Abstract
Botulinum neurotoxins (BoNTs) are used in a wide variety of medical applications, but there is limited pharmacokinetic data on active BoNT. Monitoring BoNT activity in the circulation is challenging because BoNTs are highly toxic and are rapidly taken up by neurons and removed from the bloodstream. Previously we reported a sensitive BoNT “Assay with a Large Immunosorbent Surface Area” that uses conversion of fluorogenic peptide substrates to measure the intrinsic endopeptidase activity of bead-captured BoNT. However, in complex biological samples, protease contaminants can also cleave the substrates, reducing sensitivity and specificity of the assay. Here, we present a novel set of fluorogenic peptides that serve as BoNT-specific substrates and protease-sensitive controls. BoNT-cleavable substrates contain a C-terminal Nle, while BoNT-non-cleavable controls contain its isomer ε-Ahx. The substrates are cleaved by BoNT subtypes A1–A3 and A5. Substrates and control peptides can be cleaved by non-BoNT proteases (e.g., trypsin, proteinase K, and thermolysin) while obeying Michaelis-Menten kinetics. Using this novel substrate/control set, we studied BoNT/A1 activity in two mouse models of botulism. We detected BoNT/A serum activities ranging from ~3600 to 10 attomol/L in blood of mice that had been intravenously injected one hour prior with BoNT/A1 complex (100 to 4 pg/mouse). We also detected the endopeptidase activity of orally administered BoNT/A1 complex (1 µg) in blood 5 h after administration; activity was greatest 7 h after administration. Redistribution and elevation rates for active toxin were measured and are comparable to those reported for inactive toxin.
Botulinum neurotoxins (BoNTs) are produced by several strains of spore-forming anaerobic Gram-positive bacteria of the genus Clostridium and are among the most toxic compounds known.1–5 The 150-kDa BoNT heterodimeric holotoxin molecule is composed of a 50 kDa light chain (LC) and a 100 kDa heavy chain.6–7 The enormous toxic potency of BoNT arises from its zinc-dependent metalloprotease activity, which is located on the LC and is responsible for proteolytically degrading BoNT's neuronal target proteins.4,8 There are seven botulinum neurotoxin serotypes (A–G), of which BoNT/A is considered the most toxic, having an estimated lethal dose in humans of only 1–2 ng/kg body weight when intravenously injected.9 BoNT/A is currently organized into five subtypes (A1 – A5)10 and other reported BoNT/A variants could be accepted as additional subtypes.11–12 BoNT/A inactivates the soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) complex by cleaving one of its components, the 25-kDa synaptosomal-associated protein (SNAP-25).13–14 Proteolysis of SNAP-25 prevents neurotransmitter vesicles from fusing with the plasma membrane, thereby inhibiting neuronal signal transduction and causing flaccid paralysis.
BoNTs are classified as biological warfare agents, because of the relative simplicity of production, and their lethality in minute quantities.9, 15 Despite its toxicity, BoNT/A is used in many human therapeutic and cosmetic applications.16 BoNT/A-containing medication is approved by the U.S. Food and Drug Administration for the treatment of strabismus, blepharospasm, hemifacial spasm, underarm sweating, and muscle pain disorders. Mistakes in dosing of BoNT/A, however, may cause severe adverse effects, including botulism.17–18 It is feared that patients could receive higher than recommended doses of BoNT, for example, from multiple injections or from unlicensed, unsuitable preparations of botulinum toxins.17,19 Highly sensitive, simple, rapid methods to quantitatively detect systemic BoNT in patient specimens would be useful for studying the mechanism of toxin absorption, distribution, metabolism, and elimination as well as the use of therapeutic interventions with neutralizing antitoxins.
The current gold standard for measuring BoNT toxicity is a live-mouse bioassay, which detects as little as 5 pg of holotoxin.20 However, this assay requires continuous availability of substantial numbers of mice, is expensive, labor-intensive, and time consuming, requiring up to 72 h. Other BoNT detection methods are reported to have sensitivities comparable to or better than the mouse bioassay, but use complex methods or require expensive instrumentation.21–24 Assays that detect the intrinsic proteolytic activity of BoNT are potentially very sensitive, quantitative, and simple; however, the accuracy of protease-based detection assays is limited by interference from non-specific protease contaminants with the samples.25–29 We developed the BoNT Assay with a Large Immunosorbent Surface Area (ALISSA)30–31 based on use of a BoNT/A-specific affinity matrix to purify and concentrate BoNT LC from complex biological samples. The ALISSA detects the catalytic activity of BoNTs by monitoring the extent of cleavage of fluorogenic peptide substrates. Using the ALISSA, we have detected sublethal amounts (attomolar concentrations) of BoNT/A, spiked into milk, serum, carrot juice, and gelatin-phosphate diluents. However, depending on the sample matrix, the presence of non-specific proteases co-extracted with BoNT, may lead to false positive signals.
Here we show how we improved our ALISSA methodology by introducing SNAP-25-based peptide controls that are structurally similar to the ALISSA BoNT/A substrate, cannot be cleaved by BoNT, but by other proteases. Thus, these peptides can be used as control substrates to monitor for non-BoNT/A-related protease activity. We describe the design, construction, and catalytic and kinetic properties of fluorogenic peptides for BoNT/A ALISSA to selectively detect and quantify BoNT LC-A endoprotease activity in the presence of other proteases. Furthermore, we demonstrate the application of these novel sets of peptide substrates and controls in pharmacokinetic studies of botulism in oral and intravenous mouse models.
EXPERIMENTAL SECTION
Reagents were purchased from Sigma (St. Louis, MO) unless indicated otherwise. 5-carboxyfluorescein (5-Fam), 7-hydroxy-4-methylcoumarin-3-acetic acid (4-MU) and N-α-Fmoc-N-ε-(4,4-dimethylazobenzene-4’carbonyl)-L-lysine (Fmoc-Lys(DABCYL)-OH) were obtained from AnaSpec (Fremont, CA). Dimethyl sulfoxide (DMSO) was from Thermo Fisher Scientific (Waltham, MA), and acetonitrile (CH3CN) from Fisher Chemical (Lafayette, CO). BoNT/A, B, C, E and F complexes were purchased from Metabiologics, Inc. (Madison, WI). All toxin work was conducted under BSL2 conditions, and toxin stocks were handled under a bio-safety cabinet. SNAPtide (FITC/DABCYl, product #521) fluorogenic peptide was from List Biological Laboratories (Campbell, CA). Protein A/G agarose beads were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The proteases used were: thermolysin from Bacillus thermoproteolyticus Rokko (Sigma-Aldrich), proteinase K (Roche Applied Science, Indianapolis, IN) and trypsin (Promega, San Luis Obispo, CA). Pooled human serum was from Innovative Research, Inc. (Novi, MI).
