Abstract
The human inducible nitric oxide synthase (hiNOS) gene is expressed in several disease states and is also important in the normal immune response. Previously, we described a cytokine-responsive enhancer between −5.2 and −6.1 kb in the 5′-flanking hiNOS promoter DNA, which contains multiple nuclear factor κβ (NF-κB) elements. Here, we describe the role of the IFN-Jak kinase-Stat (signal transducer and activator of transcription) 1 pathway for regulation of hiNOS gene transcription. In A549 human lung epithelial cells, a combination of cytokines tumor necrosis factor-α, interleukin-1β, and IFN-γ (TNF-α, IL-1β, and IFN-γ) function synergistically for induction of hiNOS transcription. Pharmacological inhibitors of Jak2 kinase inhibit cytokine-induced Stat 1 DNA-binding and hiNOS gene expression. Expression of a dominant-negative mutant Stat 1 inhibits cytokine-induced hiNOS reporter expression. Site-directed mutagenesis of a cis-acting DNA element at −5.8 kb in the hiNOS promoter identifies a bifunctional NF-κB/Stat 1 motif. In contrast, gel shift assays indicate that only Stat 1 binds to the DNA element at −5.2 kb in the hiNOS promoter. Interestingly, Stat 1 is repressive to basal and stimulated iNOS mRNA expression in 2fTGH human fibroblasts, which are refractory to iNOS induction. Overexpression of NF-κB activates hiNOS promoter–reporter expression in Stat 1 mutant fibroblasts, but not in the wild type, suggesting that Stat 1 inhibits NF-κB function in these cells. These results indicate that both Stat 1 and NF-κB are important in the regulation of hiNOS transcription by cytokines in a complex and cell type-specific manner.
Keywords: signal transduction, NO synthase, iNOS
Regulation of human inducible nitric oxide synthase (hiNOS) expression involves both transcriptional and posttranscriptional control. Human iNOS gene transcription is controlled in a cell type-specific manner by extracellular cytokines (1). Aberrant iNOS expression and excessive nitric oxide (NO) production are observed in a large variety of pathophysiologic conditions (2). Important differences have been described between human and rodent species in the regulation of iNOS expression (2). Therefore, it is critical to delineate the mechanisms that control hiNOS gene transcription so that therapeutic strategies can be developed to regulate hiNOS expression in various disease conditions.
Cytokine or bacterial lipopolysaccharide (LPS)-inducible NO production was originally identified by Stuehr and Marletta in LPS-stimulated murine macrophages (3), which led to the initial cloning of the murine iNOS (miNOS) cDNA (4–6). In contrast, it was difficult to demonstrate induced NO production from cytokine-stimulated human macrophages. However, NO production was elicited from primary human hepatocytes stimulated with the same combination of cytokines that was effective in inducing rat hepatocyte iNOS expression (7, 8). This allowed for the subsequent molecular cloning of the hiNOS cDNA (9) and determination of the hiNOS genomic organization (10). Early studies in both mouse and humans indicated that the regulation of iNOS expression by cytokines occurs predominantly at the level of transcription (11, 12). Transfection of miNOS promoter–reporter plasmids demonstrated that LPS or cytokine-responsive regulatory DNA elements exist within 1.0 kb of 5′-flanking DNA of the mouse iNOS gene (12, 13). In contrast, hiNOS promoter–reporter plasmids require DNA sequences farther upstream in the 5′-flanking region between −4.7 and −16 kb to exhibit cytokine-inducibility (11).
In the miNOS promoter, LPS and/or cytokine-inducible nuclear factor κβ (NF-κB) elements have been identified at −76 to −85 bp and at −962 to −971 bp upstream from the transcription start site (5, 14, 15). In marked contrast, critical NF-κB elements have been localized far upstream in the hiNOS promoter region from −5.2 to −6.1 kb (1), and at −8.3 kb (16). A recent review of the NF-κB signaling pathways is provided (17).
The induction of miNOS expression by LPS and IFN-γ has also been shown to involve the IFN–Jak–Stat (signal transducer and activator of transcription) pathway. IFN-γ activates Jak 1 and Jak 2 kinases, which phosphorylate Stat 1. Stat 1 homodimers then interact with IFN-γ-activating sequence (GAS) elements (18, 19). Significantly, IFN-γ (20), IFN-γ receptor (21), and Stat 1 (22) knockout mice are defective for the induction of NO synthesis by LPS and/or cytokines. Functional ISRE and GAS elements were identified in the miNOS promoter (23–25). Recent experiments in the murine system indicate a complex role for IFN-α/β and Jak 2 kinase in cytokine-induced miNOS transcription (26–28). Cotransfection of a dominant-negative Jak 2 kinase with a miNOS promoter–reporter revealed that Jak 2 functions as an activator or a repressor of miNOS transcription depending on the cell type.
Pharmacological inhibition of cytokine-induced hiNOS transcription has implicated Jak 2 in the regulation of hiNOS transcription by IFN-γ (29). However, the precise mechanisms by which the IFN-γ–Jak–Stat 1 pathway regulates hiNOS gene expression have not been described. The present study investigates the direct involvement of the IFN-γ–Jak 2 kinase–Stat1 and NF-κB pathways in the regulation of hiNOS gene transcription. Through the use of pharmacological inhibitors, NF-κB and Stat 1 expression plasmids, Stat 1 mutant cell lines, and mutations of hiNOS promoter–reporter plasmids, we demonstrate a direct and complex role for both Stat 1 and NF-κB in regulating hiNOS gene transcription.
