Abstract
Self-assembled monolayers (SAMs) of alkanethiolates on gold are chemically defined substrates that can be used to evaluate the effects of an immobilized biomolecule. However, the types of biomolecules that can influence stem cell behavior are numerous and inter-related, and efficient experimental formats are a critical need. Here we employed a SAM array technology to investigate the effects of multiple, distinct peptides and peptide combinations on human mesenchymal stem cell (hMSC) behavior. Specifically, we characterized the conjugation of peptide mixtures to SAM arrays and then investigated the combined effects of a bone morphogenic protein receptor-binding peptide (BR-BP), a heparin proteoglycan-binding peptide (HPG-BP), and varied densities of the integrin-binding ligand Gly-Arg-Gly-Asp-Ser-Pro (GRGDSP) on hMSC surface coverage and alkaline phosphatase activity. Results indicate that an amine reactive fluorescent probe can be used to characterize peptide composition after immobilization in SAM array spots. Furthermore, hMSC response to BR-BP and HPG-BP is dependent on GRGDSP density and at day 7, hMSC alkaline phosphatase expression is highly dependent on GRGDSP density. Taken together, we demonstrate how a SAM array approach can be used to probe the combinatorial effects of multiple peptides and motivate further investigations into potential synergies between cell adhesion and other bioactive peptides.
Introduction
Stem cell behavior can be influenced by a variety of insoluble biomolecules present in the local microenvironment, including those in the extracellular matrix (ECM). For example, stem cells in culture are significantly influenced by peptide ligands derived from ECM proteins, including fibronectin-derived,1–8 laminin-derived,6,9–11 and collagen-derived12 ligands. In addition, recent studies demonstrate that immobilized peptide ligands that mimic growth factors,13–16 growth factor-binding sites,17 and proteo-glycan-binding sites2,4,18–20 can each significantly influence stem cell behavior. Collectively, the insoluble biomolecules that have been shown to influence stem cell behavior are copious and interrelated, which suggests a need for high throughput experimental formats to probe the effects of immobilized biomolecules. In response to this need, investigators have developed a series of high-throughput experimental formats to probe for the effects of biomolecules on stem cell behavior. Micro-spotting,21,22 lithography,23 microcontact printing,24 and microfluidics25–27 techniques can each create spatially patterned surfaces for cell culture, enabling efficient measurement of cell–ECM interactions. However, it remains challenging to directly relate immobilized biomolecules to changes in stem cell behavior, as ECM-derived signals are often presented to stem cells in a complex and poorly defined signaling context.28 Thus, there remains a critical need for experimental formats that expose stem cells to specific ECM-derived biomolecules in an efficient and well-defined manner.
Chemically well-defined cell culture substrates are a particularly attractive experimental format, as they allow for specific biomolecules to be presented to stem cells in a well-defined signaling context.29 In particular, self-assembled monolayers (SAMs) of alkanethiolates on gold have a series of unique properties that are well-suited for stem cell culture. First, SAMs terminated with oligo(ethylene glycol) groups are inert to nonspecific protein adsorption and cell adhesion, providing a “bio-inert” background for presentation of reactive functional groups. Second, peptide ligands can be covalently linked to functional groups on otherwise bio-inert SAMs at controllable density, resulting in control over the ligand identity and density at the cell–substrate interface. Finally, recent studies from our group2,7,30,31 and others32 indicate that SAMs can be spatially patterned to generate SAM arrays, in which each array spot exposes cells to a specific ligand identity and density. These SAM arrays have enabled efficient screening for ligands that influence stem cell self-renewal,32 as well as discovery of new biological phenomena (e.g. unexpected phenotypic similarities30 and context-dependent growth factor signaling31). To date, SAM arrays have focused on exposing stem cells to individual peptide ligands or limited pairwise comparisons, while the combined influence of two or more peptide ligands has typically not been systematically explored.
This paper describes a SAM array system in which multiple peptide ligands are presented to stem cells in a systematic and controllable fashion. Specifically, we used an elastomeric stencil approach to create arrays of alkanethiolate SAMs, in which specific combinations of peptide ligands were immobilized in each spot. The peptide ligands included a cell adhesion peptide (GRGDSP), a bone morphogenetic protein receptor-binding peptide (BR-BP), and a heparin proteoglycan binding peptide (HPG-BP) (Fig. 1), as well as scrambled versions of each of these peptides (6 total peptides). Results demonstrate that multiple, distinct peptides can be presented to hMSCs simultaneously, with control over ligand identity and density. In addition, both individual ligands and their combinations significantly influenced hMSC attachment, proliferation, and alkaline phosphatase activity. Mechanisms governing proliferation and lineage-specific hMSC differentiation were not characterized in detail here, as the goal of this study was to describe an efficient hMSC screening system. However, these results demonstrate the utility of a multi-peptide screening platform, and provide interesting biological insights that suggest further mechanistic studies of hMSC proliferation and osteogenic differentiation.
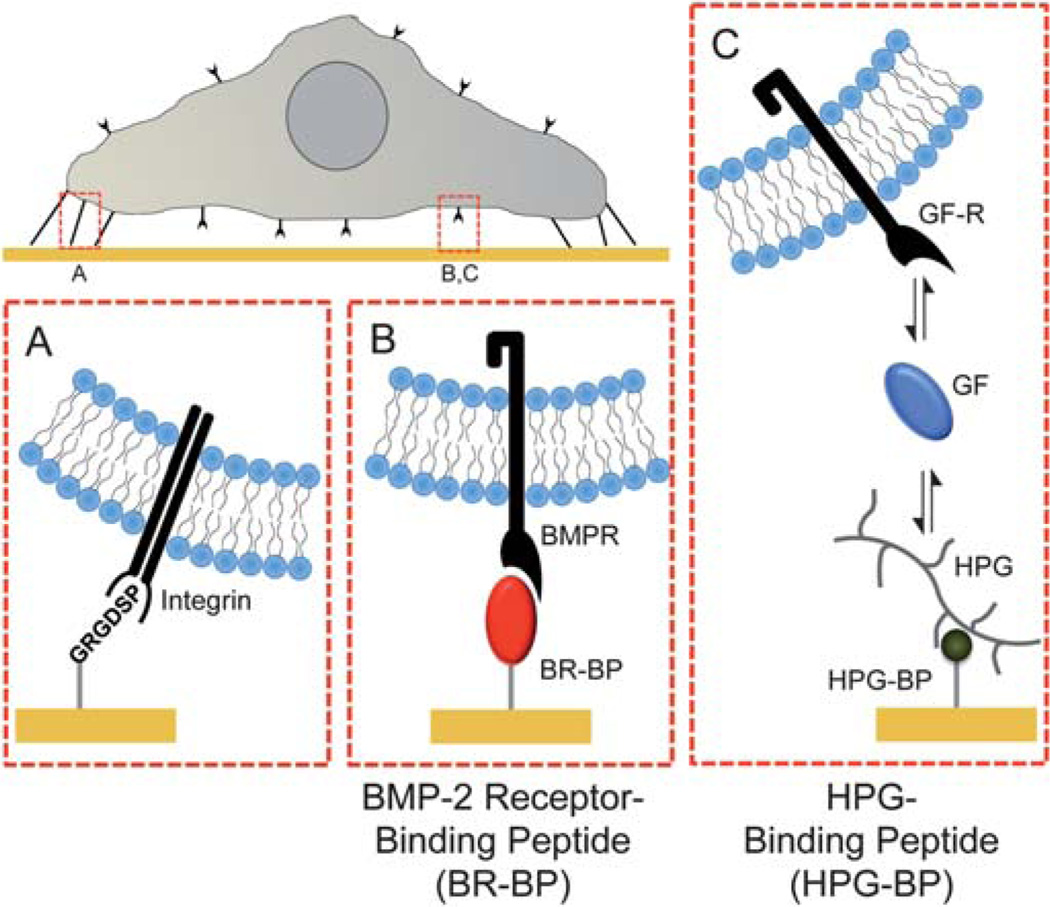
Fig. 1.
