Abstract
Kidney development is a paradigm of how multiple cell types are integrated into highly specialized epithelial structures via various inductive events. A network of transcription factors and signaling pathways have been identified as crucial regulators. The recent discovery of a group of small, non-coding RNAs, microRNAs (miRNAs), has added a new layer of complexity. Studies using the pronephric kidney of Xenopus and the metanephric kidney of mouse have demonstrated that a tight regulation of mRNA stability and translation efficiency by miRNAs is very important as well. The interplay between miRNAs and the transcriptional network provides plasticity and robustness to the system. Importantly, miRNAs are not only necessary for early aspects of kidney development, but also later in life. As such they may provide a mean to maintain/modulate kidney function during homeostasis and injury.
Post-transcriptional regulation is an important, but long neglected aspect in the control of gene expression. microRNAs (miRNAs), ~22 nucleotide long, non-coding RNAs have emerged as key regulators in this process. They bind to the 3’UTR of mRNAs and regulate translational efficiency and mRNA stability. A plethora of work has explored several aspects of miRNAs, such as, their biogenesis, mode of action and binding site specificity.1–3 Even then, the biological function of most miRNAs is still unknown. This is - in part - due to the large number of identified miRNAs, their numerous predicted targets and their rather modulatory role.1–3 But it is also hampered by the fact that miRNA function can often not be adequately studied in cell lines and requires in vivo approaches.
The kidney is an essential organ required for removal of metabolic waste products from the body and conservation of water, electrolytes and metabolites. Notably, the kidney is not a static organ, but has to be adaptable to changes in environmental conditions such as aging and dietary inputs. Failure in doing so causes kidney malfunction and can result in serious repercussions that may ultimately lead to death. Interestingly, many aspects of kidney diseases involve processes that are initially used during kidney development and are recapitulated in disease initiation/progression. For example, mesenchymal-epithelial transition (MET) is involved in the formation of renal tubules, but is perturbed during fibrotic events such as nephrotic syndrome.4 Similarly, proliferation is required to establish the final organ size, but is absent in adult kidneys and is only reinitiated under disease conditions such as polycystic kidney disease.5 It is not surprising that these processes are targeted by the regulatory role of miRNAs. Indeed, many miRNAs are expressed in the kidney and their importance is supported by mouse knockout studies.6–10 Eliminating Dicer, a key enzyme in miRNA biogenesis, from podocytes, a cell type required for the formation of the size exclusion barrier in the glomerulus, results in progressive loss of podocyte function.8–10
One disadvantage of the mouse studies is the complexity of the kidney. In the course of evolution the kidney has evolved into an organ with millions of functional units, the nephrons. This high redundancy protects the metanephros, the kidney present in all higher vertebrates, from malfunction. As long as a sufficient number of nephrons are functional, problems in a few nephrons often remain undetected. For example, the elimination of Dicer from podocytes8–10 did not equally affect all podocytes. The mutant kidneys actually exhibited a mixture of diseased, pre-diseased and normal glomeruli, a fact that greatly hampered the analysis of the underlying mechanism causing podocyte malfunction. In contrast, primitive animal models such as Xenopus or zebrafish offer alternative insights due to their simple kidney structure. The mammalian kidney develops through three successive and increasingly complex renal structures, the pro-, meso- and metanephros.11,12 While the three kidney forms appear morphologically different their nephrons are evolutionarily conserved.13–16 However, the pronephric kidney avoids the redundancy present in the more complex forms. Its two nephrons develop synchronously on the left and right side of the body making it an ideal system to study cell fate decisions. As a consequence, the Xenopus and zebrafish pronephros has emerged as a valuable model to study multiple aspects of kidney development and disease.17,18 This includes the recent report by Agrawal et al.6 that touches on several aspects of miRNA regulation (Figure 1) and will further be expanded on in the remainder of this point-of-view.