Antibodies
Rabbit polyclonal antibodies raised against BoNT/A toxoid were obtained from Abcam (ab20641, Cambridge, MA). Rabbit anti-SNAP-25 IgG was from Sigma. IR-Dye 680-conjugated polyclonal donkey anti-rabbit, highly cross adsorbed IgG was purchased from Li-Cor (Lincoln, NE). The 5A20.4 monoclonal antibody was a gift from Dr. James Marks.32
For the cloning, expression, and purification of recombinant LCs of BoNT/A subtypes A1 to A5. LC subtypes A1 to A5, containing residues 1–438 were generated by PCR using genomic clostridial DNA as template (Genbank ID: LC-A1, M30196; LC-A2, EF028393; LC-A3, 6030483; LC-A4, DQ185901; LC-A5, EU679004) and the forward primer CTCTGGATCCCCATTTGTTAATAAACAATTTAATTATAAAG and the reverse primer CTCTCCCGGGTTTAAAAGGTATTATCCCTCTTACACATAGC. Using BamHI and XmaI restriction, the LC-A PCR products were cloned into the pAR1133 vector 24 which encodes a Strep-tag II. Nucleotide sequences of all mutants were verified by DNA sequencing. Recombinant LC-As were expressed in the E. coli strain M15pREP4 (QIAGEN, Hilden, Germany) for 16 h at 22°C and purified on StrepTactin-superflow beads (IBA GmbH, Goettingen, Germany) according to the manufacturer’s instructions. Fractions containing the LCs were dialyzed against 150 mM K-glutamate, 10 mM HEPES-KOH, pH 7.2, shock frozen in liquid N2 and stored at −70 °C.
Recombinant SNAP-25
SNAP-25 was cloned as variant B from human adult normal tissue brain cDNA (BioChain Institute, Newark, CA) using primers ACCATGGCCGAAGACGCAGACATG and ACAGCTGACCACTTCCCAGCATCT, with restriction sites NcoI and Pvull, into pET Blue2 (Novagen) using established methods.34 The protein was then expressed in Tuner(DE3)pLacI E. coli cells (Novagen/EMD Millipore, Billerica, MA) and purified using a His Trap HP column (GE Healthcare, Life Science, Pittsburgh, PA).
Synthesis of fluorogenic peptides
The fluorogenic peptides f0, fN, fA, m0, mN, mA (Figure 1A) had amidated C-termini and were synthesized with Rink Amide AM resin LL (100–200 mesh, 300 mg, 75 µmoles, Novabiochem/EMD Millipore [Billerica, MA]) on an in-house built synthesizer as described previously with some modifications.35 Briefly, amino acid couplings were conducted using solid phase chemistry with fluorenylmethoxycarbonyl (Fmoc) aminoprotecting groups,35 but using a solution of piperazine (6% w/v) and ethanol (6.5%) in dimethylformamide as the deblocking reagent. Fmoc amino acids had the following side-chain protecting groups: Thr (t-Bu), Arg (Pbf), Asn and Gln (Trt), Asp and Glu (OtBu). After coupling of the amino terminal threonine, the peptide resins were split in two. Each half of the resin was coupled separately to either 5-(6) carboxy fluorescein or to 7-hydroxy-4-methylcoumarin-3-acetic acid, followed by removal of the Fmoc groups. Average coupling yields were ~99.5% per step. Peptide resins were cleaved in a mixture of water (300 µL) and triisopropyl silane (200 µL) in trifluoroacetic acid (TFA, 5 mL) for one hour at 40°C in a sonicator. The resin suspensions were then filtered and dichloromethane (2–5 mL) was added to the resins on the filters. Filtrates were collected and extracted with n-hexane (7–10 mL) three times, which removed most of the TFA and reduced the peptide solutions to ~2.5 mL each. Peptides were then precipitated at −20°C by addition of cold methyl t-butyl ether (10 mL). The precipitate was collected by centrifugation, washed with methyl t-butyl ether (10 mL), lyophilized, and dissolved in DMSO (2.5 mL). After test separations on an analytical HPLC, the bulk peptides were purified on a 250 × 10 mm HPLC column with Hamilton PRP-1 12–20 µm polymeric support, using a 40 min gradient of water:acetonitrile, 90:10 to 50:50, with constant TFA (0.1%).
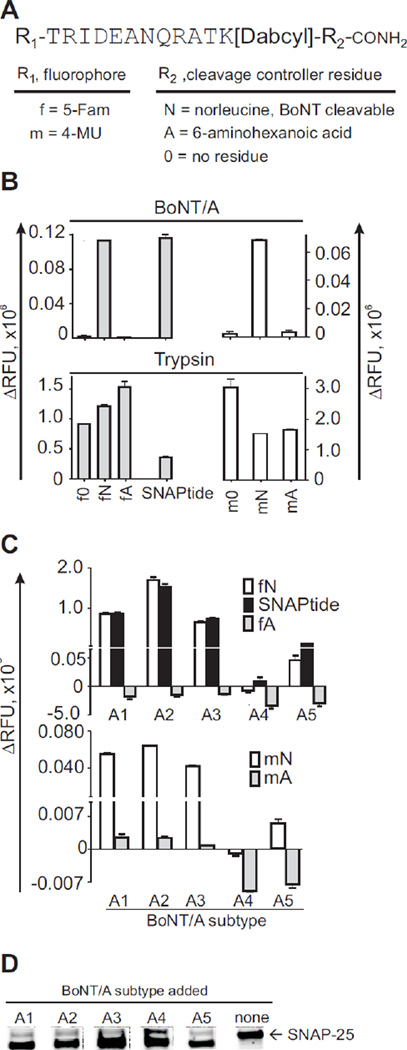
Figure 1.
Naming scheme and proteolytic behavior of fluorogenic peptides and recombinant SNAP-25 protein. (A) All combinations of 5-Fam (f) or 4-MU (m) fluorophores (R1) bound to the N-terminal amino acid of the indicated peptide sequence were tested with various C-terminal residues (R2) that control cleavability by BoNT/A (i.e., no residue (0), norleucine (N), or 6-aminohexanoic acid (A)). For example, “fN” is a BoNT/A-cleavable peptide substrate, flanked by 5-Fam and norleucine, while “mA” is a BoNT/A-non-cleavable peptide control containing 4-MU and 6-aminohexanoic acid. K[DABCYL], lysine residue with DABCYL quencher at the ε-amino group. (B) Fluorescence signal of the peptides (5 µM) after 2 h treatment with BoNT/A1 complex or trypsin (10 nM each). Commercial SNAPtide was used as a control. (C) Enzymatic reaction of fluorogenic peptides with BoNT/A subtype LC-A1 to A5. (D) Western blot of recombinant SNAP-25 treated with LCs of BoNT/A subtypes, or no treatment (none).