Chemically defined SAMs substrates presenting peptides that bind (A) integrins via cell adhesion peptides (Gly-Arg-Gly-Asp-Ser-Pro, GRGDSP), (B) BMP receptors via BMP receptor-binding peptides (BR-BP) and (C) heparin proteoglycans via heparin proteoglycan-binding peptides (HPG-BP). In this approach SAMs were designed to minimize the effects of non-specific protein adsorption via oligo(ethylene glycol) moieties that limit protein–surface interaction.
Experimental methods
Materials
Carboxylic acid-capped hexa(ethylene glycol) undecanethiol (HS–C11–(O–CH2–CH2)6–O–CH2–COOH) (referred to herein as “HS–C11–EG6–COOH”), was purchased from Prochimia (Sopot, Poland). 11-Tri(ethylene glycol)-undecane-1-thiol (HS–C11–(O–CH2–CH2)3–OH) (referred to herein as “HS–C11–EG3– OH”) was synthesized as described elsewhere.33 Fmoc-protected amino acids and Rink amide MBHA peptide synthesis resin were purchased from NovaBiochem (San Diego, CA). Hydroxybenzotriazol (HOBt) was purchased from Advanced Chemtech (Louisville, KY). Diisopropylcarbodiimide (DIC) was purchased from Anaspec (San Jose, CA). N-Hydroxysuccinimide (NHS), n-(3–dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), sodium dodecyl sulfate (SDS), trifluoroacetic acid (TFA), diethyl ether, and deionized ultrafiltered water (DIUF H2O) were purchased from Fisher Scientific (Fairlawn, NJ). Triisopropylsilane (TIPS), piperidine, dimethylformamide (DMF), acetone, hexanes, and acetonitrile were purchased from Sigma-Aldrich (St. Louis, MO). Absolute ethanol (EtOH) was purchased from AAPER Alcohol and Chemical Co. (Shelbyville, KY). All purchased items were of analytical grade and used as received. Thin films of 100 Å Au 〈111〉, 20 Å Ti on 1″ × 3″ × 0.040″ glass were purchased from Platypus Technologies, LLC (Madison, WI. Cat. no. AU.0100.ALSI).
Peptide synthesis
Standard solid phase Fmoc-peptide synthesis (Fmoc SPPS) was performed using a 316c automated peptide synthesizer (C S Bio, Menlo Park, CA). Rink amide MBHA resin was used as the solid phase, and HOBt and DIC were used for amino acid activation and coupling. After coupling the final amino acid, a 4 h incubation in TFA, TIPS, and DIUF H2O (95 : 2.5 : 2.5) released the peptide from resin and removed protecting groups. Released peptide was extracted from the TFA–TIPS–DIUF H2O cocktail via precipitation in cold diethyl ether. Lyophilized peptides were analyzed using matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry with a Bruker Reflex II (Billerica, MA). The purity of synthesized peptides was evaluated via HPLC using a C18 analytical column (Shimadzu, Kyoto, Japan) with a gradient of 0–70% H2O + 0.1% TFA/acetonitrile and a flow rate of 0.9 ml min−1. GWGGRGDSP and GWGGRGESP adhesion and mutant peptides were synthesized with tryptophan-bearing spacers to aid in determination of peptide concentration via UV/Vis. Peptide concentrations were determined by absorbance at 280 nm using extinction coefficients outlined by Gill and von Hippel.34 HPG-BP and BR-BP were synthesized using the published sequences Lys-Arg-Thr-Gly-Glu-Tyr-Lys-Leu (KRTGQYKL),35,36 and Lys-Ile-Pro-Lys-Ala-Ser-Ser-Val-Pro-Thr-Glu-Leu-Ser-Ala-Ile-Ser-Thr-Leu-Tyr-Leu (KIPKASSVPTELSAISTLYL),37 respectively. Scrambled versions of HPG-BP and BR-BP peptides (TYRKKGLQ and EPPSIATSYKLALKTSIVSL, respectively) were designed by rearranging the amino acids in the published sequences but maintaining a balance of hydrophilic and hydrophobic amino acids throughout the sequence to prevent difficulties during synthesis. Additionally, HPG-BP, BR-BP, and scrambled peptides were synthesized with an N-terminal tri-glycine spacer to increase peptide spacing away from the SAM substrate.
Fabrication of elastomer wells
Elastomeric stencils containing arrays of wells were created using soft lithography.38,39 Briefly, master molds containing arrays of 1100 µm diameter posts were fabricated from SU-8 (Microchem, Newton, MA) spin-coated silicon wafers using conventional photolithography techniques. Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, Midland, MI) was prepared by mixing a 10 : 1 ratio of base/curing agent (w/w) followed by degassing for ~30 min. The degassed mixture was cast over the mold and cured for 4 h at 85 °C. Following curing, PDMS stencils were removed from molds and cleaned in hexanes using an overnight Soxhlet extraction.40 After cleaning, stencils were placed in vacuo to remove residual solvent from the Soxhlet extraction process.