Methods
Cell Lines and Reagents.
The A549 human lung epithelial cell line was obtained from the ATCC and cultured in F-12K medium (GIBCO/BRL) supplemented with 10% heat inactivated, low endotoxin FBS (GIBCO/BRL), 100 units/ml penicillin, 100 μg/ml streptomycin, and 15 mM Hepes, pH 7.4. The human fibroblastic cell lines, 2fTGH and the Stat 1 mutant derivative U3A, were kindly provided by Timothy Wright of the University of Pittsburgh and are described (30). The 2fTGH and U3A cell lines were cultured in DMEM media (BioWhittaker) supplemented with 5% heat-inactivated, low endotoxin FCS (HyClone), 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 15 mM Hepes, pH 7.4. Unless indicated, cells were stimulated with a cytokine mixture consisting of 1,000 units/ml human TNF-α (R & D Systems), 100 units/ml IL-1β (provided by Craig Reynolds of the National Cancer Institute, Bethesda), and human 250 units/ml IFN-γ (R & D Systems or Roche Molecular Biochemicals), which were purified–recombinant proteins. Tyrphostin A25 and B42 drugs were purchased from Calbiochem and were used at concentrations indicated. All other chemicals were purchased from Sigma.
Plasmid Constructs.
The hiNOS promoter–reporter plasmid piNOS(7.2)Luc contains −7.2 kb of upstream 5′-flanking DNA linked to the luciferase reporter gene and has been described (1, 11). The NF-κB mutant derivative plasmids, piNOS(m5.2)Luc and piNOS(m5.8)Luc, contain NF-κB mutations in the context of piNOS(7.2)Luc (1, 11). Highly selective mutations of the −5.8 kb NF-κB and Stat 1 elements were generated from the piNOS(7.2)Luc reporter plasmid by using the QuickChange mutagenesis kit according to manufacturer recommendations (Stratagene). Confirmations of all mutations were accomplished with DNA sequence analysis by the University of Pittsburgh DNA Sequencing Facility and are shown in Fig. 1. Expression plasmids encoding the human p50 and p65 NF-κB proteins we kindly provided by Joseph DiDinato of the University of California at San Diego, La Jolla, CA. Expression plasmids pCAGGS encoding wild-type Stat 1 (pStat 1) and dominant-negative Stat 1 (pStat1 F) were kindly provided by Koichi Nakajima, Osaka City University Medical School, Osaka. All plasmids were amplified according to standard procedures and purified on low endotoxin maxi prep columns (Qiagen, Chatsworth, CA).
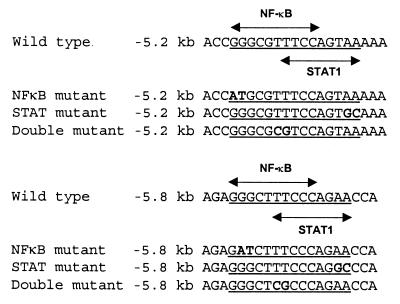
Figure 1.
Overlapping NF-κB and Stat 1 elements at −5.2 kb and −5.8 kb in the hiNOS promoter.
Transient Transfections and Activity Assays.
DNA transfections of cells were carried out in six-well plates (Corning), using Lipofectamine (GIBCO) as described (11). Briefly, cells were exposed to serum-free medium containing 1 μg of DNA and 20 μg of liposomes for 4 h, washed, and replenished with medium supplemented with 5% calf serum. Preliminary transfection experiments showed optimal transfection efficiency and low toxicity with a DNA:liposome ratio of 1:20. To control for transfection efficiency between groups, 0.5 μg of a plasmid containing a cytomegalovirus promoter-driven β-galactosidase gene (pIEP-Lacz) was added to each well. As a positive control, cells were transfected with PRSV-Luc while transfection of the promoterless plasmid pXP2 served as a negative control. Cells were lysed with Reporter lysis buffer (Promega) or buffer containing 1% Triton X-100, 5 mM dithiotreitol, 50% glycerol, 10 mM EDTA, and 125 mM Tris-phosphate (pH 7.8). Luciferase activity was assayed with 20 μl of lysate in a Berthold (Nashua, NH) AutoLumat LB 953 luminometer using a commercially available kit (Promega). β-Galactosidase activity was determined as recommended (Promega), using a 96-well multiplate reader with SOFTMAX software (Molecular Devices). Luciferase activity was normalized to β-galactosidase activity. Cotransfection experiments with the NF-κB or Stat 1 expression vectors included an additional 1.0 μg of the indicated expression plasmid.
Northern and Western Blotting.
Northern and Western blot experiments were performed as described according to established protocol (9).
Preparation of Nuclear and Nonnuclear Protein Fractions.
Briefly, the cytokine-stimulated cells were washed and scraped into phosphate-buffered solution and centrifuged at 4,500 rpm for 5 min in a microfuge. The pelletted cells were suspended in buffer A [10 mM Tris (pH 7.5)/1.5 mM MgCl2/10 mM KCl/0.5% Nonidet P-40] at ≈10 × the packed cell volume and lysed by gentle pipetting. Nuclei were recovered by microcentrifugation at 7,000 rpm for 5 min. The supernatant was collected and stored at −80°C and represents the cytoplasmic and membrane protein fraction. Nuclear proteins were extracted at 4°C by gentle resuspension of the nuclei (at ≈2 × the packed nuclear volume) of buffer containing 20 mM Tris (pH 7.5), 10% glycerol, 1.5 mM MgC12, and 420 mM NaCl, 0.2 mM EDTA, followed by 30 min of platform rotation. The nuclear protein suspension was cleared by microcentrifugation at 13,000 rpm for 15 min. The supernatants were collected and frozen at −80 or directly used in gel shift assays. All buffers contained the following additions: 1–2 μg/ml each of aprotinin, chymostatin, leupeptin, pepstatin, 0.2 mM PMSF, 0.5 mM DTT, and 0.1 mM Na-vanadate. All steps were carried out on ice or at 4°C. Protein concentrations were measured by the Bio-Rad protein assay, using BSA as a standard.