Peptide structure validation
Molecular weights of newly synthesized peptides were confirmed by matrix-assisted laser-desorption time-of-flight mass spectrometry on a prO-TOF 2000 (Perkin Elmer, Fremont, CA) with α-cyano-4-hydroxycinnamic acid matrix (Waters, Milford, MA). Excitation, emission and absorption spectra of peptides in 20 mM HEPES (pH 7.8) and 0.1 % Tween-20 were measured on a SpectraMax M2 Microplate Reader (Molecular Devices, Sunnyvale, CA). Fluorescence intensity was measured in 100-µL half area polystyrene 96-well black microplates (Corning Life Sciences, Lowell, MA) with a Victor X2 Multilabel Plate Reader (Perkin Elmer) using excitation/emission wavelengths of 355 nm/460 nm and 485 nm/535 nm for 4-MU and 5-Fam-containing peptides, respectively.
Kinetics with BoNT/A and other proteases
All buffers and solutions were prepared using 18 MΩ/cm-conductivity, autoclaved, protease-free water. Peptide stocks (2.5 mM) were prepared in DMSO, aliquoted and stored at −20°C. A convenient stock solution (250 µM in 30% acetonitrile) was then used to prepare the reaction buffer (see below) with peptide concentrations ranging from 0.5–25 µM. Kinetic parameters of peptides were calculated from nonlinear regression of the initial velocities as a function of a series of eight stepwise increasing substrate concentrations, with a fixed concentration of BoNT/A, trypsin, thermolysin, or proteinase K. The change of fluorescence intensity (ΔRFU) was followed every 2 min for > 1 h at 37°C. Data analysis was performed with Prism 6.0 software (Graph-Pad Software).
BoNT/A ALISSA
The BoNT/A ALISSA was performed with antibodies immobilized on protein A/G agarose beads30–31 or onto CNBr-activated sepharose beads, for higher stability.36 Briefly: The CNBr-activated sepharose beads were washed and allowed to swell in ice-cold HCl (1 mM) for 30 min (330 mg beads in 1 mL suspension). Subsequently, CNBr-activated beads were washed with distilled water and coupling buffer (0.1 M NaHCO3, pH 8.3, 0.5 M NaCl). Antibodies (250 – 500 µg), types as indicated elsewhere in the text, were dissolved in the coupling buffer, mixed with the beads (1 mL), and rotated for 16 hours at 4°C. Unbound antibody was removed by washing once with the coupling buffer, after which the beads were blocked with Tris base (0.2 M, pH 8.0) for two hours at 22°C. Beads were washed at alternating pHs, first, with basic coupling buffer (pH 8.3) followed by a wash with a mixture of sodium acetate (0.1 M) and NaCl (0.5 M), at pH 4.0. These washes were repeated four times. Finally, beads were washed with PBS, re-suspended in ammonium bicarbonate (0.1 M), lyophilized and stored at 4°C.
To generate ALISSA standard curves, dilution series (10−18 – 10−11 M) of BoNT were generated in pooled human or mouse serum (100 µL), and adjusted to a final 1-mL volume with IP buffer (0.025M Tris, pH 7.4, 0.15M NaCl, 1% NP-40, 5% glycerol). Antibody-coupled beads (~5–7 µg of antibody per assay, 200 µL suspension) were transferred into each BoNT dilution sample and to the toxin-negative control. The bead suspension was gently rotated on a rotisserie for 2 hours at 22 °C or overnight (~16 h) at 4°C. The beads were then washed once with IP buffer, twice with Tris-buffered saline (25 mM Tris, 150 mM NaCl, pH 7.2; 0.5 mL/wash), once with conditioning buffer (5 mM Tris, 30 mM NaCl, pH 6.5; 0.5 mL/wash), once with 1 M NaCl (0.5 mL/wash) and three times with protease-free autoclaved water.
Toxin was activated with DTT in the following manner: after incubation with holotoxin or BoNT complex, beads (100 µL) were transferred into reaction buffer (20 mM HEPES, 0.1% Tween-20, 5 µM ZnSO4, 10 mM DTT, pH 7.5; 250 µL) and incubated (30 min, 22°C). The fluorogenic peptides (10 µM) were dissolved in reaction buffer and subsequently added to the DTT-activated toxin, and incubated in the dark for 2 h at 37° C or for 16 hours at 22° C under gentle mixing on a rotary shaker (250 rpm). When the BoNT ALISSA was performed with LC-A1, reduction with DTT was omitted. Instead, to enhance the enzymatic activity of LC-A1, the reaction buffer was supplemented with bovine serum albumin (0.5%).8
Detection of BoNT/A in serum of intoxicated mice. Groups of three to four female Swiss Webster mice (18–21 g) for each dose studied, were treated with active BoNT/A complex either through intravenous injection (i.v., 100 µL), or through oral gavage using Popper feeding needles (100 µL volumes). BoNT/A1 complex doses for each group were 4, 20, and 100 pg/mouse in the i.v. model, and 1 and 4 µg/mouse in the oral model. At indicated time points, blood was collected from intoxicated mice by submandibular bleeding into BD Microtainer serum collection tubes. Blood samples were refrigerated for 1 h at 4°C, and then centrifuged for 10 min at 3000 g to separate sera from cell fractions. Sera were then aliquoted and frozen at − 80°C until use. Animal studies were performed according to approved animal use protocols by the Animal Use and Care Committee of the USDA.
Serum samples were split equally and each portion was analyzed with one set of BoNT-cleavable peptide substrates and their controls: peptide fN combined with peptide mA, or mN combined with fA. Standard curves were measured as described above (Figure S-2 in the Supporting Information). Pharmacokinetic parameters such as the biologic alpha and beta half-lives (t1/2α and t1/2β) of the toxin were calculated using BoNT/A1 serum concentrations, measured 7–8 h post oral administration of the BoNT/A1 complex (1 µg/mouse). Calculations used the following: t1/2 = ln 2/ke,37 where ke is the toxin elimination rate constant. ke was computed with: ke= − (ln C1− ln C2) / (t1− t2), where C1 is the concentration at time = t1, and C2 is the concentration at time = t2.37
RESULTS
Design and performance of fluorogenic peptide substrates and controls for BoNT endoprotease assays
We synthesized a set of six SNAP-25-analogous fluorogenic peptide substrates that used either 5-Fam (green) or 4-MU (blue) as fluorophores. Donor/acceptor pairs of fluorophores and quenchers were chosen to minimize background fluorescence. To make the fluorogenic peptides more stable, we covalently attached 5-carboxyfluorescein (5-Fam) and 4-methylumbelliferone (4-MU) fluorophores to the amino terminus.38–39 The Stokes shift of 4-MU at 116 nm is more than twice as large as the 50 nm-Stokes shift of fluorescein,40 and the emission spectrum of the 4-MU-conjugated peptides overlaps perfectly with the absorption spectrum of the peptide bond-conjugated quencher, 4-((4-(dimethylamino) phenyl) azo) benzoic acid (DABCYL), leading to low background fluorescence (Figure S-1 in the Supporting Information). The fluorescence quantum yield for the 4-MU blue fluorophore, however is 70%,41 which is significantly lower than that of the green 5-Fam fluorophore at 92%.42 We then evaluated the role of the C-terminal amino acid of these peptides, which corresponds to M202 in the native human SNAP-25 sequence, in their ability to be cleaved by BoNT and other proteases. This amino acid is replaced with non-oxidizable norleucine (Nle) in the commercial SNAPtide substrate.43 All peptides had identical sequences, except for their amidated C-terminal residue, which was either Nle (peptides fN and mN), 6-aminohexanoic acid (ε-Ahx)/amide (peptides fA and mA), or lacked that terminal residue (f0 and m0, Figure 1). Peptides with SNAP-25 sequence that are C-terminally truncated before M202 (corresponding to Nle here) are not cleaved by BoNT/A.44 ε-Ahx was chosen as a non-branched isomer of Nle.