Surface preparation and array fabrication
Gold slides (3″ × 1″) were placed into a 150 mm glass Petri dish, covered with EtOH, and sonicated for ~1 min using an ultrasonic bath (Bransonic 1510, Branson, Danbury, CT). Sonicated gold chips were then rinsed with EtOH and blown dry with N2. SAM arrays were fabricated as previously described (Fig. S1†).30 Briefly, an elastomeric stencil containing arrays of 1.1 mm holes was placed on a bare gold surface to form an array of wells on the gold substrate. Wells were then filled with 1 mM ethanolic alkanethiolate solution and incubated for 10 minutes in a chamber containing a laboratory wipe soaked with ethanol to prevent evaporation during local SAM formation. Alkanethiolate solutions were then aspirated and wells were rinsed with DIUF H2O. Carboxylate groups were then converted to active ester groups by adding a solution of 100 mM NHS and 250 mM EDC in DIUF H2O pH 5.5 to wells and incubated for 10 minutes. After an additional rinse with DIUF H2O, 100 µM solutions of peptide in 1 : 10 DMSO : PBS at pH 7.4 were added to each well and incubated for 1 h in a humidity controlled chamber to covalently couple peptides to each array spot. After a final rinse with DIUF H2O, regions surrounding array spots were backfilled with HS–C11–EG3–OH. This was achieved by submerging the gold substrate and attached elastomeric stencil in an aqueous 0.1 mM HS–C11–EG3–OH solution (pH 2.0), removing the stencil, and incubating for 10 minutes. Following backfilling, the array was rinsed with 0.1 wt% SDS in DIUF H2O, DIUF H2O, and EtOH and then dried under a stream of N2. Arrays were stored in sterile DIUF H2O at 4 °C and used within 24 h.
In this SAM array approach, each spot was designed to contain the same total molar density (mol cm–2) of peptide from spot to spot. Therefore, control over individual peptide density was achieved by mixing scrambled BR-BP, scrambled HPG-BP, and mutant GRGESP peptides with functional BR-BP, HPG-BP or GRGDSP peptides, respectively. Therefore, in a typical SAM array, SAMs were locally formed within spots using an alkanethiolate mixture of 95% HS–C11–EG3–OH and 5% HS–C11– EG6–COOH to create substrates with a total of 5% carboxylate groups for peptide conjugation. Here, “X%” refers to the mole percent of alkanethiolate present during SAM formation and subsequently the approximate amount of an alkanethiolate present on the surface after SAM formation. Therefore, to create a spot presenting a “high” density of GRGDSP with BR-BP and HPG-BP, a 100 µM peptide solution with 50 µM GRGDSP, 25 µM BR-BP, and 25 µM HPG-BP was used during peptide conjugation to generate a spot with 2.5% GRGDSP, 1.25% BR-BP, and 1.25% HPG-BP. Likewise, to create a spot presenting a “low” density of GRGDSP and BR-BP, a 5.6 µM GRGDSP, 44.4 µ.M GRGESP, 25 µ.M BR-BP, and 25 µ.M scrambled HPG-BP was used during peptide conjugation. In our experiments, high, medium, and low GRGDSP densities correspond to 2.50%, 0.83%, and 0.27% GRGDSP with 0%, 1.67%, and 2.23% GRGESP, respectively. In this manner, the amount of active peptide could be varied between spots while holding total peptide content constant. As mentioned previously, peptide concentrations were easily measured using UV/Vis since all peptides either contained residues that absorbed strongly at 280 nm, or were engineered to contain tryptophan residues in the poly-glycine tail.
Monitoring peptide incorporation
Peptide solutions containing 50 µM GRGDSP and 50 µM of mixtures of BR-BP, HPG-BP, and scrambled peptides were conjugated to SAM array spots formed with 5% HS–C11–EG6– COOH. More specifically, we mixed together pairs of peptides including (i) BR-BP + HPG-BP; (ii) BR-BP + scrambled HPG-BP; (iii) scrambled BR-BP + HPG-BP; and (iv) scrambled BR-BP + scrambled HPG-BP at ratios of 100 : 0, 80 : 20, 60 : 40, 40 : 60, 20 : 80, and 0 : 100. To visualize and evaluate peptide conjugation in SAM array spots, Alexa Fluor® 488 sulfodichlorophenol ester (AlexaFluor488 5-SDP ester, Invitrogen, Eugene, OR) was used to label epsilon primary amine groups present in lysine residues of immobilized peptides. To achieve this, a reaction solution of 100 µM AlexaFluor488 5-SDP ester in 0.15 M sodium bicarbonate solution at pH 8.3 reaction solution was prepared from a 10 mM stock of AlexaFluor488 5-SDP ester in DMSO. SAM arrays were immersed in freshly prepared reaction solution and incubated for 1 h at RT. After labeling, SAM arrays were rinsed with 0.1% SDS, DIUF H2O, and EtOH and then dried under a stream of N2. A GE Healthcare Typhoon Trio Variable Mode Imager was used to scan SAM arrays containing fluorescently labeled peptide. Fluorescent intensity was quantified using Image J (ImageJ, Freeware, NIH, Bethesda, MD) imaging software.
Cell culture and assays on SAM arrays
Bone marrow-derived human mesenchymal stem cells (56 year old, male) were expanded at low density on tissue culture polystyrene plates to maintain multipotency as described by Sotiropoulou et al41 and used by passage 7. During hMSC expansion and SAM array experiments performed in “growth medium” (GM), hMSCs were cultured in low glucose Dulbecco’s Modified Eagles Medium supplemented with 10% Fetal Bovine Serum (Atlanta Biologicals-Lawerenceville GA) and Penicillin (10 000 IU ml–1)/Streptomycin (10 000 µg ml–1)/Amphotericin B (25 µg ml–1) antibiotic (CellGro, MediaTech – Manassas, VA). For experiments performed in “osteogenic medium” (OM), hMSCs on SAM arrays were cultured in growth medium supplemented with 1% β-glycerolphosphate, 0.1% ascorbic acid, 0.01% dexamethasone (Sigma, St. Louis, MO), and 0.01 µM vitamin D3 (Enzo Life Science, Farmingdale, NY). For experiments on SAM arrays, hMSCs were removed from plastic culture plates using a 0.05% trypsin solution, resuspended in medium, and seeded onto SAM arrays in 4-well polystyrene culture plates (Nunc/Thermo Scientific, Rochester, NY). After allowing cells to attach for ~1 h in a humidified incubator at 37 °C and 5% CO2, arrays were dipped in warm medium to remove loosely attached cells and then transferred to a rectangular multidish (Thermo Scientific/Nunc, Rochester, NY) with warm GM or OM. Furthermore, it is important to point out that all cell experiments for comparison between GM and OM conditions were run simultaneously and media were replaced every 48 hours.
Alkaline phosphatase (ALP) staining
After 7 days in culture, media were removed and then hMSCs on SAM arrays were rinsed with PBS and fixed via treatment with a 1% paraformaldehyde in PBS for 5 min. To detect the activity of ALP, cells were then stained with a BCIP/NBT alkaline phosphatase substrate kit (Vector Laboratories, Burlingame, CA) according to the manufacturer’s protocol using a 1 h incubation at room temperature. After staining, chips were rinsed with PBS and then imaged.