EMSA Assays.
All oligonucleotides were purchased from (GIBCO/BRL), the sequences of which are listed in Fig. 1, except for the SIE oligonucleotide, which was purchased from Santa Cruz Biotechnology. DNA probes were prepared by end-labeling with [γ-32P]dATP (DuPont/NEN) and T4 polynucleotide kinase (Boehringer Mannheim) and purified in TEN by using G-50 resin columns (Whatman). Typically, 5 μl (10–20 μg) of nuclear proteins was incubated with ≈100,000 cpm of 32P-labeled oligonucleotides (≈0.5 ng) for 2 h at room temperature. The nuclear proteins and various oligonucleotide probes were incubated in a buffer containing 10 mM Tris (pH 7.5), 10% glycerol, and 0.2% Nonidet P-40. Additionally, 2–4 μg of poly (dI-dC) (Boehringer Mannheim) was included as a nonspecific competitor DNA. Protein–DNA complexes were resolved on 4% nondenaturing polyacrylamide gels in 0.4× TBE running buffer (450 mM Tris borate and 1 μM EDTA, pH 8.0). After electrophoresis, gels were dried and subjected to autoradiography. Antibody supershift experiments included the addition of 2 μl of various antibodies, all of which were purchased from Santa Cruz Biotechnology. Purified, recombinant p50 NF-κB protein was purchased from Promega.
Results
Pharmacological Inhibitors of the IFN-γ–Jak Kinase–Stat 1 Pathway Inhibit Cytokine-Induced hiNOS Gene Expression.
Previously, we described a novel enhancer between −5.2 and −6.1 kb of hiNOS promoter DNA that contained multiple NF-κB elements (1). The NF-κB element located at −5.8 kb was the most critical motif required for cytokine-induced hiNOS transcription as mutation of this site abrogated all inducible promoter activity. Surprisingly, this NF-κB element at −5.8 kb overlapped with a predicted Stat 1 DNA-binding sequence (Fig. 1). Additionally, the functional element at −5.2 kb also contained a putative overlapping NF-κB–Stat 1 DNA-binding sequence suggesting a possible role for Stat 1 in controling hiNOS transcription. To assess the role of the Jak–Stat pathway in mediating cytokine induction of hiNOS gene expression, we used pharmacological inhibitors of the Jak 2 kinase in Northern and Western blot experiments to assess iNOS mRNA and protein expression in response to cytokine mixture. As depicted in Fig. 2A, incubation with tyrphostin A25 inhibited cytokine-induced iNOS mRNA and protein expression in a dose-dependent manner in the human A549 lung cell line. A similar effect was seen with JAK 2–Stat inhibitor tyrphostin B42 (data not shown). Tyrphostin B42 also inhibited cytokine-induced Stat 1 DNA-binding activity to the consensus Stat 1 binding oligo (SIE) in gel shift experiments (Fig. 2B). Tyrphostin B42 had no significant negative effect on NF-κB DNA-binding. These data suggest that the Jak–Stat pathway is involved in regulating cytokine-induced hiNOS expression in the A549 cells.
Figure 2.
The effect of Jak kinase inhibitors on hiNOS expression and on cytokine induction of nuclear NF-κB or Stat 1 DNA-binding. (A) Northern and Western blot analysis of hiNOS mRNA and protein induced by CM. The Jak kinase inhibitor (tyrophostin A25) inhibits cytokine-induced iNOS mRNA and protein expression in a dose-dependent fashion. (B) The gel shift experiment shows the effects of the Jak 2 kinase inhibitor tyrophostin B42 on the nuclear DNA-binding activities of both NF-κB and Stat 1. Nuclear protein extracts were prepared from cells exposed for 2 h to a cytokine mixture in the absence or presence of tyrophostin B42 as indicated. The nuclear proteins were subjected to EMSA, using the consensus NF-κB or hSIE oligonucleotide probes. Blots shown are representative of three similar experiments.
Expression of a Dominant-Negative Stat 1 Mutant Inhibits Cytokine-Induced hiNOS Reporter Gene Transcription.
To further implicate a functional role for Stat 1 in hiNOS transcription, we used a dominant-negative Stat 1 mutant expression plasmid in a cotransfection experiment analyzing cytokine-induced hiNOS reporter expression in the A549 epithelial cells. Cytokine mixture induced a 4- to 5-fold increase in hiNOS promoter–luciferase reporter activity (data not shown). In cells cotransfected with empty or wild-type Stat 1 expression plasmid, no significant change was observed in hiNOS reporter induction. However, cotransfection with the dominant-negative Stat 1 expression vector significantly inhibited cytokine-induced hiNOS reporter expression by ≈50%, implicating Stat 1 as a positive regulator of hiNOS gene transcription in these cells.
Identification of a Bifunctional NF-κB-Stat 1 Element at −5.8 kb in the hiNOS Promoter That Binds Both NF-κB and Stat 1 in a Protein–DNA Complex.
