptake through its interaction with HemO. To further test this hypothesis we determined the BVIX metabolite profile of the ΔphuS/ΔhemO strain in the presence of both low (0.5 μM) and high (5 μM) extracellular heme levels. At 0.5 μM heme the loss of PhuS results in inefficient heme uptake as seen by the low level of BVIXα derived from extracellular heme (m/z 301.1) (Supplementary Figure S5). The inability to effectively utilize heme is reflected in the elevated mRNA levels of PhuR and BphO in an effort to compensate for the loss of both PhuS and HemO (Figure 1D and Supplementary Figure S2). Similar to the ΔphuS strain at the higher heme concentration the elevated PhuR and BphO mRNA levels appear compensate to some degree for the loss of PhuS with the conversion of 13C-heme to BVIXα (Figure 6).
Figure 6. LC-MS/MS BVIX isomer fragmentation patterns for PAO1 ΔphuS/ΔhemO strain supplemented with 5 μM 13C-heme.
(A) HPLC analysis of BV isomers following extraction from the extracellular media. BVIX isomer peaks as marked. *Indicates a non-BVIX contaminant; (B) MS/MS fragmentation of 13C-BVIXα (red line) and 12C-BVIXα (black); (C) As in (B) for BVIXδ; (D) As in (B) for BVIXβ. LC-MS/MS was performed as described in the Methods with multiple reaction monitoring.
CONCLUSIONS
Based on the previous in vitro characterization of a specific PhuS-HemO driven heme transfer (9, 20, 21) and the current studies we propose a model where the equilibrium between apo-PhuS and holo-PhuS acts as the “control-valve” regulating heme flux through HemO. In such a model at low heme concentrations the flux of heme from PhuS to HemO drives the kinetic equilibrium in the direction of apo-PhuS as heme is transferred to HemO (Scheme 2A). In contrast at higher heme levels the repression of HemO on release of iron shifts the equilibrium toward holo-PhuS shutting down extracellular heme uptake (Scheme 2B). Consistent with the decreased heme flux the mRNA levels of PhuR, PhuS, and HemO are down regulated in a heme dependent manner (Figure 2A). Similarly, in the absence of HemO the rapid shift in the equilibrium from apo-PhuS to holo-PhuS shuts down extracellular heme uptake as no extracellular 13C-BVIX metabolites are detected at either low or high heme concentrations.
Scheme 2.

Proposed model for the regulation of heme uptake by the PhuS-HemO two-component system.
Consistent with the model of PhuS acting as a “control-valve” for heme flux into the cell deletion of phuS leads to inefficient heme uptake at low heme levels, which can be overcome at higher heme concentrations (Supplementary Figure S3 and Figure 4). In this scenario at high heme levels the lack of PhuS leads to “leaky uptake” and degradation via the catalytic action of both HemO and BphO, which under normal conditions would not degrade exogenous heme (Scheme 2C). The current model is consistent with previous studies on the genetic knockouts of the phuS homologs in Shigella dysenteria and Yersinia entercolitica that were inefficient at utilizing heme at low concentration, whereas higher heme concentrations induced a heme toxicity phenotype (18, 19). The heme toxicity is presumably a consequence of free heme being released into the cell on the uncoupling of the Phu-HemO relay.
Interestingly, in the ΔphuS strain the inability to efficiently utilize extracellular heme is reflected in the increased mRNA levels of PhuR, HemO and BphO over that of wild type PAO1 (Supplementary Figure S2). The mechanism by which PhuS directly or indirectly affects the expression levels of PhuR and HemO is unknown. This may merely be an indirect effect of inefficient heme utilization resulting in a “compensatory” up regulation of PhuR, HemO and BphO. However it is interesting that the cytoplasmic heme binding protein PhuS and the previously characterized tandem iron-regulated small-RNA’s PrrF1 and PrrF2, are found in the same locus and only in the pathogenic P. aeruginosa strains (16, 22, 23). The expression of the PrrF’s is negatively regulated by Fur and thus de-repressed under iron-starvation. PrrF RNAs bind to complementary sequences of their target RNAs causing RNAseE and Hfq-dependent RNA degradation. Recently, the unique tandem arrangement of prrF1 and prrF2 was shown to encode an over-lapping non-coding RNA PrrH, whose expression is repressed by heme (17). Although the specific target RNAs of PrrH have not yet been identified, transcriptional analysis of the ΔprrF/H strain shows significant overlap with respect to the differentially transcribed genes when compared to the phuS deletion strain. It is therefore intriguing to speculate that the unique tandem arrangement of prrF/H reflects a functional heme-dependent regulatory link between extracellular heme flux and the heme-dependent regulation of PrrH. Coupling the metabolic flux of heme through PhuS-HemO to the regulatory RNA network would provide a novel mechanism for bacteria to rapidly respond and adapt to sudden changes in heme levels. One scenario is that the equilibrium between apo-PhuS and holo-PhuS is somehow involved in the heme dependent regulation of PrrH. Potential regulatory mechanisms may involve apo-PhuS acting as an anti-terminator in regulating PrrH transcription or in modulating the stability of PrrH mRNA. Collaborative studies to address the target RNAs and potential link between PrrH and PhuS in the post-transcriptional regulation of the heme uptake system are ongoing.