Array imaging
Cells in culture on SAM arrays were imaged using a Nikon Eclipse Ti inverted microscope equipped with the Perfect Focus System. For phase contrast imaging, a 10 × PhL objective and a Photometrics HQ-2 was used to capture 4 images of each spot, which were automatically stitched together using the Nikon NIS Elements software. For color images of stained arrays, a Nikon DS-Fi1 was used to capture images and exposure time, lamp intensity, and white balance were kept constant from array to array.
Analysis, quantification, and statistics
Analysis of hMSC array spot coverage in phase contrast images was achieved using Nikon NIS Elements Software (Melville, NY). Briefly, the “edge detection” operation in NIS Elements was applied to stacked images of array spots and then a standard threshold was applied to all images. Automated measurements of area with respect to a region of interest (ROI) sized to the same dimensions of an array spot were used to acquire a surface coverage percentage for each spot (Fig. S2†). Analysis of ALP staining was performed in a similar manner in which color images were thresholded and then automated measurements using an ROI were used to determine staining coverage. Furthermore, ALP staining coverage was divided by hMSC coverage to account for differences in hMSC spot coverage from spot to spot. Statistical analysis of all quantitative datasets was performed using a two-tailed Student’s t-test, where p < 0.05 is used to denote statistical significance.
Results and discussion
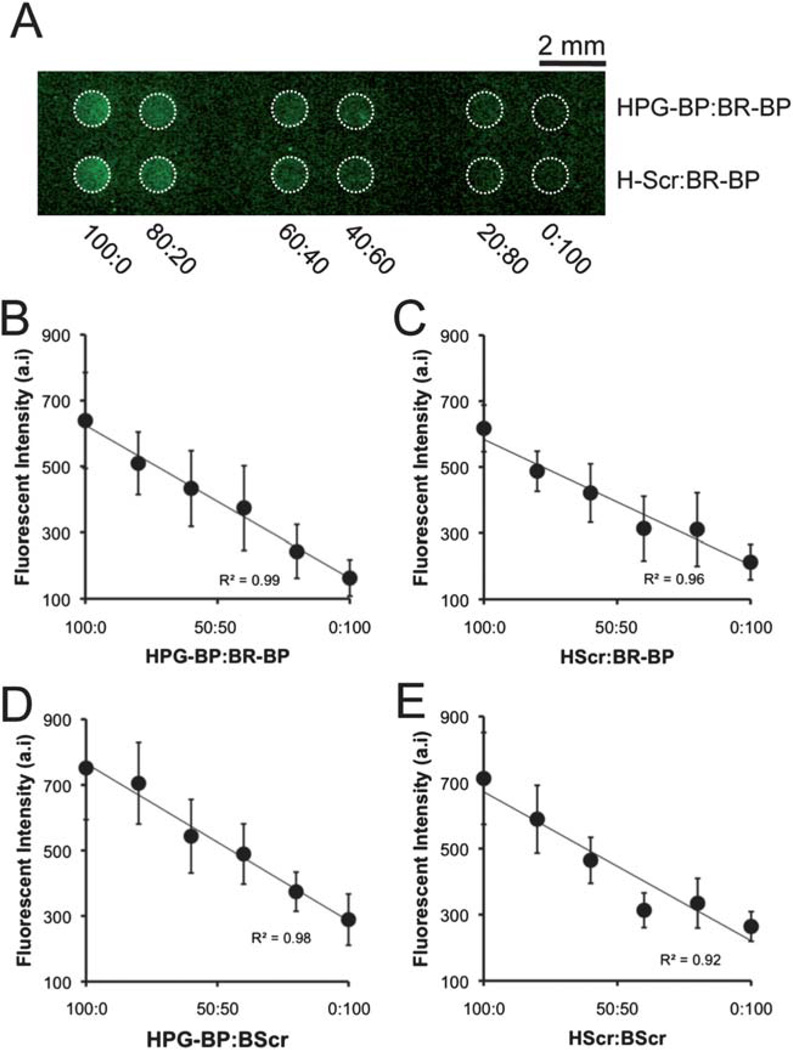
On array spots presenting the same total density of peptide, differences in immobilized peptide content (i.e., the fractional amount of each peptide linked to the surface) between spots could be visualized using a fluorescent amine-reactive probe (Fig. 2). Paired mixtures of BR-BP, HPG-BP, scrambled BR-BP, scrambled HPG-BP, and GRGDSP were conjugated to array spots presenting a fixed density of reactive carboxylic acid moieties, reacted with the amine-reactive probe, and then imaged using a fluorescent surface scanner (Fig. 2A). In all combinations, HPG-BP (or scrambled HPG-BP) exhibited increased fluorescent signal compared to BR-BP (or scrambled BR-BP), and peptides serially mixed together at different ratios yielded linear changes in fluorescent intensity (Fig. 2B–E). This linear relationship allowed for a direct correlation between measured fluorescence intensity and relative peptide content in each individual spot, and covalently immobilized peptide content reflected the compositions present in solution during coupling. It is noteworthy that peptide labeling was performed after covalent immobilization onto the SAM array, and this analysis method eliminates the potential for confounding influences (e.g. changes in peptide immobilization efficiency) that can occur when fluorescent tags are introduced before peptide coupling to a substrate. Additionally, when similar experiments were performed on standard (non-patterned) SAM chips and analyzed via infrared spectroscopy (Fig. S3†), we observed changes in amide II peak characteristics that were dependent on peptide composition during coupling, similar to the differences in fluorescence intensity measured in the SAM array spots. This approach for detecting differences in peptide content between spots presenting the same total peptide density relied on being able to detect differences in fluorescent signal following labeling with the amine-reactive probe. Interesting, all peptides contained 2 lysine amino acids per chain, yet there are clear differences in fluorescent intensity between HPG-BP and BR-BP (Fig. 2B), and similar differences between scrambled HPG-BP and scrambled BR-BP (Fig. 2E). These differences in fluorescent intensity could be due to factors such as probe reactivity with specific lysine resides or differences in probe fluorescence when conjugated to certain lysine resides. Regardless, the inherent differences in fluorescent intensity with respect to HPG-BP and BR-BP allowed us to monitor relative peptide content in spots presenting the same total peptide density.
Fig. 2.