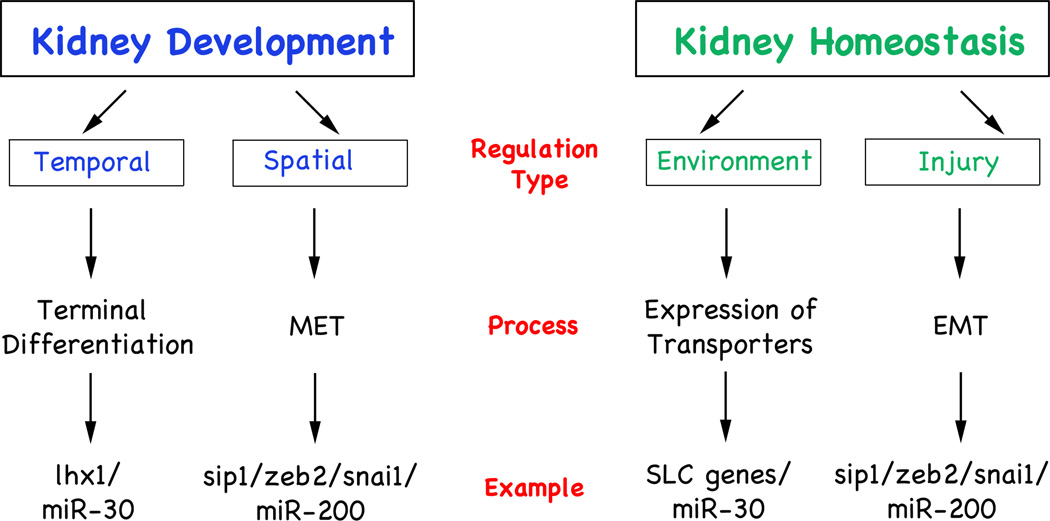
Fig. 1. miRNA Activity in Kidney Development and Physiology.
Flow diagrams depicting the role of miRNAs in kidney development and during homeostasis showing the type of regulation, the process involved and an example of each.
Temporal Regulation of Gene Expression by miRNAs
miRNAs bind to the 3’UTR of a given gene and repress its activity. This regulation is not absolute, since most native miRNA binding sites only result in a modest (30–50%), but not complete reduction of gene expression.1,6 However, a 50% decrease in expression is often insufficient to change developmental fates. This fact is exemplified by the observation that most genetic mutations are recessive and do not result in overt phenotypes in the heterozygous state. So what is the function of miRNAs if they are unable to promote such decisions? One often-neglected aspect in gene regulation is the inactivation of an existing input. To proceed along the path of differentiation, genes have to be turned on and off at the proper place and time. The latter occurs at several levels, the cessation of de novo transcription or translation, as well as the degradation of mRNAs and proteins. miRNAs play an important role in two of these aspects. By binding to the 3’UTR they interfere with translation and promote mRNA degradation. Importantly, in this process a 50% down-regulation is sufficient to lower the concentration of e.g. transcription factors beyond threshold levels and sharpen the borders.
The work of Agrawal et al.6 offers a paradigm for this type of regulation. We found that early specification of the pronephric field is miRNA-independent. At times when kidney-specific transcription factors are first expressed, no miRNAs are yet detected in this domain. Moreover, knockdown of individual miRNA families or interfering with miRNA biogenesis does not impair the initial specification of the pronephric kidney. In contrast, subsequent steps are miRNA-dependent. During terminal differentiation when the first wave of transcription factors is down-regulated, several miRNAs are detected in renal epithelial cells. Moreover, lack-of-miRNA activity causes defects such as decreased proliferation, aberrant nephron patterning and delayed terminal differentiation of kidney tubules.6
At the molecular level, the expression of lhx1, a key transcription factor involved in several aspects of kidney development19–21, is normally down-regulated upon terminal differentiation. However, in the absence of miRNAs, lhx1 expression levels are maintained. This regulation is dependent on the miR-30 family and is partially responsible for the phenotype. Indeed, close inspection of the lhx1 3’UTR revealed two pairs of miRNA binding sites, miR-30 and miR-96/182, both of which are expressed in the developing kidney of Xenopus and mouse. Gain-of-function studies with a lhx1 construct lacking the 3’UTR showed that ectopic lhx1 protein is equivalent to the loss-of-miR-30 activity and mimicked the phenotype.6 Using antisense morpholino oligomers to block both miR-30 binding sites on the lhx1 3’UTR also delayed terminal differentiation (our unpublished observations). Together, these data suggest that the miR-30/lhx1 interaction is integral in regulating the timing of pronephros development. Interestingly, other key transcription factors involved in kidney development, such as, WT1, Pax2, Pax8 and Hnf1b16 also harbor multiple miRNA binding sites in their 3’UTR. Given their highly regulated expression it seems likely that miRNAs also influences these transcription factors. Thus, the temporal control of gene expression is clearly a very important aspect in the activity of miRNAs during kidney development.