All peptides were exposed to BoNT/A complex or trypsin. Only peptides with the Nle residues were efficiently cleaved by BoNT/A, while trypsin cleaved all six peptides, including commercial SNAPtide (Figure 1B). We detected negligible BoNT/A-mediated cleavage of 4-MU-containing peptide substrates that had ε-Ahx (mA) or no terminal residue (m0). This cleavage was less than 2.78–5.44 % of the extent of cleavage obtained with the mN peptide, and was only present after extended incubation times (20 h). At shorter incubation times, such cleavage was not detectable (not shown). No cleavage by BoNT/A was detected for 5-Fam-conjugated peptide substrates that lacked the C-terminal residue (f0) or had ε-Ahx in its place (fA, Figure 1B).
Because the peptides containing Nle and ε-Ahx shared almost identical sequences, were isomers, and were produced with either blue (4-MU) or green (5-Fam) fluorophores, we investigated if the BoNT-non-cleavable peptides (fA or mA) could be multiplexed with BoNT-cleavable substrates (fN and mN), to serve as internal controls to identify non-BoNT-related protease activity. Such use of controls and substrates requires that the controls do not compete for binding at the BoNT/A catalytic site, which would inhibit BoNT’s proteolytic activity. To assess the potential inhibitory effect of the BoNT-non-cleavable peptides (control peptides), BoNT LC-A was incubated with mixtures of control and substrate peptides, in which concentrations of the control peptides (mA or fA) ranged from 1 nM to 1000 µM, while the concentration of BoNT/A-cleavable peptides (fN or mN) was kept constant at 5 µM. The mA and fA control peptides had IC50 values of 215 µM and 160 µM, respectively (Figure S-2 in the Supporting Information). However, in all BoNT detection experiments we used the control or substrate peptides at working concentrations from 1–10 µM, in which both control peptides fA and mA do not inhibit BoNT/A activity.
Substrates and BoNT/A subtypes
To determine if the BoNT/A-cleavable peptides were hydrolyzed by BoNT/A subtypes, we exposed combinations of cleavable substrates and non-cleavable controls (fN with mA, and mN with fA), and commercial SNAPtide to the LCs of subtypes A1–A5 (Figure 1C). fN, mN, and SNAPtide were cleaved by LC-A1, A2, A3, and A5, but not by LC-A4. LC-A4, however, did cleave recombinant SNAP-25, albeit to a much lesser extent than did A1-3 and A5, indicating that LC-A4 had catalytic activity (Figure 1D). The control peptides were not cleaved by any of the A subtypes. For some subtypes, particularly A4 and A5, negative ΔRFU values were obtained for the BoNT-non-cleavable controls. This effect likely arose from quenching of fluorescence by a non-catalytic interaction between the BoNT LCs and the peptides, because the ΔRFU values were always determined by subtracting the background fluorescence of a BoNT-free control buffer. None of the other BoNT serotypes tested (B, C, E, and F) cleaved the substrates or the control peptides (data not shown), indicating that our set of substrates and controls are specific for detecting BoNT/A activity.
Performance of a substrate/control pair in the bead-based ALISSA
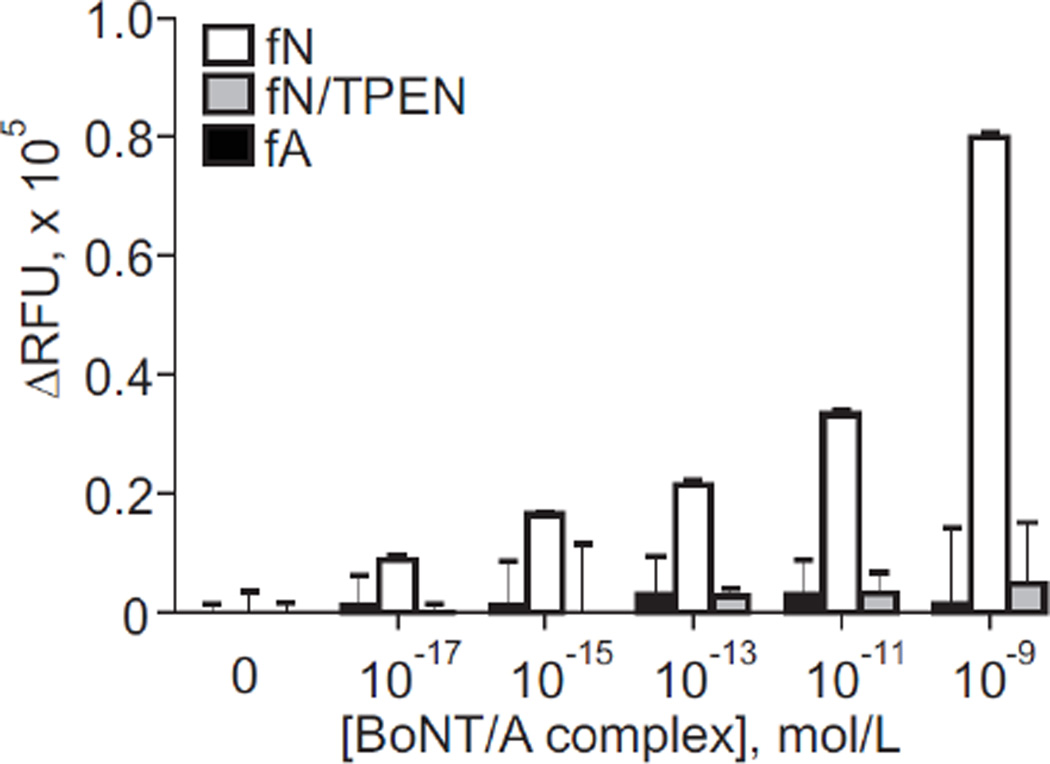
After determining the catalytic properties of the peptides, we tested their performance in a bead-based reaction using protein A/G agarose beads and rabbit polyclonal antibodies to BoNT/A toxoid, as done in our initial ALISSAs.30–31 As a control, in which BoNT/A activity was inhibited, we incubated BoNT/A for one hour with the Zn2+-chelator TPEN (50 µM), one of the most effective metalloprotease inhibitors.45 We then performed enzymatic reactions with the BoNT/A-cleavable (fN) and control peptides (fA). Reactions performed with TPEN showed a significant suppression of fluorescent signal (Figure 2) as compared to those done without TPEN, confirming that the concentration-dependent increase in fluorescent signal seen in TPEN-free reactions was generated by the enzymatic activity of BoNT immobilized on antibody-coupled beads. We also observed a negligible fluorescent signal produced by cleavage of the fA control peptide, likely indicating the presence of non-specific protease activity (Figure 2).