Fluorescent detection of peptide incorporation. Peptide solutions containing a fixed concentration of GRGDSP and pairs of peptides including (i) HPG-BP + BR-BP; (ii) scrambled HPG-BP + BR-BP; (iii) HPG-BP + scrambled BR-BP; and (iv) scrambled HPG-BP + scrambled BR-BP were conjugated to SAM array spots containing 5% HS–C11–EG6–COOH. Each pair of peptides was mixed at varied ratios including 100 : 0, 80 : 20, 60 : 40, 40 : 60, 20 : 80, and 0 : 100. After peptides were coupled to SAM array spots, lysine side chains were labeled using an Alexa Fluor 488 sulfodichlorophenol ester and imaged using a fluorescent scanner. (A) A representative image of spots presenting a series of HPG-BP and BR-BP mixtures (top) and scrambled HPG-BP and BR-BP mixtures (bottom). (B–E) Fluorescent intensity was quantified for each spot and paired mixtures were plotted on in the same graph (HScr = scrambled HPG-BP, BScr = scrambled BR-BP. Error bars indicate standard error of the mean).
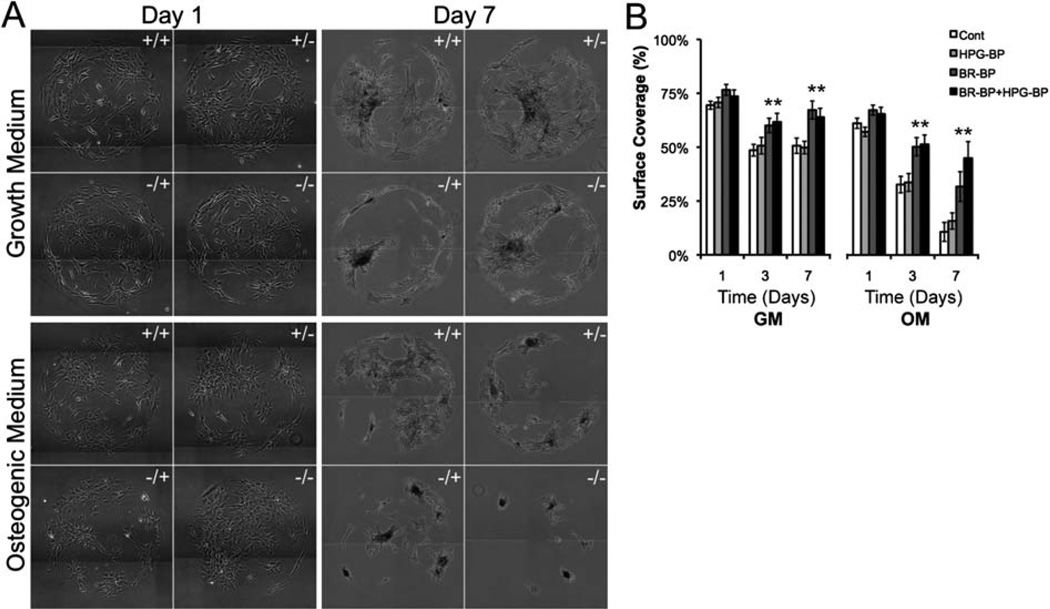
The combination of peptides within SAM array spots significantly influenced the “surface coverage” of hMSC populations within each spot (Fig. 3), where surface coverage is defined as the fractional area occupied by hMSCs. The GRGDSP cell adhesion ligand increased hMSC surface coverage as expected, since this peptide has been previously shown by us and others3,4,7,29,30 to support hMSC spreading. Specifically, hMSCs were confluent at day 1 on all spots presenting medium or high GRGDSP densities (Fig. S4†), but were not confluent on spots presenting low GRGDSP density (Fig. 3). At day 1, hMSCs on all low GRGDSP spots exhibited greater than 70% coverage in GM and greater than 57% coverage in OM (Fig. 3B). After day 1 and at low GRGDSP density, control conditions and HPG-BP conditions each exhibited decreases in hMSC coverage, while each condition containing BR-BP exhibited higher surface coverage compared to control conditions (Fig. 3B). These data indicated that BR-BP significantly increased hMSC coverage, while HPG-BP did not. These observed effects of BR-BP on surface coverage were primarily due to cell spreading rather than mitogenesis, as BR-BP did not significantly influence hMSC proliferation in growth medium (Fig. S5†). Indeed, the only condition that significantly increased hMSC proliferation was the HPG-BP condition at high GRGDSP density, which is consistent with a pro-mitogenic effect of HPG-BP we observed in a previous study.2 The effects of BR-BP are distinct from those reported in previous studies, which have not investigated cell surface coverage in response to BR-BP.37,42 However, BMP-2 signaling has previously been shown to synergize with integrin signaling,43 and several studies have suggested that integrin signaling is essential for BMP-2 receptor activation.44–47 Additionally, Lai and Cheng have demonstrated that αvβ1 integrins directly interact with BMP-2 receptors, and that BMP-2 signaling upregulates expression of αvβ1 integrin.44 Taken in the context of our current work, we can speculate that signals arising from immobilized BR-BP may increase integrin expression and promote increased hMSC surface coverage on surfaces presenting low densities of the GRGDSP cell adhesion ligand.
Fig. 3.
hMSC surface coverage on SAM arrays presenting mixtures of BR-BP and HPG-BP and a low density of GRGDSP in growth medium (GM) and osteogenic medium (OM). (A) Images of hMSCs on SAM array spots and (B) quantification of spot coverage. 1 × 106 hMSCs were seeded onto each SAM array and media were changed every other day (“−/−” Control, “+/−” BR-BP, “−/+” HPG-BP, “+/+” BR-BP and HPG-BP. Error bars indicated standard error of the mean and asterisk indicates significance difference compared to control, p < 0.05, n = 9).