Spatial Regulation of Gene Expression by miRNAs
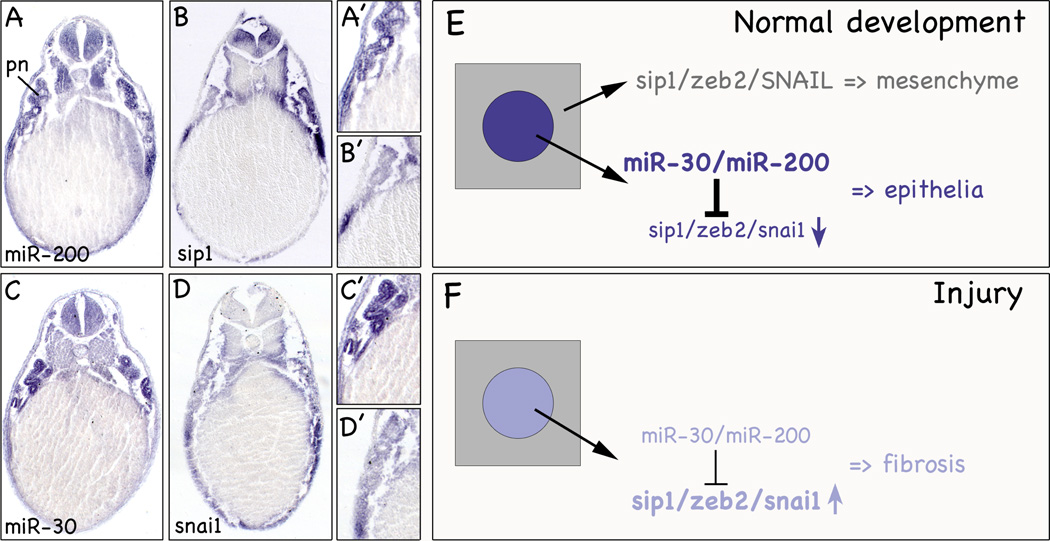
Another concept that has been brought forth early on was that a miRNA and its target are mutually exclusive and do not share the same expression domains.22,23 This means that in a tissue that is high in a given miRNA, its target is expressed at low levels. Due to the differences in abundance, the miRNA ascertains that the low expression of the target mRNA would not result in any significant translation. Our studies provide data supporting such a mechanism. In addition to miR-30, the miR-200 family was one of the most abundant miRNAs in the kidney.6 miR-200 is expressed in the developing pronephros, the skin and part of the somites (Figure 2A,A’). Interestingly, miR-200 has been implicated in the regulation of epithelial-mesenchymal transition (EMT) by means of regulating the E-box genes Zeb1 and Zeb2, transcriptional repressors of E-cadherin.24 These proteins are among the key genes upregulated in skin tumors. As such, it is not surprising that in situ hybridization in Xenopus did not detect zeb1 and zeb2 in the epidermis, whereas miR-200 is highly expressed (Figure 2A-B’ and our unpublished data). Therefore, this pair fulfills the criteria for spatial regulation. High levels of miR-200 in the skin prevent the translation of zeb1 and zeb2 proteins and protect the integrity of the epidermis. The pronephros, however, did not follow this rule since both miRNA and target are expressed in the developing pronephros. This can be explained by two ways: first, zeb1 and zeb2 are expressed in the developing pronephros, but do not play an important role in its formation. This seems unlikely since Xenopus embryos injected with an antisense morpholino oligomer targeting zeb2 mRNA results in EMT defects in the pronephros (our unpublished observations). Second, the renal epithelial cells present in the pronephros are in the process of epithelialization, but have not yet turned off zeb1/zeb2. Upon terminal differentiation the expression of zeb1/zeb2 will cease. At this moment the high levels of miR-200 will protect the renal epithelial cells from spontaneous de-differentiation. While such a hypothesis still needs to be experimentally tested, data from another E-box protein involved in MET/EMT, Snai1, support this scenario. In Xenopus, Snai1 is expressed during kidney development and has a predicted miR-30 binding site in its 3’UTR (Figure 2C-D’ and our unpublished data). In mouse, Snai1 regulates the differentiation of renal epithelial tubules.25 Snai1 is expressed early, but is subsequently turned off and is nearly undetectable in adult kidneys. During injury Snai1 expression is re-activated to facilitate the repair process. While this scenario is normally transient, excessive injury results in sustained Snai1 expression, the accumulation of extracellular matrix and ultimately the impairment of renal function.26 The direct involvement of E-box proteins was shown in transgenic mice activating Snai1 in adults leading to renal fibrosis presumably by the activation of an EMT program in the renal epithelial cells.25 Similarly, it has been shown that miR-192 also targets Zeb1 and may contribute to its up-regulation in diabetic kidneys.27 As such miRNAs may have an important modulatory role in kidney homeostasis. Transcriptional regulation of these E-box proteins acts as an on/off switch, while miRNAs provide a means to fine tune expression and allow proper responses to minor injury.