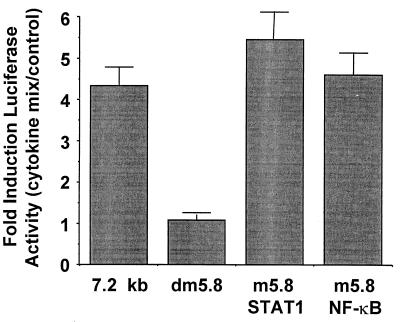
Analysis of the DNA sequence at −5.8 kb in the hiNOS promoter indicates the possibility of overlapping NF-κB and Stat 1 cis-acting elements (Fig. 1). By chance, the original site-directed mutagenesis construct at −5.8 kb in the context of the −7.2 kb human iNOS promoter actually altered both of the NF-κB and Stat 1 DNA-binding sequences (labeled double mutant, Fig. 1). Therefore, we devised a more specific mutagenesis strategy to determine the individual contributions of NF-κB or Stat 1 to hiNOS promoter–reporter expression. The highly selective mutant hiNOS promoter–reporter plasmids were transfected into A549 cells and tested for cytokine inducibility by luciferase assay. As expected, the cytokine mixture of TNF-α + IL-1β + IFN-γ induced a 4- to 5-fold induction in luciferase activity with the −7.2 kb wild-type hiNOS promoter construct (Fig. 3). Mutation of both the NF-κB and Stat 1 elements at −5.8 kb (double mutant) completely eliminated the cytokine-induced luciferase expression. Mutation of either the NF-κB or Stat 1 sites individually failed to inhibit inducible promoter activity, suggesting that binding of either transcription factor at this vicinity was sufficient for transcriptional activity. Individual cytokines did not produce any significant increase in human iNOS promoter activity in the wild-type or mutant constructs at −5.8 kb, consistent with previous work in human liver cells (11).
Figure 3.
The cis-acting transcription element at −5.8 kb in the hiNOS promoter is a bifunctional, composite NF-κB/GAS element. The graph depicts the cytokine-induced expression of the −7.2-kb hiNOS promoter–luciferase reporter and various mutant derivatives shown in Fig. 1. Mutation of both the NF-κB and the Stat 1 elements eliminate cytokine induction of the hiNOS reporter plasmid, whereas mutation of the NF-κB or Stat 1 sequence individually had no effect on hiNOS promoter activity (n = 4–6 transfections per condition).
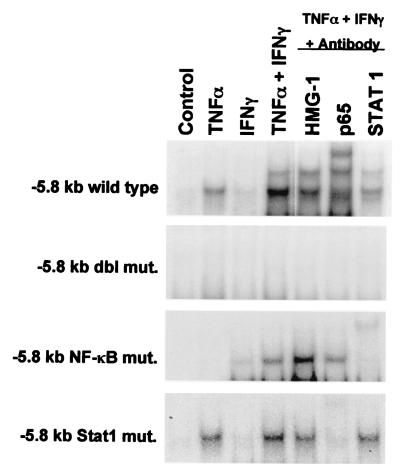
To demonstrate that NF-κB and Stat 1 can interact directly with the DNA sequence at −5.8 kb, gel shift experiments were performed with wild type, double mutant, NF-κB mutant, or Stat 1 mutant specific oligos as described in Fig. 1. As shown in Fig. 4, TNF-α (and to a lesser extent IFN-γ), induced a protein–DNA complex at the wild-type −5.8 kb element. The combination of TNF-α + IFN-γ induced two distinct protein–DNA complexes. Interestingly, either p65 NF-κB or Stat 1 (but not HMG-1) antibodies eliminated or retarded the migration of the protein–DNA complex. No protein–DNA binding was observed with the double mutant oligo at −5.8 kb, consistent with the lack of inducible hiNOS promoter activity when using the double mutant promoter plasmid in the transfection experiments. With the NF-κB mutant oligo, TNF-α alone failed to induce a complex. IFN-γ alone or with TNF-α induced a protein–DNA complex that was only supershifted by antibody to Stat 1. With the Stat 1 mutant oligo, TNF-α alone or with IFN-γ yielded an inducible complex that was supershifted by antibody to NF-κB. The supershift results support the notion that TNF-α signals through NF-κB, whereas IFN-γ signals through Stat 1. The gel shift findings, taken together with the mutant promoter transfection studies, indicate that the element at −5.8 kb in the hiNOS promoter is a composite, bifunctional NF-κB/Stat 1 element.
Figure 4.
Inflammatory cytokines induce distinct NF-κB or Stat 1–DNA complexes at the −5.8 kb hiNOS promoter element. The figure is a gel shift assay analyzing the induction of nuclear DNA-binding proteins in response to either TNF-α, IFN-γ, or a combination in nuclear extracts from human A549 lung epithelium. Antibody supershift assays indicate that TNF-α induces a protein–DNA complex containing NF-κB protein, whereas IFN-γ induces a Stat 1–DNA complex. Blots shown are representative of two similar experiments.
The Functional Element at −5.2 kb in the hiNOS Promoter Is a Stat 1 Binding Sequence.