Importantly, no cell attachment was observed on array spots presenting the mutant cell adhesion peptide GRGESP in combination with any of the other peptides or peptide mixtures (data not shown). Therefore, hMSC adhesion to SAM arrays was chemically defined, in the sense that it was mediated by specific peptide–receptor interactions. Furthermore, this observation indicates that cell adhesion required the GRGDSP ligand, and that hMSCs could not adhere to BR-BP and/or HPG-BP. The lack of cell adhesion to BR-BP in this well-defined context provides additional support for previous studies,14,15,37,42 which have attributed the effects of immobilized BR-BP to BMP receptor activation rather than cell adhesion. The lack of cell adhesion to HPG-BP is consistent with our previous studies, which have shown little or no cell adhesion to heparin-binding peptides.2,4,20
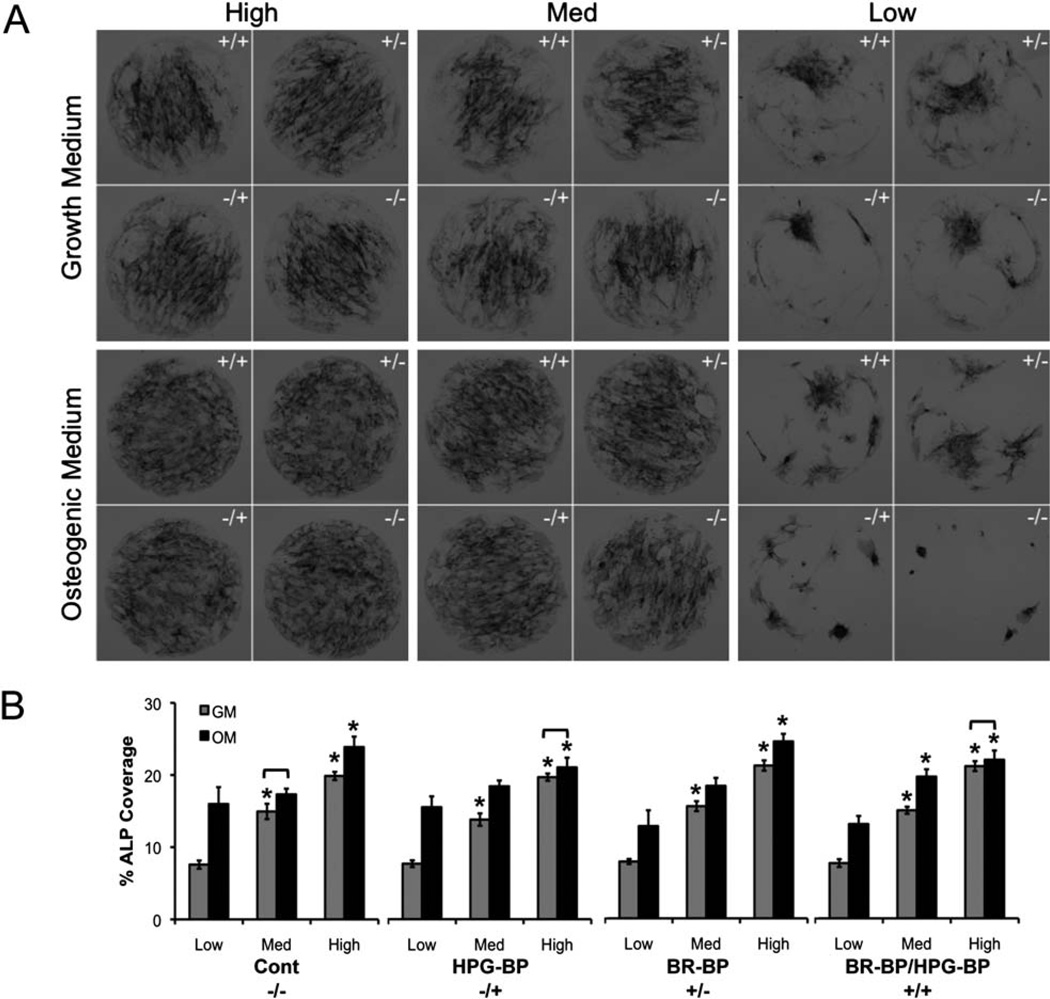
To screen for potential changes in hMSC phenotype on the various peptide-presenting array spots we stained hMSC populations for ALP activity, a hallmark of osteogenic differentiation (Fig. 4). The purpose of this analysis was to demonstrate an initial screen for changes in hMSC phenotype using an efficient experimental method, not to perform a comprehensive analysis of stem cell phenotype. On high and medium GRGDSP densities, hMSCs in growth medium exhibited ALP staining that was most concentrated in the middle of each spot, while hMSCs in osteogenic medium exhibited more uniform staining patterns throughout an entire spot (Fig. 4A). Interestingly, the uniform staining patterns observed in osteogenic medium on high and medium GRGDSP densities were similar to patterns observed in a recent study by Ruiz and Chen, in which hMSCs were cultured on 1 mm diameter microcontact printed spots of fibronectin.48 However, in the same work by Ruiz and Chen, hMSCs on fibronectin spots cultured in growth medium did not show significant ALP staining, and this differs from our observation of ALP staining toward the center of spots in growth medium (Fig. 4). This distinction between the Ruiz and Chen study and our current study could be explained by a difference in the nature of the ligand presented to the cells (GRGDSP versus fibronectin), a difference in the hMSC characteristics in each study (e.g. donor characteristics, expansion conditions), or a difference in the specific medium conditions used to grow the cells. If this distinction is due to a difference in the nature of the ligand presented to the cells, it may suggest that the GRGDSP fragment of fibronectin preferentially promotes hMSC ALP activity in the absence of soluble osteogenic supplements, but further studies will be needed to explore this mechanism.
Fig. 4.
hMSC alkaline phosphatase (ALP) staining on SAM arrays presenting mixtures of BR-BP and HPG-BP and varied densities of GRGDSP (low, medium, high) in growth and osteogenic media formulations. (A) hMSCs were stained for ALP activity after 7 days in culture on SAM arrays. (B) ALP staining was normalized by hMSC surface coverage (error bars represent standard error of the mean and asterisk indicates significance compared to low GRGDSP within a medium condition and bracket indicates no significant difference between growth (GM) and osteogenic (OM) medium conditions, p < 0.05, n = 9).
In all cases, high GRGDSP density significantly increased ALP staining coverage compared to low density GRGDSP (Fig. 4B). These ALP activity increases in response to the fibronectin-derived peptide GRGDSP are consistent with previous studies that have shown a pro-osteogenic influence of fibronectin or GRGDSP in multiple contexts.13,48–54 In addition, osteogenic medium resulted in significant increases in ALP staining in most conditions (Fig. 4B), as expected. However, within any single GRGDSP density there were no significant effects of HPG-BP and BR-BP on ALP staining. This result is surprising, as previous studies have shown that HPG-BP and BR-BP can each promote increases in mesenchymal stem cell alkaline phosphatase production.2,14,37 There are multiple potential explanations for this discrepancy between previous studies and our current work. First, the peptide densities used in this current work differ from those used in previous studies,2 and the density of a peptide ligand can significantly influence its activity. The differences could also be explained by differing signaling contexts. In previous studies, Lee and coworkers demonstrated increased ALP activity when hMSCs were cultured on hydroxyapatite substrates coated with a modular version of BR-BP.14,16 Similarly, work by Saito and coworkers observed that C3H10T1/2 cells increased ALP activity when cultured on BR-BP-coated tissue culture plates.37 Additionally, He and coworkers also observed increases in ALP activity when bone marrow stromal cells were cultured on hydrogels containing pendent polymer chains modified with BR-BP.13,54 In these previous studies, in which BR-BP could slowly release from a substrate or was tethered to a long PEG linker, BR-BP may function as a soluble molecule. However, in our current study, it is unlikely that peptides presented on a SAM substrate via a relatively short linker would behave like a soluble molecule. Therefore, the covalently immobilized context in which the peptide is presented on the SAM surface may result in different effects compared to previous studies.