Fig. 2. miRNAs and Epithelial Differentiation.
(A-D’) Sections of whole mount in situ hybridizations of Xenopus embryos at stage 39 with miR-200 (A,A’), zeb2 (B,B’), miR-30 (C,C’) and snai1 (D,D’). Close-ups show the pronephric kidney. e, epidermis; pn, pronephros. (E,F) Schematic diagrams illustrating the role of E-box transcription factors and miR-30/miR-200 in the formation of renal epithelial tubules during kidney development (E) and their imbalance in response to injury (F).
Together, these data further strengthen the idea that miRNAs are important in controlling the spatial gene expression and that this mechanism is instrumental in kidney development and disease.
Homeostasis and miRNA function
One intriguing aspect of our miRNA profiling in mouse kidneys is that the levels of several miRNAs (including the miR-30 family) increase as kidney development progresses (our unpublished data). While their expression levels are relatively low in embryonic kidneys, they are very high in adult ones. At this time point renal epithelial cells are terminally differentiated and cell fate determination has concluded. This high expression suggests other roles for miRNAs besides the two above-mentioned concepts. The kidney has to be able to adapt to environmental influences and some of these processes may be mediated by miRNAs. Injury response is obviously one example as it provides plasticity under adverse conditions. Indeed, miRNA profiling of patients with IgA nephropathy or hypertensive nephrosclerosis showed changes in miRNA levels.28,29 This is also supported by the elimination of Dicer from differentiated podocytes.8–10 These mice exhibited progressive loss of podocytes leading to proteinuria and a decline in kidney function ultimately leading to death. Interestingly, kidneys exhibited both healthy and diseased podocytes long after miRNA biogenesis had been disrupted. This suggests that one of the functions of miRNAs is to enable the podocyte to react to continuous external influences and to maintain its structural plasticity. Among the leading candidate for this phenotype is miR-30a-5p, the very same miRNA that was implicated in our study on the early development of the Xenopus pronephric kidney.6
A bivalent role for the miR-30 family is also supported by in silico target gene analysis that identified more than 20 solute carrier proteins (SLCs). These are integral membrane proteins involved in the transport of organic molecules and inorganic ions across the cell membrane. SLCs are important to maintain urine homeostasis in the kidney and their expression levels have to be adaptable to the environment. Although not experimentally shown yet, miRNAs would ideally be suited for this purpose. They result in relatively small changes in expression (i.e. 30–50% reduction at most) and target several genes simultaneously. As such, changes in miRNA levels could coordinate complex responses. For example, pH control of the urine involves the concerted action of several proteins and it would be intriguing if changes in miRNA levels were a means to prevent acidosis or alkalosis.