Previously, we demonstrated that the element at −5.2 kb in the hiNOS promoter is important for cytokine-induced iNOS transcription (1). Like the element at −5.8 kb, the element at −5.2 kb was predicted to contain putative overlapping NF-κB and Stat 1 DNA-binding sequences when compared with known consensus binding sites (31, 32). To identify which proteins interact with the −5.2 kb sequence, gel shift assays were performed by using the wild-type or highly selective mutant oligos from the DNA sequence at −5.2 kb in the hiNOS promoter. In nuclear extracts from cytokine-stimulated A549 cells, only IFN-γ alone or as part of a cytokine mixture induced a protein–DNA complex (Fig. 5A). Selective mutation of the putative NF-κB sequence had no effect on protein–DNA binding, but mutation of the Stat 1 site abolished all binding. These results suggest that Stat 1, but not NF-κB, binds at −5.2 kb. To confirm the identity of the transcription factor recognizing the DNA element at −5.2 kb, antibody supershift experiments were performed. Only antibodies to Stat 1 protein produced a supershift in the IFN-γ-induced protein–DNA complex (Fig. 5B). Antibodies raised against five members of the NF-κB family, as well as other known IFN-inducible nuclear proteins, did not alter the protein–DNA complex (Fig. 5B). Finally, in an attempt to determine whether we could “force” the binding of NF-κB to the DNA site at −5.2 kb, purified recombinant p50 NF-κB protein failed to recognize the −5.2 kb sequence or a consensus Stat 1 (SIE) sequence (Fig. 5C). As positive control, the recombinant p50 protein did bind to a consensus NF-κB oligo in a concentration-dependent manner. These data indicate that the element at −5.2 kb in the hiNOS promoter is a strict Stat 1 DNA-binding sequence and not an NF-κB element.
Figure 5.
The cytokine responsive element at −5.2 kb in the hiNOS promoter is a functional Stat 1 DNA-binding sequence. (A) Only IFN-γ alone or as part of CM induces a protein/DNA-binding complex with the −5.2-kb element. Mutation of the NF-κB domain does not alter binding, but mutation of the Stat 1 site abolishes all binding. (B) Antibody supershift shows that the protein–DNA complex at −5.2 kb is recognized exclusively by Stat 1 antibody. (C) Purified recombinant NF-κB protein binds to a consensus NF-κB oligo, but not to the −5.2 kb or to a consensus Stat 1 (hSIE) element. Blots shown are representative of three similar experiments.
The IFN–Stat 1 Pathway Is a Repressor of iNOS Gene Expression in a Human Fibroblast Cell Line.
To further address the role of Stat 1 in the regulation of human iNOS mRNA expression, we performed Northern blot experiments in the Stat 1-null human fibroblast cell line U3A, and in its parental wild-type derivative 2fTGH (30). In the wild-type 2fTGH fibroblasts, very-low-level iNOS mRNA was detected in resting cells (Fig. 6A). TNF-α caused only a minimal increase in iNOS mRNA, whereas either IFN-β or IFN-γ decreased the low-level basal iNOS mRNA expression that we observed. Repression of basal iNOS mRNA expression by IFNs was not observed in the Stat 1 mutant U3A cells, which expressed a higher basal level of iNOS mRNA. In addition, in the Stat 1-null cells, either TNF-α, IFN-γ, or combinations of TNF-α and IFNs were able to induce a modest increase in iNOS mRNA. Two distinct iNOS mRNA bands were observed, possibly reflecting alternatively spliced mRNA in these cells. These data suggest that the IFN–Jak–Stat 1 pathway can serve as a repressor of basal and stimulated iNOS mRNA expression in human fibroblasts.
Figure 6.
Interferons and Stat 1 repress hiNOS mRNA and NF-κB-mediated hiNOS transcription in a human fibroblast cell line. (A) The Northern blot shows hiNOS mRNA expression in response to various combinations of cytokines as labeled. IFN-β or IFN-γ inhibit basal and stimulated hiNOS mRNA expression in the wild-type 2fTGH human fibroblasts, but not in the Stat 1-null U3A cells. (B) Analysis of hiNOS promoter–luciferase reporter plasmids in human 2fTGH and U3A fibroblasts indicate that Stat 1 can function as a repressor of NF-κB-induced iNOS transcription. The figure illustrates the fold induction of a hiNOS–luciferase reporter in cells that were cotransfected with empty vector or vectors that express either p50 + p65 NF-κB protein, wild-type Stat 1, or dominant-negative mutant Stat 1 protein. Notice that NF-κB overexpression will induce a significant hiNOS expression only in the Stat 1 mutant U3A cells. Blots shown are representative of three similar experiments.
Endogenous Stat 1 Is a Repressor of NF-κB-Induced iNOS Transcription in Human Fibroblasts.
The increased level of iNOS mRNA in the human Stat 1 mutant fibroblast cell line (U3A) was a surprising finding. Because NF-κB is critical for rodent and human iNOS expression, we sought to determine whether NF-κB overexpression could induce the hiNOS–luciferase reporter activity in the wild-type (2fTGH) and Stat 1 mutant (U3A) human fibroblasts. Cotransfection of ectopic expression plasmids for p50 and p65 NF-κB proteins in wild-type 2fTGH cells was minimally effective at inducing hiNOS reporter expression (1.5-fold). Interestingly, overexpression of NF-κB in the Stat 1 mutant U3A cells induced hiNOS reporter expression ≈7-fold (Fig. 6B). Addition of cytokines had no effect on hiNOS promoter activity, consistent with the low-level of cytokine inducible iNOS mRNA seen on Northern blot. Overexpression of either Stat 1 or the dominant-negative Stat 1 mutant minimally repressed the hiNOS reporter activity in either cell line. These data suggest that endogenous Stat 1 functions as an inhibitor of NF-κB activity in the human 2fTGH fibroblasts. Deletion of Stat 1 in the U3A cells results in higher basal and stimulated hiNOS expression. Inhibition of NF-κB function by the Stat 1 pathway provides a possible mechanism by which IFNs may inhibit iNOS mRNA expression and why certain cells are refractory to iNOS induction. This appears to be a cell-type-specific phenomenon as cotransfection of the p50/p65 expression plasmids in the A549 human lung cells led to a 5-fold induction in hiNOS promoter activity (data not shown), similar to that seen in the Stat 1 mutant U3A cells.