Conclusion
Here we described the use of a chemically defined SAM array to examine the effects and combined effects of multiple, distinct peptides on hMSC behavior. Our results demonstrated that fluorescently tagging peptides after conjugation to the SAM array can be used to monitor incorporation of peptide mixtures within array spots. SAM arrays allowed for efficient screening of hMSC behavior in response to a broad range of peptide identities and densities, at multiple time points, using multiple analytical methods. Specifically, on a standard size gold-coated microscope slide, we generated SAM arrays with 120 spots to probe the effects of 16 distinct peptide combinations with up to 9 replicates per condition on hMSC behavior. Furthermore, identical SAM arrays were used to probe hMSC response in growth and osteogenic media conditions at several time points. The results validated a series of prior assertions, including that the cell adhesion ligand GRGDSP significantly increases hMSC cell spreading and ALP activity. The results also suggested novel effects, including that BR-BP can significantly increase hMSC surface coverage. In addition, the effects of BR-BP or HPG-BP on hMSC behavior were dependent on the density of the GRGDSP cell adhesion ligand. Therefore, by using SAM arrays to carefully probe combinations of peptides, we were able to identify specific contexts in which stem cells responded to bioactive peptides. Taken together, these results highlight how substrates designed to systematically probe the individual and combined effects of peptide ligands can be useful to study stem cell behavior.
Supplementary Material
Acknowledgements
The authors would like to acknowledge funding from the National Institutes of Health (R01HL093282 and the Biotechnology Training Program NIGMS 5 T32GM08349) and the National Science Foundation (DMR 0906123). Fluorescent scans were obtained using a GE Healthcare Typhoon Trio Variable Mode Imager at the Scientific Instrumentation Facility of the UW Carbone Cancer Center, Madison, WI. PM-IRRAS was performed in the National Science Foundation-funded UW-MRSEC facility.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2jm32242k
References
- 1.Garcia AS, Dellatore SM, Messersmith PB, Miller WM. Effects of supported lipid monolayer fluidity on the adhesion of hematopoietic progenitor cell lines to fibronectin-derived peptide ligands for alpha5beta1 and alpha4beta1 integrins. Langmuir. 2009;25(5):2994–3002. doi: 10.1021/la802772y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hudalla GA, Kouris NA, Koepsel JT, Ogle BM, Murphy WL. Harnessing endogenous growth factor activity modulates stem cell behavior. Integr. Biol. 2011;3(8):832–842. doi: 10.1039/c1ib00021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudalla GA, Murphy WL. Using "click" chemistry to prepare SAM substrates to study stem cell adhesion. Langmuir. 2009;25(10):5737–5746. doi: 10.1021/la804077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudalla GA, Murphy WL. Immobilization of peptides with distinct biological activities onto stem cell culture substrates using orthogonal chemistries. Langmuir. 2010;26(9):6449–6456. doi: 10.1021/la1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jongpaiboonkit L, King WJ, Lyons GE, Paguirigan AL, Warrick JW, Beebe DJ, Murphy WL. An adaptable hydrogel array format for 3-dimensional cell culture and analysis. Biomaterials. 2008;29(23):3346–3356. doi: 10.1016/j.biomaterials.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jongpaiboonkit L, King WJ, Murphy WL. Screening for 3D environments that support human mesenchymal stem cell viability using hydrogel arrays. Tissue Eng. A. 2009;15(2):343–353. doi: 10.1089/ten.tea.2008.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koepsel JT, Murphy WL. Patterning discrete stem cell culture environments via localized self-assembled monolayer replacement. Langmuir. 2009;25(21):12825–12834. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martino MM, Mochizuki M, Rothenfluh DA, Rempel SA, Hubbell JA, Barker TH. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials. 2009;30(6):1089–1097. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakaji-Hirabayashi T, Kato K, Iwata H. Improvement of neural stem cell survival in collagen hydrogels by incorporating laminin-derived cell adhesive polypeptides. Bioconjugate Chem. 2012;23(2):212–221. doi: 10.1021/bc200481v. [DOI] [PubMed] [Google Scholar]
- 10.Saha K, Irwin EF, Kozhukh J, Schaffer DV, Healy KE. Biomimetic interfacial interpenetrating polymer networks control neural stem cell behavior. J. Biomed. Mater. Res., Part A. 2007;81(1):240–249. doi: 10.1002/jbm.a.30986. [DOI] [PubMed] [Google Scholar]
- 11.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303(5662):1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 12.Liu SQ, Tian Q, Hedrick JL, Po Hui JH, Ee PL, Yang YY. Biomimetic hydrogels for chondrogenic differentiation of human mesenchymal stem cells to neocartilage. Biomaterials. 2010;31(28):7298–7307. doi: 10.1016/j.biomaterials.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 13.He X, Yang X, Jabbari E. Combined effect of osteopontin and BMP-2 derived peptides grafted to an adhesive hydrogel on osteogenic and vasculogenic differentiation of marrow stromal cells. Langmuir. 2012;28(12):5387–5397. doi: 10.1021/la205005h. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Lee JS, Wagoner-Johnson A, Murphy WL. Modular peptide growth factors for substrate-mediated stem cell differentiation. Angew. Chem. Int. Ed. 2009;48(34):6266–6269. doi: 10.1002/anie.200901618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore NM, Lin NJ, Gallant ND, Becker ML. Synergistic enhancement of human bone marrow stromal cell proliferation and osteogenic differentiation on BMP-2-derived and RGD peptide concentration gradients. Acta Biomater. 2011;7(5):2091–2100. doi: 10.1016/j.actbio.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Lee JS, Lee JS, Murphy WL. Modular peptides promote human mesenchymal stem cell differentiation on biomaterial surfaces. Acta Biomater. 2010;6(1):21–28. doi: 10.1016/j.actbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Supramolecular design of self-assembling nanofibers for cartilage regeneration. Proc. Natl. Acad. Sci. U. S. A. 2010;107(8):3293–3298. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat. Methods. 2010;7(12):989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levenstein ME, Berggren WT, Lee JE, Conard KR, Llanas RA, Wagner RJ, Smith LM, Thomson JA. Secreted proteoglycans directly mediate human embryonic stem cell-basic fibroblast growth factor 2 interactions critical for proliferation. Stem Cells. 2008;26(12):3099–3107. doi: 10.1634/stemcells.2007-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudalla GA, Koepsel JT, Murphy WL. Surfaces that sequester serum-borne heparin amplify growth factor activity. Adv. Mater. 