METHODS
Chemicals, media bacterial strains and growth conditions
Liquid chromatography (LC)-mass spectrometry (MS) grade formic acid and acetone were obtained from Sigma Aldrich. Hemin was obtained from Frontier Scientific. Luria-Bertani (LB) broth was used for culture and maintenance of bacterial strains. All bacterial strains used in these studies are listed in Supplementary Table S1. Strains of wild type and mutant PAO1 strains were maintained on Pseudomonas Isolation Agar (PIA) (BD Biosciences) or LB plates with 50 μg/mL gentamicin (Gm). PAO1 ΔhemO and phuS/ΔhemO deletion strains were maintained on PIA plates containing 50 μg/mL gentamicin. For bacterial growth under iron limiting conditions Pseudomonas aeruginosa was first grown overnight in LB and sub-cultured into fresh M9 minimal media (42 mM Na2HPO4, 24 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 2% glucose, 0.5 μg/mL thiamine). The cultures were grown for a further 6 hours and used to inoculate fresh M9 cultures (30 mL) to an optical density at 600 nm of 0.08. The cultures were grown for 8 hours at 37°C with shaking (200 rpm). The resulting supernatant and cells were stored at −20°C until needed. Growth curves were obtained under the same media conditions on a Bioscreen C MBR (Growth Curves USA) with continuous shaking at 37°C. The OD600 was taken every 30 min over a 24 hour period. Heme stocks were prepared in 0.1 M sodium hydroxide and buffered to pH 7.4 with 1 M Tris-HCl (pH 7.4) and the concentration determined by pyridine hemochrome assay (24).
Preparation and isolation of 12C and 13C-heme
δ-Aminolevulinic acid (ALA) or [4-13C]-δ-ALA was used as a biosynthetic precursor to produce 12C- or 13C-labeled heme, respectively. Unlabeled δ-ALA was purchased from Sigma and [4-13C]M δ-ALA from Cambridge Isotope Laboratories. 13C-heme was prepared by a slight modification of the method described by Rivera and Walker (25). Expression of cytochrome b5 in the presence of 1 mM δ-ALA induces heme biosynthesis with the resulting heme being captured by the over-expressed apo-cytochrome b5. Cytochrome b5 was expressed in E. coli BL21(DE3) and following harvesting and lysis the cell debris was removed by centrifugation. The resulting supernatant was applied to a Q-Sepharose column (3 × 10 cm) equilibrated in 50 mM Tris-HCl (pH 7.4) containing 50 mM NaCl. The Q-sepharose column was washed (5–10 column volumes) with equilibration buffer and the protein eluted in 50 mM Tris-HCl (pH 7.4) containing 350 mM NaCl. Heme was extracted from purified cytochrome b5 by the acid-butanone method (26). An aliquot of the heme following extraction was taken and the final yield calculated by the pyridine hemochrome assay (24). The 13C-heme labeling pattern obtained with [4-13C]-δ-ALA is shown in Scheme 1
Extraction of BVIX isomers from wild type and mutant PAO1 supernatants
Analysis of the BVIX products was performed as previously described with the following modifications (15). PAO1 wild type or mutant strains were grown overnight in LB-media and sub cultured into M9 minimal media (25 mL) to an OD600 of 0.2. The culture was grown for a further 6 hours and used to inoculate a fresh 50 mL M9 culture to a starting OD600 of 0.08. 12C- or 13C-heme was added to a final concentration of 0.5 or 5 GM. Cells were grown for 8 hours at 37°C in a 250 mL baffled flask with shaking (200 rpm) and harvested by centrifugation (20 minutes at 6000 × g). The supernatant was acidified to ~pH 3.0 with the addition of 10% trifluoroacetic acid (TFA). The BVIX isomers were purified over a C18 SepPak with an additional wash step of methanol, 0.1% TFA (50:50) prior to the final 450 μL methanol wash. The BVIX isomers were eluted in 650 μL methanol and dried under air prior to LC-MS/MS analysis.