SUMMARY
The already available data on miRNAs in kidney development suggest profound roles for this novel class of regulators. Obviously, - to really appreciate their role in development and homeostasis of the kidney - the information of multiple miRNAs has to be integrated into a higher order gene regulatory network. From a therapeutic standpoint, such a network would help to identify miRNAs that can induce self-sustainable cell populations. These parameters will allow reprogramming and/or trans-differentiation of renal cells to cure fibrotic diseases or other kidney ailments.
ACKNOWLEDGEMENTS
The authors would like to thank Drs. S. El-Dahr, J. Larraín, T. Obara, S. Piccolo and D. Romaker for critically reviewing the manuscript. We apologize that we could not cite all pertinent literature due to space constraints. This work was supported by a grant from NIH/NIDDK (#5R21DK077763-03) to O.W.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10(2):141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machuca E, Benoit G, Antignac C. Genetics of nephrotic syndrome: connecting molecular genetics to podocyte physiology. Hum Mol Genet. 2009;18(R2):R185–R194. doi: 10.1093/hmg/ddp328. [DOI] [PubMed] [Google Scholar]
- 5.Torres VE, Harris PC. Polycystic kidney disease: genes, proteins, animal models, disease mechanisms and therapeutic opportunities. J Intern Med. 2007;261(1):17–31. doi: 10.1111/j.1365-2796.2006.01743.x. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal R, Tran U, Wessely O. The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development. 2009;136(23):3927–3936. doi: 10.1242/dev.037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32(22):e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19(11):2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19(11):2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19(11):2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxén L. Organogenesis of the Kidney. Cambridge, UK: Cambridge University Press; 1987. [Google Scholar]
- 12.Vize P, Woolf A, Bard J. The Kidney: From Normal Development to Congenital Diseases. San Diego: Academic Press; 2002. [Google Scholar]
- 13.Mobjerg N, Larsen EH, Jespersen A. Morphology of the kidney in larvae of Bufo viridis (Amphibia, Anura, Bufonidae) J Morphol. 2000;245(3):177–195. doi: 10.1002/1097-4687(200009)245:3<177::AID-JMOR1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 14.Raciti D, Reggiani L, Geffers L, Jiang Q, Bacchion F, Subrizi AE, Clements D, Tindal C, Davidson DR, Kaissling B, Brandli AW. Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 2008;9(5):R84. doi: 10.1186/gb-2008-9-5-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Vize PD. Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric kidney tubules. Dev Biol. 2004;271(2):322–338. doi: 10.1016/j.ydbio.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 17.Drummond IA. Kidney development and disease in the zebrafish. J Am Soc Nephrol. 2005;16(2):299–304. doi: 10.1681/ASN.2004090754. [DOI] [PubMed] [Google Scholar]
- 18.Jones EA. Xenopus: a prince among models for pronephric kidney development. J Am Soc Nephrol. 2005;16(2):313–321. doi: 10.1681/ASN.2004070617. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132(12):2809–2823. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- 20.Chan TC, Takahashi S, Asashima M. A role for Xlim-1 in pronephros development in Xenopus laevis. Dev Biol. 2000;228(2):256–269. doi: 10.1006/dbio.2000.9951. [DOI] [PubMed] [Google Scholar]
- 21.Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374(6521):425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 22.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123(6):1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 24.Cano A, Nieto MA. Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition. Trends Cell Biol. 2008;18(8):357–359. doi: 10.1016/j.tcb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Boutet A, De Frutos CA, Maxwell PH, Mayol MJ, Romero J, Nieto MA. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J. 2006;25(23):5603–5613. doi: 10.1038/sj.emboj.7601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int. 2006;69(2):213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- 27.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104(9):3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. Am J Hypertens. 2010;23(1):78–84. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab Invest. 2010;90(1):98–103. doi: 10.1038/labinvest.2009.118. [DOI] [PubMed] [Google Scholar]