2011;23(45):5415–5418. doi: 10.1002/adma.201103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004;22(7):863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 22.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 23.Chin VI, Taupin P, Sanga S, Scheel J, Gage FH, Bhatia SN. Microfabricated platform for studying stem cell fates. Biotechnol. Bioeng. 2004;88(3):399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 24.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, Kumacheva E, Zandstra PW. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 2007;26(22):4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5(4):401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 26.Kim L, Vahey MD, Lee HY, Voldman J. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab Chip. 2006;6(3):394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 27.Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, Wikswo JP, Cooper JM. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip. 2009;9(18):2659–2664. doi: 10.1039/b902083g. [DOI] [PubMed] [Google Scholar]
- 28.Vroman L, Adams AL. Findings with recording ellipsometer suggesting rapid exchange of specific plasma proteins at liquid/solid interfaces. Surf. Sci. 1969;16:438. [Google Scholar]
- 29.Hudalla GA, Murphy WL. Chemically well-defined self-assembled monolayers for cell culture: toward mimicking the natural ECM. Soft Matter. 2011;7(20):9561–9571. doi: 10.1039/C1SM05596H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koepsel JT, Loveland SG, Le NN, Schwartz MP, Murphy WL. A chemically defined screening platform reveals behavioral similarities of primary human mesenchymal stem cells and endothelial cells. Submitted. 2012 doi: 10.1039/c2ib20029e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koepsel JT, Nguyen EH, Murphy WL. Differential effects of a soluble or immobilized VEGFR-binding peptide. Integr. Biol. 2012 doi: 10.1039/c2ib20055d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derda R, Musah S, Orner BP, Klim JR, Li L, Kiessling LL. High-throughput discovery of synthetic surfaces that support proliferation of pluripotent cells. J. Am. Chem. Soc. 2010;132(4):1289–1295. doi: 10.1021/ja906089g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prime KL, Whitesides GM. Adsorption of proteins onto surfaces containing end-attached oligo(ethylene oxide) – a model system using self-assembled monolayers. J. Am. Chem. Soc. 1993;115(23):10714–10721. [Google Scholar]
- 34.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 35.Thompson LD, Pantoliano MW, Springer BA. Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry. 1994;33(13):3831–3840. doi: 10.1021/bi00179a006. [DOI] [PubMed] [Google Scholar]
- 36.Li L-Y, Safran M, Aviezer D, Boehlen P, Seddon AP, Yayon A. Diminished heparin binding of a basic fibroblast growth factor mutant is associated with reduced receptor binding, mitogenesis, plasminogen activator induction, and in vitro angiogenesis. Biochemistry. 1994;33(36):10999–11007. doi: 10.1021/bi00202a020. [DOI] [PubMed] [Google Scholar]
- 37.Saito A, Suzuki Y, Ogata S, Ohtsuki C, Tanihara M. Activation of osteo-progenitor cells by a novel synthetic peptide derived from the bone morphogenetic protein-2 knuckle epitope. Biochim. Biophys. Acta. 2003;1651(1–2):60–67. doi: 10.1016/s1570-9639(03)00235-8. [DOI] [PubMed] [Google Scholar]
- 38.Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 39.Jo BH, Van Lerberghe LM, Motsegood KM, Beebe DJ. Three-dimensional micro-channel fabrication in polydimethylsiloxane (PDMS) elastomer. J. Microelectromech. Syst. 2000;9(1):76–81. [Google Scholar]
- 40.Thibault C, Severac C, Mingotaud A-F, Vieu C, Mauzac M. Poly(dimethylsiloxane) contamination in microcontact printing and its influence on patterning oligonucleotides. Langmuir. 2007;23(21):10706–10714. doi: 10.1021/la701841j. [DOI] [PubMed] [Google Scholar]
- 41.Sotiropoulou PA, Perez SA, Salagianni M, Baxevanis CN, Papamichail M. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells. 2006;24(2):462–471. doi: 10.1634/stemcells.2004-0331. [DOI] [PubMed] [Google Scholar]
- 42.Zouani OF, Chollet C, Guillotin B, Durrieu MC. Differentiation of pre-osteoblast cells on poly(ethylene terephthalate) grafted with RGD and/or BMPs mimetic peptides. Biomaterials. 2010;31(32):8245–8253. doi: 10.1016/j.biomaterials.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 43.Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling crosstalk. Cytokine Growth Factor Rev. 2005;16(3):251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Lai CF, Cheng SL. Alphavbeta integrins play an essential role in BMP-2 induction of osteoblast differentiation. J. Bone Miner. Res. 2005;20(2):330–340. doi: 10.1359/JBMR.041013. [DOI] [PubMed] [Google Scholar]
- 45.Tamura Y, Takeuchi Y, Suzawa M, Fukumoto S, Kato M, Miyazono K, Fujita T. Focal adhesion kinase activity is required for bone morphogenetic protein—Smad1 signaling and osteoblastic differentiation in murine MC3T3-E1 cells. J. Bone Miner. Res. 2001;16(10):1772–1779. doi: 10.1359/jbmr.2001.16.10.1772. [DOI] [PubMed] [Google Scholar]
- 46.Jikko A, Harris SE, Chen D, Mendrick DL, Damsky CH. Collagen integrin receptors regulate early osteoblast differentiation induced by BMP-2. J. Bone Miner. Res. 1999;14(7):1075–1083. doi: 10.1359/jbmr.1999.14.7.1075. [DOI] [PubMed] [Google Scholar]
- 47.Su JL, Chiou J, Tang CH, Zhao M, Tsai CH, Chen PS, Chang YW, Chien MH, Peng CY, Hsiao M, Kuo ML, Yen ML. CYR61 regulates BMP-2-dependent osteoblast differentiation through the {alpha}v{beta}3 integrin/integrin-linked kinase/ERK pathway. J. Biol. Chem. 2010;285(41):31325–31336. doi: 10.1074/jbc.M109.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26(11):2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107(11):4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Bhadriraju and C. S. Chen, Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 51.Hidalgo-Bastida LA, Cartmell SH. Cartmell, Mesenchymal stem cells, osteoblasts and extracellular matrix proteins: enhancing cell adhesion and differentiation for bone tissue engineering. Tissue Eng., Part B: Rev. 2010;16(4):405–412. doi: 10.1089/ten.TEB.2009.0714. [DOI] [PubMed] [Google Scholar]
- 52.Hosseinkhani H, Hosseinkhani M, Tian F, Kobayashi H, Tabata Y. Osteogenic differentiation of mesenchymal stem cells in self-assembled peptide-amphiphile nanofibers. Biomaterials. 2006;27(22):4079–4086. doi: 10.1016/j.biomaterials.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 53.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26(30):5991–5998. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 54.He X, Ma J, Jabbari E. Effect of grafting RGD and BMP-2 protein-derived peptides to a hydrogel substrate on osteogenic differentiation of marrow stromal cells. Langmuir. 2008;24(21):12508–12516. doi: 10.1021/la802447v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.