Discussion
Previously, promoter regions required for cytokine-induced hiNOS transcription were mapped far upstream in the 5′-flanking region of the hiNOS gene from −4 to −16 kb (11). Functionally important NF-κB-like sequences have been identified at −5.2, −5.5, −5.8, −6.1 kb (1), and −8.2 kb (16) in the hiNOS promoter. In addition, inducible AP-1 binding sites have been reported at −5.1 and −5.3 kb in the hiNOS promoter (16). However, no information exists as to the precise function of Stat 1 in governing hiNOS transcription. Therefore, the purpose of this study was to define the molecular role for the IFN–JAK–STAT 1 pathway in regulating hiNOS gene expression. The major and novel findings of these experiments are the following: (i) identification of a specific cis-acting DNA element at −5.2 kb in the hiNOS promoter that bind Stat 1 in response to IFN-γ; (ii) determination that the DNA element at −5.8 kb in the hiNOS promoter is actually a critical bifunctional motif that binds either Stat 1 and/or NF-κB in response to cytokines; and (iii) recognition that Stat 1 signaling is complex and may actually serve as a positive or negative regulator of hiNOS transcription, depending on the cell type.
In the current study, pharmacological inhibitors of IFN-γ-induced Jak kinase activity inhibited Stat 1 DNA-binding activity and inhibit cytokine-induced iNOS mRNA and protein expression in A549 human lung epithelial cells. Similar observations were reported in DLD-1 human colon epithelial cells (29). However, it should be noted that the tyrphostin drugs have been shown to inhibit NF-κB in some situations (33); therefore, additional experiments were done to precisely delineate the role of Stat 1 in regulating hiNOS transcription. Expression of dominant-negative Stat 1 protein inhibited cytokine-induced hiNOS promoter–reporter activity indicating a positive role for Stat 1 in regulating hiNOS transcription. Importantly, functional GAS elements that bind to Stat 1 were identified at −5.2 and at −5.8 kb in the hiNOS promoter, highlighting the marked differences from the rodent iNOS promoter where only ≈1.0 kb of 5′-flanking DNA is required to confer LPS and cytokine-inducibility (12, 13).
Nuclear extracts from human lung cells stimulated with IFN-γ alone or IFN-γ as part of a cytokine mixture produced strong gel-shift protein binding to the wild-type DNA oligo at −5.2 kb in the hiNOS promoter. Previously, we identified the site at −5.2 kb as being an NF-κB response element (1) based on the sequence “resemblance” to the NF-κB response element (8/10 match). Mutation of the TT to CG decreased cytokine-inducible promoter activity by over 50% (1). Our interpretation of the data were that we were inhibiting NF-κB binding. What we failed to initially recognize was that the −5.2 kb site actually contained overlapping NF-κB and Stat 1 response motifs. Therefore, we generated highly selective mutant oligos and found that mutation of the NF-κB domain did not alter protein–DNA binding, but mutation of the Stat 1 domain abolished all binding. Antibody supershifts confirmed that the protein binding at −5.2 kb was actually Stat 1 and not NF-κB. The importance of the DNA element at −5.2 kb was also shown in vivo in living cells where mutation of this site within the −7.2 kb hiNOS promoter construct significantly decreased cytokine-induced luciferase activity in transfection experiments in human liver and lung cells (1). These data demonstrate that Stat 1 functions directly in the regulation of hiNOS transcription by binding to a GAS element at −5.2 kb in the hiNOS promoter DNA.
Interestingly, the DNA element at −5.8 kb was shown to be a bifunctional composite NF-κB/Stat 1 binding site. A two-point mutation that changed both cis-acting motifs (double mutant) abolished all inducible DNA binding in vitro in the gel shifts and blocked all inducible hiNOS promoter activity in vivo in the cell transfections, indicating that this −5.8 kb site is indeed critical for hiNOS transcription. One interpretation of the data is that both NF-κB and Stat 1 bind in a protein-protein–DNA complex. This interaction could provide a molecular basis for the cytokine synergy required to achieve significant hiNOS expression where TNF-α or IL-1β signal through NF-κB (1), and IFN-γ signals through Stat 1 for hiNOS transcription. An alternative interpretation of the data are that binding of NF-κB and Stat 1 are mutually exclusive at −5.8 kb, and that binding of either nuclear factor is permissive for the transcriptional machinery. In favor of this view is the observation that the double mutation completely abrogates inducible promoter activity, but mutation of either site alone does not diminish cytokine-driven hiNOS reporter expression.
Surprisingly, we show that IFN-γ and IFN-α/β are repressive to basal and stimulated iNOS mRNA expression in the 2fTGH human fibroblasts, and that this repression is Stat 1-dependent because it was lost in the Stat 1-null U3A cells. Further, we show that endogenous Stat 1 in the 2fTGH cells represses the 7-fold increase in hiNOS promoter activity driven by overexpression of NF-κB in the U3A cells. Additionally, IFN-γ can repress TNF-α-induced NF-κB–luciferase reporter expression in a Stat 1-dependent manner (data not shown). We believe that Stat 1-dependent repression of NF-κB function may contribute to the lack of iNOS induction in human fibroblasts and other human cell types. These data indicate that the interactions between TNF-α and IFN-γ and between NF-κB and Stat 1 are complex, cell type-specific, and can be cooperative or antagonistic to various functions within a single cell type.