Figure 2.
ALISSA performance of 5-Fam-containing BoNT/A-cleavable substrate (fN) and control peptide (fA) in the presence or absence of the Zn2+-metalloprotease inhibitor TPEN. Toxin was immobilized with a BoNT/A toxoid-generated polyclonal rabbit antibody on prA/G agarose beads.
Kinetic properties of fluorogenic peptides
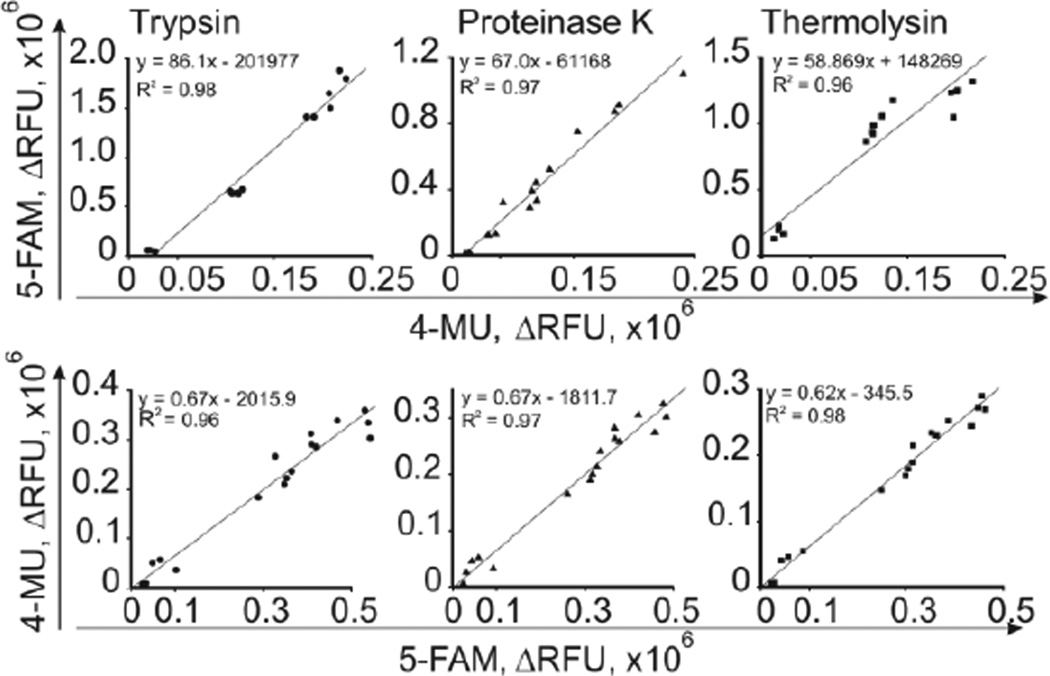
BoNT/A-dependent hydrolysis of peptide substrates produced an increase in fluorescence when measured at excitation and emission wavelengths of 485 and 535 nm for fN and at 355 and 460 nm for mN. We examined the suitability of combining the peptides in the ALISSA and studied their detailed kinetic behavior when exposed to BoNT/A or other proteases such as trypsin, proteinase K, and thermolysin (Table S-1, Figure S-3 in the Supporting Information). The activities of all proteases obeyed Michaelis-Menten kinetics. Although kcat and Km values varied among enzymes and peptide substrates, the catalytic efficiency rates kcat/Km were similar and ranged between 1.50 µM−1s−1 to 18.85 µM−1s−1. Comparable values were reported by Poras et al.27 for cleavage of BoNT/A-cleavable PL50 and PL51 fluorogenic peptide-substrates. For example, trypsin cleaved fN and mN at a higher kcat rate, but at increased Km values, resulting in kcat/Km values, comparable to those for BoNT/A. The control peptides fA and mA were cleaved by trypsin, thermolysin, and proteinase K at kinetic parameters similar to those of the BoNT/A-cleavable peptides fN and mN. Combining BoNT/A-cleavable and control peptides in the same assay appeared to be feasible, and would allow us to distinguish between true and false-positive ALISSA signals.
To compare signals from fluorophores that have different quantum yields (see above), we correlated signals from fA versus those of mA. Each peptide-substrate with a specific fluorophore was combined with a BoNT/A non-cleavable control peptide that was conjugated with the other fluorophore. For example, the 5-Fam-conjugated peptide-substrate fA was combined with the 4-MU-conjugated control peptide mA (Figure 3, upper panels) and vice versa (Figure 3, lower panels), and then incubated with trypsin, proteinase K, or thermolysin at varying concentrations (10 fM to 100 pM). The relationship between the fluorescent signals produced by 5-Fam and 4-MU-conjugated peptides was linear (Figure 3), which allows comparison of fluorescent signals from the different peptides and fluorophores.
Figure 3.
Linear correlation of fluorescent responses produced by 4-MU- and 5-Fam-conjugated BoNT/A-cleavable and control peptides, hydrolyzed by trypsin, thermolysin, or proteinase K. Equimolar mixtures of peptides (5 µM), mN and fA (upper panel), or fN and mA (lower panel), were used.