LC-MS/MS Analysis of BVIX isomers
Samples were resuspended in 10 μL DMSO and diluted to 40 μL with the mobile phase (Acetone: 20 mM formic acid (50:50, v/v)) and filtered through a 0.45 Gm PTFE syringe. The BVIX isomers (10 μl) were separated and analyzed by LC-tandem Mass Spectrometry (MS/MS) (Waters TQD triple quadrupole mass spectrometer with AQUITY H-Class UPLC). BVIX isomers were separated on a reverse phase Phenomenex Ultracarb 5 GM ODS analytical column (4.6 × 250 mm) at a flow rate of 0.6 mL/min and detected at 377 nm. Fragmentation patterns of the parent ions at 583.21 (12C-BVIX) and 591.21 (13C-BVIX) were analyzed using multiple reaction monitoring (MRM). The source temperature was set to 150°C, the capillary voltage to 3.30 kV and the cone voltage to 72V. The collision energy was set to 34 V for BVIXα, 30 V for BVIXδ and 36 V for BVIXβQ respectively.
RNA Extraction and Isolation
PAO1 wild type or mutant strains were grown as described previously for the BVIX metabolite analysis. Samples (1 mL) were taken from cultures and harvested at 13,000 rpm for 10 minutes. The supernatant was discarded and total RNA was extracted from the cells using the RNAeasy mini spin columns (Qiagen). To remove contaminating DNA the column was treated with RNase-free DNaseI (New England Biolabs). RNA was eluted in 40 μL RNase-free water and any remaining DNA removed by incubating with RNase free-DNaseI for 2 hours at room temperature. Samples were precipitated overnight at −20°C with the addition of 5 μL 3M sodium acetate (pH 5.5) and 150 μL 100% ethanol. RNA was pelleted, washed with cold ethanol and resuspended in 50 μL RNase-free water. RNA was quantified by reading the absorbance at 260 nm (A260 of 1.0 = 40 μg/mL) and adjusted to 50 ng/μL.
Expression studies
cDNA from wild type or mutant PAO1 cells was prepared from 50 ng RNA using the ImPromII Reverse Transcription System (Promega) as follows; Random Primer (0.5 μg in 1 μL) was added to 1 μL RNA (50 ng) and adjusted to a total volume of 5 μL with RNAse-free water. The sample was incubated at 65°C for 10 minutes and then cooled on ice. Master mixture (4 μL 5X Buffer, 4 μL MgCl2, 1 μL dNTPs, 1 μL Reverse Transcriptase, 5 μL RNase-free water) was added to the priming reaction and the samples were incubated at 25°C for 5 minutes, 42°C for 1 hour and 70°C for 15 minutes. qRT-PCR reactions were carried out on a Light Cycler 480 (Roche) using the TaqMan Gene Expression Master Mix (Life Technologies). CT values were calculated and relative amounts cDNA were normalized by dividing the expression values by the relative amounts of the constitutively expressed gene omlA. All primers and probes used in the studies are listed in Supplementary Table S2.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grant number AI55192 to AW. The authors would like to thank A. Oglesby-Sherrouse and M. Kane for helpful discussions with regard to qRT-PCR and LC-MS methods, respectively.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 2.Wilks A, Burkhard KA. Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007;24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 3.Wilks A, Barker KD. Mechanism of heme uptake and utilization in bacterial pathogens. In: Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science. 1. World Scientific; Singapore: 2011. pp. 357–398. [Google Scholar]
- 4.Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology (Reading, England) 2000;146(Pt 1):185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 5.Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 6.Letoffe S, Deniau C, Wolff N, Dassa E, Delepelaire P, Lecroisey A, Wandersman C. Haemophore-mediated bacterial haem transport: evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol Microbiol. 2001;41:439–450. doi: 10.1046/j.1365-2958.2001.02530.x. [DOI] [PubMed] [Google Scholar]