Ohmori reported (34) that synergy between TNF-α and IFN-γ for induction of ICAM-1, IP-10, and MIG-1 transcription was mediated by cooperative interactions between NF-κB and Stat 1 cis-regulatory elements (34). Others have shown a direct interaction between NF-κB and Stat 6 proteins (35). In the murine system, IFN-γ has been shown to activate miNOS expression directly via Stat 1 protein–miNOS promoter DNA interaction as well as indirectly, through the IFN-γ-induced, Stat 1-dependent induction of IRF-1 gene expression (23–25, 36). Importantly, macrophages from Stat 1 knockout mice are totally defective for induction of NO production by LPS plus IFN-γ (22). Splenic derived macrophages from IRF-1 knockout mice are partially defective for miNOS induction by LPS and cytokines. However, IRF-1 has no effect on miNOS induction in peritoneal macrophages from the same mouse (25, 37). Murphy's group has shown that LPS alone can induce miNOS expression in macrophages, and this requires de novo synthesis and paracrine functions of IFN-α/β (27). Likewise, IL-1β alone is effective for induction of hepatocyte iNOS transcription (38) and this induction requires de novo synthesis and paracrine function of IFN-γ (39). We propose that Stat 1 is important for miNOS and hiNOS gene expression, whereas IRF-1 serves as a cell type-specific modulator of high-level miNOS gene transcription. A role for the IRF-1 protein in the regulation of hiNOS gene expression remains undefined.
Supporting our finding that Stat 1 can serve as a negative regulator of NF-κB-mediated transcription is the recent report by Wang where Stat 1 is inhibitory to NF-κB function ascribed to a Stat 1-TRADD interaction at the TNF-α receptor complex (40). In this study, over-expression of Stat1 decreased cytokine-stimulated hiNOS mRNA levels in the U3A cells, consistent with the inhibition of NF-κB driven hiNOS promoter–reporter activity that we have shown. In addition, IFN-α has been shown to inhibit NF-κB activation in multiple cell types (41). Sedger has shown that IFN-γ treatment inhibits basal but not induced NF-κB reporter expression in primary human fibroblasts (42). Further, Gao has shown that prolonged exposure of mouse macrophages to IFN-β suppressed miNOS transcription because of altered availability of phosphorylated Stat-1α (43). We postulate that IFN- and Stat 1-dependent antagonism of NF-κB represents an important cell type-specific mechanism to down-regulate the inflammatory response. Identification of the factors that determine cooperativity verses antagonism between these two signal transduction pathways could identify an important target for the regulation of hiNOS gene expression during chronic inflammatory conditions.
Although the regulatory circuits governing both human and murine iNOS gene transcription involve the NF-κB and Stat 1 signal transduction pathways, our results highlight the marked differences between them. The relative ease of inducing miNOS expression may reflect that functionally important NF-κB, IRSE, and GAS elements are localized within 1.0 kb of 5′-flanking promoter DNA, whereas in humans the critical elements are localized much further upstream between −5.0 and −8.3 kb of 5′-flanking promoter DNA. The manner by which the critical NF-κB and Stat 1 elements communicate with the general transcription machinery may differ because of the distance involved for the human system. Perhaps multiple, physically interacting NF-κB and Stat 1 elements are required to function from such a distance and would dictate that a rather strong NF-κB and Stat 1 inducer would be required for cooperative induction of iNOS transcription in humans.
Acknowledgments
This work was supported by National Institutes of Health Grants R01 GM-52021 (to D.A.G.) and NRSA F32 GM-19877 (to R.W.G.), and the George H. A. Clowes, Jr., M.D., F.A.C.S. Memorial Research Career Development Award of the American College of Surgeons (to D.A.G.).