ALISSA quantification of BoNT/A activity in mouse models of botulism
We further investigated the utility of our novel peptide-substrates and their controls in a pharmacokinetic study involving two mouse models of botulism. BoNT/A ALISSA with the new peptides was used to evaluate serum samples from mice that were i.v. or orally administered with BoNT/A. Mice were injected with BoNT/A complex (100 pg), and within one hour after injection we detected a serum activity equivalent to a concentration of approximately 2879.4 ± 156.83 aM active BoNT/A with the combination of mN and fA peptides and 4343.0 ± 803.18 aM active BoNT/A with the combination of fN and mA peptides. Mice injected with 20 pg of BoNT/A had a serum concentrations of 77.04 ± 12.24 aM (mN/fA) and 74.85 ± 14.29 aM (fN/mA), while those injected with 4 pg of BoNT/A complex had serum concentrations of 12.85 ± 1.96 aM (mN/fA) and 7.15 ± 0.73 (fN/mA) (Figure 4A and Table S-2 in the Supporting Information). A time course analysis of mice injected with 100 pg of BoNT/A1 complex showed serum concentrations of 4.34 ± 0.80 fM (mN/fA) and 2.88 ± 0.16 fM (fN/mA) one hour after injection. There was no detectable BoNT/A at time points greater than three hours (Figure 4B), indicating that BoNT/A is rapidly removed from the circulation.
Figure 4.
BoNT/A ALISSA quantification of BoNT/A in mouse serum measured with 5A20.4 antibody on CNBr-activated sepharose beads. (A) BoNT/A activity in serum from mice (n=4) that were i.v. injected with 100, 20 or 4 pg/mouse dose of BoNT/A. A blood was withdrawn 1 h after intoxication. (B) Time course analysis of BoNT/A serum activity in mice (n=3) that were i.v. injected with 100 pg BoNT/A complex. (C) BoNT/A serum concentration measured over time in mice (n=3) that were orally administered 4 µg or (D) 1 µg of BoNT/A complex. Pre-injection serum (t = 0h) was used for background subtraction.
For orally administered BoNT, a substantial amount of time was required for the toxin to reach the blood stream. In mice administered 4 µg BoNT/A1 complex, toxin blood levels rose slowly and first became detectable 5 h after administration. BoNT was present at serum equivalent concentrations of 165.14 ± 68.15 aM (mN/fA) and 234.94 ± 38.12 aM (fN/mA) (Figure 4C and Table S-3 in the Supporting Information). BoNT concentrations peaked 7 h after administration at 647. 27 ± 172.69 aM (mN/fA) and 723.06 ± 196.81 aM (fN/mA). Mice died at around 7 h after intoxication and the experiment was therefore not long enough to observe eventual absorption and clearance of the toxin from the blood. Thus, we performed another pharmacokinetic study of orally administered BoNT/A to follow the progression of active toxin in the bloodstream. For this study, we used as little as 1 µg of BoNT/A and observed mice for 48 h. BoNT/A again became detectable in the blood 5 h after gavage and peaked 7 h after gavage at serum equivalent concentrations of 72.42 (mN/fA) and 74.69 aM (fN/mA) (Figure 4D and Table S-4 in the Supporting Information). Detection of BoNT/A activity by ALISSA at 24 h after intoxication showed a decreased level of toxin in blood serum because of its clearance from the circulation.
The resulting decrease in BoNT/A serum concentration followed a biphasic pattern. The phase one t1/2α value of the BoNT/A1 complex, representing the distribution from the circulation to the neurons, was approximately 46 ± 12 min. For phase two, the elimination phase and clearance of the toxin from the blood circulation, we determined t1/2β to be approximately 458 ± 57 min. The t1/2α and t1/2β obtained in this study were consistent with pharmacokinetic constants previously determined in mice or rats through the use of catalytically inactive or radiolabeled BoNT/A and B serotypes,46–48 indicating that the BoNT/A ALISSA with our novel set of substrates and control peptides produces sound results, but with much lower doses of active toxin.
DISCUSSION
Blood plasma or serum is among the most convenient and commonly used diagnostic matrices, but botulinum toxin may be present only transiently or at low levels in such samples. Several high affinity BoNT/A synthetic and recombinant substrates have been reported for BoNT enzymatic activity assays.26–29 However, non-specific cleavage of substrates by serum proteases may significantly decrease the sensitivity methods that use conventional peptide substrates to measure the proteolytic activity of BoNT in blood samples. Although the specificity of the assays can be increased by using antibody-capture of toxin from complex biological mixtures, prior to assessing toxin activity,26,30,49 nonspecific binding of protease sample contaminants to the antibodies can lead to false positive results. Pre-incubation of the specimen with non-metalloprotease inhibitors can enhance the assay’s specificity,28 but its sensitivity may suffer. For example, phenylmethylsulfonyl fluoride, a serine protease inhibitor, can react with serine proteases and also with aspartic and metalloproteases, and causes non-specific inhibition of the metalloprotease activity of BoNT.50 Although the accuracy of the assay for the analysis of complex matrices that have interfering concentrations of nonspecific proteases was significantly improved by including a stringent high salt washing during toxin purification,24,51 this method is limited because it substantially decreases the assay sensitivity by also washing away less tightly bound toxin.
Our modified ALISSA is able to overcome these problems. BoNT/A-cleavable peptides contain a non-oxidizable Nle residue instead of an oxidizable methionine residue within the peptide sequence. Control peptides were constructed by replacing the Nle with its isomer, ε-Ahx. These peptides can be used in in vitro assays that measure the proteolytic activity of BoNT. The Nle and ε-Ahx residues appear to take the function of a “cleavage controller residue” (Figure 1A). The M202 residue that these amino acids replace in SNAP-25 was reported to establish substrate/enzyme contact by packing into the β exosite of LC-A.52 Therein, M202, and presumably the Nle residue of a BoNT-cleavable peptide, lies in a hydrophobic cavity formed by two loops of LC-A1, referred to as the “250” and “370 loops”.52 The ε-Ahx residue lacks a side chain, and extends the peptide’s backbone structure linearly, which presumably causes steric hindrance with the 250 loop, preventing the substrate from entering the catalytic site.
LC-A4, differing from A1 by 11% in amino acid sequence, did not cleave any of the substrate peptides tested here. BoNT/A4 was previously reported as cleaving peptide substrates inefficiently,53 potentially because of steric hindrance with the DABCYL quencher at the P4’ (K201) residue of the substrate, which may block interaction with LC-A’s S4’ binding pocket within the toxin’s active site.54 All other BoNT/A subtypes, including LC-A1, A2, A3 and A5, successfully cleave the same peptides, so this effect seems to be specific for A4. It was suggested that the mutation of R264 in LC-A1 to I264 in LC-A4 could destabilize the coordination activity of E262 in relation to the zinc atom in the active site,53 potentially weakening LC-A4’s catalytic activity.
The toxico-pharmacokinetics of BoNT has been largely understudied because of a lack of sufficiently sensitive methods to detect and quantify active toxin in blood. Previous research seeking a window of opportunity for effective treatment of botulism with neutralizing antitoxins found that the window of opportunity and the fractional redistribution of toxin vary as a function of dose in systemic intoxication rodent models of botulism.46–48 However, in those studies the ability to provide therapeutic protection with a set of antibodies against BoNT/A was tested based on the amount of catalytically inactive BoNT/A in the serum of mouse or rat systemic and mouse oral intoxication models.46–47 In addition, because of technical limitations and the lack of sensitive methods, relatively large amounts of holo toxin were used for both the systemic and oral intoxication mouse models for real-time quantification of toxin levels over time.