- 7.Krieg S, Huche F, Diederichs K, Izadi-Pruneyre N, Lecroisey A, Wandersman C, Delepelaire P, Welte W. Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc Natl Acad Sci U S A. 2009;106:1045–1050. doi: 10.1073/pnas.0809406106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lansky IB, Lukat-Rodgers GS, Block D, Rodgers KR, Ratliff M, Wilks A. The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase. J Biol Chem. 2006;281:13652–13662. doi: 10.1074/jbc.M600824200. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill MJ, Bhakta MN, Fleming KG, Wilks A. Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO) Proc Natl Acad Sci U S A. 2012;109:5639–5644. doi: 10.1073/pnas.1121549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block DR, Lukat-Rodgers GS, Rodgers KR, Wilks A, Bhakta MN, Lansky IB. Identification of two heme-binding sites in the cytoplasmic heme-trafficking protein PhuS from Pseudomonas aeruginosa and their relevance to function. Biochemistry. 2007;46:14391–14402. doi: 10.1021/bi701509n. [DOI] [PubMed] [Google Scholar]
- 11.Friedman J, Lad L, Li H, Wilks A, Poulos TL. Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry. 2004;43:5239–5245. doi: 10.1021/bi049687g. [DOI] [PubMed] [Google Scholar]
- 12.Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. Homologues of Neisserial Heme Oxygenase in Gram-Negative Bacteria: Degradation of Heme by the Product of the pigA Gene of Pseudomonas aeruginosa. J Bacteriol. 2001;183:6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegele R, Tasler R, Zeng Y, Rivera M, Frankenberg-Dinkel N. The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J Biol Chem. 2004;279:45791–45802. doi: 10.1074/jbc.M408303200. [DOI] [PubMed] [Google Scholar]
- 14.Barkovits K, Harms A, Benkartek C, Smart JL, Frankenberg-Dinkel N. Expression of the phytochrome operon in Pseudomonas aeruginosa is dependent on the alternative sigma factor RpoS. FEMS Microbiol Lett. 2008;280:160–168. doi: 10.1111/j.1574-6968.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- 15.Barker KD, Barkovits K, Wilks A. Metabolic Flux of Extracellular Heme Uptake in Pseudomonas aeruginosa Is Driven by the Iron-regulated Heme Oxygenase (HemO) J Biol Chem. 2012;287:18342–18350. doi: 10.1074/jbc.M112.359265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oglesby AG, Farrow JM, 3rd, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oglesby-Sherrouse AG, Vasil ML. Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS One. 2010;5:e9930. doi: 10.1371/journal.pone.0009930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin- specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 19.Wyckoff EE, Lopreato GF, Tipton KA, Payne SM. Shigella dysenteriae ShuS promotes utilization of heme as an iron source and protects against heme toxicity. J Bacteriol. 2005;187:5658–5664. doi: 10.1128/JB.187.16.5658-5664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhakta MN, Wilks A. The mechanism of heme transfer from the cytoplasmic heme binding protein PhuS to the delta-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry. 2006;45:11642–11649. doi: 10.1021/bi060980l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lansky IB, Lukat-Rodgers GS, Block D, Rodgers KR, Ratliff M, Wilks A. The Cytoplasmic Heme-binding Protein (PhuS) from the Heme Uptake System of Pseudomonas aeruginosa Is an Intracellular Heme-trafficking Protein to the {delta}-Regioselective Heme Oxygenase. J Biol Chem. 2006;281:13652–13662. doi: 10.1074/jbc.M600824200. [DOI] [PubMed] [Google Scholar]
- 22.Vasil ML. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals. 2007;20:587–601. doi: 10.1007/s10534-006-9067-2. [DOI] [PubMed] [Google Scholar]
- 23.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuhrop JH, Smith KM, editors. Porphyrins and Metalloporphyrins. Elsevier; Amsterdam: 1975. pp. 804–807. [Google Scholar]
- 25.Rivera M, Walker FA. Biosynthetic preparation of isotopically labeled heme. Anal Biochem. 1995;230:295–302. doi: 10.1006/abio.1995.1477. [DOI] [PubMed] [Google Scholar]
- 26.Teale FW. Cleavage of the haem-protein link by acid methylethylketone. Biochim Biophys Acta. 1959;35:543. doi: 10.1016/0006-3002(59)90407-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.