Abbreviations
- NF-κB
nuclear factor κβ
- Stat
signal transducer and activator of transcription
- iNOS
inducible nitric oxide synthase
- hiNOS
human iNOS
- miNOS
murine iNOS
- GAS
IFN-γ-activating sequence
- LPS
lipopolysaccharide
References
- 1.Taylor B S, de Vera M E, Ganster R W, Wang Q, Shapiro R A, Morris S M, Jr, Billiar T R, Geller D A. J Biol Chem. 1998;273:15148–15156. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 2.Ganster R W, Geller D A. In: Nitric Oxide: Biology and Pathobiology. Ignarro L, editor. San Diego: Academic; 2000. pp. 129–156. [Google Scholar]
- 3.Stuehr D J, Marletta M A. Proc Natl Acad Sci USA. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Q W, Cho H J, Calaycay J, Mumford R A, Swiderek K M, Lee T D, Ding A, Troso T, Nathan C. Science. 1992;256:225–228. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 5.Lowenstein C J, Glatt C S, Bredt D S, Snyder S H. Proc Natl Acad Sci USA. 1992;89:6711–6715. doi: 10.1073/pnas.89.15.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons C R, Orloff G J, Cunningham J M. J Biol Chem. 1992;267:6370–6374. [PubMed] [Google Scholar]
- 7.Geller D A, Nussler A K, Di Silvio M, Lowenstein C J, Shapiro R A, Wang S C, Simmons R L, Billiar T R. Proc Natl Acad Sci USA. 1993;90:522–526. doi: 10.1073/pnas.90.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nussler A K, DiSIlvio M, Billiar T R, Hoffman R A, Geller D A, Selby R, Madariaga J, Simmons R L. J Exp Med. 1992;176:261–264. doi: 10.1084/jem.176.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geller D A, Lowenstein C J, Shapiro R A, Nussler A K, Di Silvio M, Wang S C, Nakayama D K, Simmons R L, Snyder S H, Billiar T R. Proc Natl Acad Sci USA. 1993;90:3491–3495. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartrain N A, Geller D A, Koty P P, Sitrin N F, Nussler A K, Hoffman E P, Billiar T R, Hutchinson N I, Mudgett J S. J Biol Chem. 1994;269:6765–6772. [PubMed] [Google Scholar]
- 11.de Vera M E, Shapiro R A, Nussler A K, Mudgett J S, Simmons R L, Morris S M, Jr, Billiar T R, Geller D A. Proc Natl Acad Sci USA. 1996;93:1054–1059. doi: 10.1073/pnas.93.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Q W, Whisnant R, Nathan C. J Exp Med. 1993;177:1779–1784. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowenstein C J, Alley E W, Raval P, Snowman A M, Snyder S H, Russell S W, Murphy W J. Proc Natl Acad Sci USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Q W, Kashiwabara Y, Nathan C. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 15.Spink J, Cohen J, Evans T J. J Biol Chem. 1995;270:29541–29547. doi: 10.1074/jbc.270.49.29541. [DOI] [PubMed] [Google Scholar]
- 16.Marks-Konczalik J, Chu S C, Moss J. J Biol Chem. 1998;273:22201–22208. doi: 10.1074/jbc.273.35.22201. [DOI] [PubMed] [Google Scholar]
- 17.Mercurio F, Manning A M. Curr Opin Cell Biol. 1999;11:226–232. doi: 10.1016/s0955-0674(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 18.Darnell J E., Jr J Interferon Cytokine Res. 1998;18:549–554. doi: 10.1089/jir.1998.18.549. [DOI] [PubMed] [Google Scholar]
- 19.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 20.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 21.Kamijo R, Shapiro D, Le J, Huang S, Aguet M, Vilcek J. Proc Natl Acad Sci USA. 1993;90:6626–6630. doi: 10.1073/pnas.90.14.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 23.Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, Le J, Koh S I, Kimura T, Green S J, et al. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 24.Gao J, Morrison D C, Parmely T J, Russell S W, Murphy W J. J Biol Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 25.Martin E, Nathan C, Xie Q W. J Exp Med. 1994;180:977–984. doi: 10.1084/jem.180.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faure V, Courtois Y, Goureau O. J Biol Chem. 1997;272:32169–32175. doi: 10.1074/jbc.272.51.32169. [DOI] [PubMed] [Google Scholar]
- 27.Gao J J, Filla M B, Fultz M J, Vogel S N, Russell S W, Murphy W J. J Immunol. 1998;161:4803–4810. [PubMed] [Google Scholar]
- 28.Lopez-Collazo E, Hortelano S, Rojas A, Bosca L. J Immunol. 1998;160:2889–2895. [PubMed] [Google Scholar]
- 29.Kleinert H, Wallerath T, Fritz G, Ihrig-Biedert I, Rodriguez-Pascual F, Geller D A, Forstermann U. Br J Pharmacol. 1998;125:193–201. doi: 10.1038/sj.bjp.0702039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenardo M J, Baltimore D. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 32.Darnell J E. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa Y, Mukaida N, Kuno K, Rice N, Okamoto S, Matsushima K. J Biol Chem. 1995;270:4158–4164. [PubMed] [Google Scholar]
- 34.Ohmori Y, Schreiber R D, Hamilton T A. J Biol Chem. 1997;272:14899–14907. doi: 10.1074/jbc.272.23.14899. [DOI] [PubMed] [Google Scholar]
- 35.Shen C H, Stavnezer J. Mol Cell Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saura M, Zaragoza C, Bao C, McMillan A, Lowenstein C J. J Mol Biol. 1999;289:459–471. doi: 10.1006/jmbi.1999.2752. [DOI] [PubMed] [Google Scholar]
- 37.Shiraishi A, Dudler J, Lotz M. J Immunol. 1997;159:3549–3554. [PubMed] [Google Scholar]
- 38.Geller D A, DeVera M E, Russell D, Shapiro R A, Nussler A K, Simmons R L, Billiar T R. J Immunol. 1995;155:4890–4898. [PubMed] [Google Scholar]
- 39.Schroeder R A, Gu J S, Kuo P C. Hepatology. 1998;27:711–719. doi: 10.1002/hep.510270312. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Wu T R, Welte T, Chin Y E. Mol Cell Biol. 2000;20:4505–4512. doi: 10.1128/mcb.20.13.4505-4512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manna S K, Mukhhopadhyay A, Aggarwal B B. J Immunol. 2000;165:4927–4934. doi: 10.4049/jimmunol.165.9.4927. [DOI] [PubMed] [Google Scholar]
- 42.Sedger L M, Shows D M, Blanton R A, Peschon J J, Goodwin R G, Cosman D, Wiley S R. J Immunol. 1999;163:920–926. [PubMed] [Google Scholar]
- 43.Gao J J, Filla M B, Lorsbach R B, Pace J L, Crespo A, Russell S W, Murphy W J. Eur J Immunol. 2000;30:1551–1561. doi: 10.1002/1521-4141(200006)30:6<1551::AID-IMMU1551>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]