We used biologically active BoNT/A1 complex to intoxicate mice since its toxicity after oral uptake is much higher than that of holotoxin.55 This oral model mimics human food-borne botulism, a common form of this illness. Lowering the oral doses of BoNT/A1 complex from 4 to 1 µg allowed the observation of BoNT activity in the serum over 48 h. The BoNT activity reached a maximum at 7 h (Figure 4C and D, and Tables S-3 and S-4 in the Supporting Information). Analysis of the biologic half-lives of active BoNT/A in the bloodstream indicated that BoNT/A administered orally declined biexponentially. Active toxin had a very short ~45 min distribution time during the early intoxication phase of t1/2α, and an extended t1/2β clearance time of about 7.6 h. Together, these results confirmed the utility of our approach to assess the pharmacokinetics of biologically active BoNT/A.
The combination of BoNT-cleavable fluorogenic peptide substrates and BoNT-non-cleavable controls allowed us to quantitatively distinguish between BoNT and non-BoNT-related protease activities and to directly quantify toxin levels in the serum of mouse models of botulism. We were able to measure an atto- to femtomolar final concentration of toxin in blood one hour after mice were injected with 4 pg of BoNT/A complex. The toxin could not be detected 3 hours post-intoxication. Our data confirm that selective accumulation of the BoNT toxin does not occur over time in formed blood elements (cells and fragments); instead the toxin is rapidly removed from the circulation, most likely due to neural absorption.47
The absorption and distribution of BoNT toxin after oral intoxication is greatly affected by digestion of the toxin by gastric enzymes,56 and detection of the toxin in blood is challenging. Nevertheless, use of the modified ALISSA with BoNT/A-cleavable and control peptides to measure the toxin’s pharmacokinetics in mouse serum identified attomolar concentrations of BoNT/A. To date, few animal studies have measured the pharmacokinetics, physiological distribution and stability of BoNT toxins.46–48,55 To our knowledge, the study presented here is the only study of active botulinum neurotoxin pharmacokinetics in an oral model of botulism. Mice treated orally with the low dose of 1 µg of BoNT/A1 complex allowed the robust detection of toxin after more than 24 h by the ALISSA.
CONCLUSIONS
Our study demonstrates the advantages of using a combination of well-defined BoNT/A-cleavable fluorogenic peptides and controls, which should greatly reduce, or even eliminate, false positive results due to protease contaminants. When integrated into the ALISSA, this system proved to be useful for the pharmacokinetic study of BoNT in mouse models of botulism. It could potentially be used for future pharmacological, toxicological, and diagnostic assessment of BoNT in humans with botulism or in patients who receive BoNT-based medical treatments.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. James Marks and Jianlong Lou from the University of California, San Francisco, for providing the monoclonal anti-BoNT LC-A antibody 5A20.4. The study was funded by the National Institutes of Health through grants AI096169-01 (MK and KB), AI065359-05 (MK, KB and LWC) and the United States Department of Agriculture, Agriculture Research Service, CRIS project 5325-42000-048-00D (LWC), and the Robert Koch Institut, 1362/I-979 (AR and JS).
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Fenicia L, Franciosa G, Pourshaban M, Aureli P. Clin. Infect Dis. 1999;29:1381–1387. doi: 10.1086/313497. [DOI] [PubMed] [Google Scholar]
- 2.Harvey SM, Sturgeon J, Dassey DE. J. Clin. Microbiol. 2002;40:2260–2262. doi: 10.1128/JCM.40.6.2260-2262.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peck MW. Adv. Microb. Physiol. 2009;55:183–265. 320. doi: 10.1016/S0065-2911(09)05503-9. [DOI] [PubMed] [Google Scholar]
- 4.Schiavo G, Matteoli M, Montecucco C. Physiol. Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 5.Summanen P. Clin Infect Dis. 1993;16(Suppl 4):S168–S174. doi: 10.1093/clinids/16.supplement_4.s168. [DOI] [PubMed] [Google Scholar]
- 6.Oguma K, Fujinaga Y, Inoue K. Microbiol. Immunol. 1995;39:161–168. doi: 10.1111/j.1348-0421.1995.tb02184.x. [DOI] [PubMed] [Google Scholar]
- 7.Singh BR. Nat. Struct. Biol. 2000;7:617–619. doi: 10.1038/77900. [DOI] [PubMed] [Google Scholar]
- 8.Montecucco C, Schiavo G. Mol. Microbiol. 1994;13:1–8. doi: 10.1111/j.1365-2958.1994.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 9.Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson MJ, Lin G, Raphael B, Andreadis J, Johnson EA. Appl. Environ. Microbiol. 2008;74:2778–2786. doi: 10.1128/AEM.02828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luquez C, Raphael BH, Maslanka SE. Appl. Environ. Microbiol. 2009;75:6094–6101. doi: 10.1128/AEM.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazuet C, Ezan E, Volland H, Popoff MR, Becher F. J. Clin. Microbiol. 2012;50:4091–4094. doi: 10.1128/JCM.02392-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellizzari R, Rossetto O, Schiavo G, Montecucco C. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1999;354:259–268. doi: 10.1098/rstb.1999.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Sudhof TC, Jahn R, Niemann H. J. Biol. Chem. 1994;269:1617–1620. [PubMed] [Google Scholar]
- 15.Wein LM, Liu Y. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9984–9989. doi: 10.1073/pnas.0408526102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster KA. Toxicon. 2009;54:587–592. doi: 10.1016/j.toxicon.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 17.Chertow DS, Tan ET, Maslanka SE, Schulte J, Bresnitz EA, Weisman RS, Bernstein J, Marcus SM, Kumar S, Malecki J, Sobel J, Braden CR. JAMA. 2006;296:2476–2479. doi: 10.1001/jama.296.20.2476. [DOI] [PubMed] [Google Scholar]
- 18.Crowner BE, Brunstrom JE, Racette BA. Clin Neuropharmacol. 2007;30:310–313. doi: 10.1097/WNF.0b013e31804b1a0d. [DOI] [PubMed] [Google Scholar]
- 19.Cote TR, Mohan AK, Polder JA, Walton MK, Braun MM. J. Am. Acad. Dermatol. 2005;53:407–415. doi: 10.1016/j.jaad.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Weingart OG, Schreiber T, Mascher C, Pauly D, Dorner MB, Berger TF, Egger C, Gessler F, Loessner MJ, Avondet MA, Dorner BG. Appl. Environ. Microbiol. 2010;76:3293–3300. doi: 10.1128/AEM.02937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer AE, Moura H, Woolfitt AR, Kalb SR, McWilliams LG, Pavlopoulos A, Schmidt JG, Ashley DL, Barr JR. Anal. Chem. 2005;77:3916–3924. doi: 10.1021/ac050485f. [DOI] [PubMed] [Google Scholar]
- 22.Parpura V, Chapman ER. Croat. Med. J. 2005;46:491–497. [PubMed] [Google Scholar]
- 23.Sachdeva A, Defibaugh-Chavez SL, Day JB, Zink D, Sharma SK. Appl. Environ. Microbiol. 2010;76:7653–7657. doi: 10.1128/AEM.00820-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wictome M, Newton K, Jameson K, Hallis B, Dunnigan P, Mackay E, Clarke S, Taylor R, Gaze J, Foster K, Shone C. Appl. Environ. Microbiol. 1999;65:3787–3792. doi: 10.1128/aem.65.9.3787-3792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt JJ, Stafford RG. Appl. Environ. Microbiol. 2003;69:297–303. doi: 10.1128/AEM.69.1.297-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruge DR, Dunning FM, Piazza TM, Molles BE, Adler M, Zeytin FN, Tucker WC. Anal. Biochem. 2011;411:200–209. doi: 10.1016/j.ab.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Poras H, Ouimet T, Orng SV, Fournie-Zaluski MC, Popoff MR, Roques BP. Appl. Environ. Microbiol. 2009;75:4382–4390. doi: 10.1128/AEM.00091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalb SR, Moura H, Boyer AE, McWilliams LG, Pirkle JL, Barr JR. Anal. Biochem. 2006;351:84–92. doi: 10.1016/j.ab.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 29.Hines HB, Kim AD, Stafford RG, Badie SS, Brueggeman EE, Newman DJ, Schmidt JJ. Appl. Environ. Microbiol. 2008;74:653–659. doi: 10.1128/AEM.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagramyan K, Barash JR, Arnon SS, Kalkum M. PLoS One. 2008;3:e2041. doi: 10.1371/journal.pone.0002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagramyan K, Kalkum M. Methods Mol. Biol. 2011;739:23–36. doi: 10.1007/978-1-61779-102-4_3. [DOI] [PubMed] [Google Scholar]
- 32.Marks JD, Lou J, Rodrigez MC, Geren IM. 0200615A1. United State Patent. 2011
- 33.Rummel A, Bade S, Alves J, Bigalke H, Binz TJ. Mol. Biol. 2003;326:835–847. doi: 10.1016/s0022-2836(02)01403-1. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook JFEF, Maniatis T. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Kaplan BE, Hefta LJ, Blake RC, 2nd, Swiderek KM, Shively JE. J. Pept. Res. 1998;52:249–260. doi: 10.1111/j.1399-3011.1998.tb01239.x. [DOI] [PubMed] [Google Scholar]
- 36.Madru B, Chapuis-Hugon F, Pichon V. Talanta. 2011;85:616–624. doi: 10.1016/j.talanta.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Garcia A, Bermejo M, Moss A, Casabo VG. J. Pharm. Sci. 2008;97:654–690. doi: 10.1002/jps.21009. [DOI] [PubMed] [Google Scholar]
- 38.Bradley M, Alexander L, Duncan K, Chennaoui M, Jones AC, Sanchez-Martin RM. Bioorg. Med. Chem. Lett. 2008;18:313–317. doi: 10.1016/j.bmcl.2007.10.075. [DOI] [PubMed] [Google Scholar]
- 39.Demant E. J. Anal. Biochem. 1999;267:366–372. doi: 10.1006/abio.1998.3019. [DOI] [PubMed] [Google Scholar]
- 40.Tsourkas A, Behlke MA, Xu Y, Bao G. Anal. Chem. 2003;75:3697–3703. doi: 10.1021/ac034295l. [DOI] [PubMed] [Google Scholar]
- 41.Chen RF. Anal. Lett. 1968;1:423–428. [Google Scholar]
- 42.Ratilainen T, Holmen A, Tuite E, Haaima G, Christensen L, Nielsen PE, Norden B. Biochemistry. 1998;37:12331–12342. doi: 10.1021/bi9808722. [DOI] [PubMed] [Google Scholar]
- 43.Shine NR, Crowford KR, Eaton LJ. 6,504,006. United States Patent. 2003
- 44.Schmidt JJ, Bostian KA. J. Protein Chem. 1995;14:703–708. doi: 10.1007/BF01886909. [DOI] [PubMed] [Google Scholar]
- 45.Adler M, Dinterman RE, Wannemacher RW. Toxicon. 1997;35:1089–1100. doi: 10.1016/s0041-0101(96)00215-2. [DOI] [PubMed] [Google Scholar]
- 46.Cheng LW, Stanker LH, Henderson TD, 2nd, Lou J, Marks JD. Infect. Immun. 2009;77:4305–4313. doi: 10.1128/IAI.00405-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravichandran E, Gong Y, Al Saleem FH, Ancharski DM, Joshi SG, Simpson LL. J. Pharmacol. Exp. Ther. 2006;318:1343–1351. doi: 10.1124/jpet.106.104661. [DOI] [PubMed] [Google Scholar]
- 48.Al-Saleem FH, Ancharski DM, Ravichandran E, Joshi SG, Singh AK, Gong Y, Simpson LL. J. Pharmacol. Exp. Ther. 2008;326:856–863. doi: 10.1124/jpet.108.136242. [DOI] [PubMed] [Google Scholar]
- 49.Kalb SR, Lou J, Garcia-Rodriguez C, Geren IN, Smith TJ, Moura H, Marks JD, Smith LA, Pirkle JL, Barr JR. PLoS One. 2009;4:e5355. doi: 10.1371/journal.pone.0005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nam KH, Kim SJ, Priyadarshi A, Kim HS, Hwang KY. Biochem. Biophys. Res. Commun. 2009;389:247–250. doi: 10.1016/j.bbrc.2009.08.123. [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Baudys J, Kalb SR, Barr JR. Anal. Biochem. 2011;412:67–73. doi: 10.1016/j.ab.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 52.Breidenbach MA, Brunger AT. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 53.Henkel JS, Jacobson M, Tepp W, Pier C, Johnson EA, Barbieri JT. Biochemistry. 2009;48:2522–2528. doi: 10.1021/bi801686b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S, Kim JJ, Barbieri JT. J. Biol. Chem. 2007;282:9621–9627. doi: 10.1074/jbc.M611211200. [DOI] [PubMed] [Google Scholar]
- 55.Ohishi I, Sakaguchi G. Infect. Immun. 1980;28:303–309. doi: 10.1128/iai.28.2.303-309.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonventre PF. Rev. Infect. Dis. 1979;1:663–667. doi: 10.1093/clinids/1.4.663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.