SUMMARY
Differences in gut commensal flora can dramatically influence autoimmune responses, but the mechanisms behind this are still unclear. We report, in a Th1 cell-driven murine model of autoimmune arthritis, that specific gut commensals, such as segmented filamentous bacteria, have the ability to modulate the activation threshold of self-reactive T cells. In the local microenvironment of gut-associated lymphoid tissues, inflammatory cytokines elicited by the commensal flora dynamically enhanced the antigen responsiveness of T cells that were otherwise tuned down to a systemic self-antigen. Together with subtle differences in early lineage differentiation, this ultimately led to an enhanced recruitment of pathogenic Th1 cells and the development of a more severe form of autoimmune arthritis. These findings define a key role for the gut commensal flora in sustaining ongoing autoimmune responses through the local fine-tuning of T cell receptor proximal activation events in autoreactive T cells.
INTRODUCTION
Commensal flora can have profound effects on the adaptive immune response and in particular on the development and plasticity of gut-associated T helper (Th) and T regulatory (Treg) cell subsets (Mazmanian et al., 2005; Gaboriau-Routhiau et al., 2009; Cerf-Bensussan and Gaboriau-Routhiau, 2010; Round and Mazmanian, 2010; Atarashi et al., 2011; Chung et al., 2012). In mouse, a single member of the gut microflora, segmented filamentous bacteria (SFB), promotes the generation of an intestinal subset of Th17 cells as well as enhances the expression of IFN-γ or Foxp3 in gut T cell subsets (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009). When properly controlled, such dialog participates in enhancing host protection against external pathogens, such as Citrobacter rodentium (Ivanov et al., 2009). In the context of a susceptible genetic background, however, SFB can influence the development of autoimmune pathologies, as demonstrated in murine models of rheumatoid arthritis (Wu et al., 2010) and multiple sclerosis (Lee et al., 2011). In both cases, the exact mechanism(s) involved in the coordination of T cell responses by proinflammatory members of the gut microflora remain poorly understood. Furthermore, analyzing the effect of the gut commensal flora solely on the basis of T cell differentiation can’t explain the broad effect that SFB can have on gut-associated immune responses (Talham et al., 1999; Gaboriau-Routhiau et al., 2009; Chung et al., 2012).
The case of T cell responses directed against self-antigen expressed in the gut and gut-associated lymphoid organs adds an additional layer of complexity. In various settings of chronic antigenic stimulation, T cells have been shown to rapidly lose their proliferative and effector potential (Carmichael et al., 1993; Rehermann et al., 1996; Oxenius et al., 1998; Zajac et al., 1998; Brooks et al., 2005). Detailed studies in double transgenic models have demonstrated the intrinsic property of mature CD4+ T cells to tune their functional avidity to the level of ambient antigenic stimulation (Tanchot et al., 2001; Singh and Schwartz, 2003), as predicted by the tunable activation threshold (TAT) model (Grossman and Paul, 1992; 2001). In contrast to in vitro clonal anergy, this tuning of T cell sensitivity, also known as adaptive tolerance, requires persistent antigenic expression and is partially reversible upon removal of the antigen (Tanchot et al., 2001; Han et al., 2010). From a molecular point of view, this unresponsive state reflects an impairment of T cell receptor (TCR) proximal signaling (Schwartz, 2003; Choi and Schwartz, 2007; 2011), a block that can be bypassed by directly stimulating the T cells with phorbol myristate acetate (PMA) and ionomycin along with fresh antigen-presenting cells (APCs) (Chiodetti et al., 2006). As this T cell intrinsic tolerance mechanism represents one of the first checkpoints in the establishment of a chronic self-specific T cell response, an impact of proinflammatory members of the gut microbiota at the level of T cell tuning could explain their potential in driving such a broad spectrum of autoimmune diseases.
We report here an example of Th1 cell-driven autoimmune arthritis for which disease incidence and severity was crucially linked to a specific flora, correlating with the colonization of hosts by SFB. Carefully analyzing the impact of the gut microbiota on the highly coordinated, chronic, self-specific T cell response observed in this in vivo model, we show that proinflammatory signals elicited by specific commensals have the ability to lower the activation threshold of T cells facing chronic antigenic stimulation in the local microenvironment of the gut-draining lymphoid tissues. By bypassing and/or reversing T cell-intrinsic tuning mechanisms, microbiota-induced signals favored chronic activation of self-reactive T cells and magnified early effects of the gut flora on distal parameters such as lineage differentiation. This work provides new insights into the sequence of events locally triggered by proinflammatory members of the gut microbiota to promote systemic autoimmune diseases.
RESULTS
SFB regulate the severity of autoimmune disease in a T cell transfer model of arthritis
We previously reported that transfer of 5C.C7 TCR transgenic, Rag2−/− CD4+ T cells, expressing a Vα11Vβ3 TCR specific for pigeon cytochrome c (PCC), into a T cell-deficient, mPCC, Cd3e−/− host leads to polyclonal hypergammaglobulinemia associated with mild arthritis (Singh et al., 2006). We noticed a delayed incidence as well as a decreased severity of the arthritis when our colony of mPCC, Cd3e−/− hosts, originally housed in a Murine Pathogen FreeTM (MPF) barrier in Taconic Farms, was rederived, colonized with the sole Altered Schaedler Flora and transferred to a separate Restricted FloraTM (RF) barrier.
Side by side experiments carefully monitoring the appearance of arthritis in RF- or MPF-housed hosts substantiated our initial observations. RF-housed hosts developed less severe arthritis (Figure 1A) and the first symptoms appeared on average 2 weeks later than in their MPF-housed counterparts. This was also reflected in the emergence of anticellular antibodies in the sera, where the late qualitative change in the autoantibody response, detectable by day 21 in MPF-housed hosts, was delayed about 10 days in RF-housed hosts (Figure 1B). Co-housing mPCC, Cd3e−/− hosts of RF and MPF origin for 3 weeks was sufficient to transfer much of the enhanced susceptibility to arthritis to mice originally raised in an RF microenvironment (cohoused-RF(MPF)) (Figure 1C–D). This confirmed that a transmissible factor from the MPF microenvironment, and not a genetic drift and selection linked to our recent rederivation, was having a dominant impact on the outcome of the autoimmune response.
Figure 1. SFB colonization of host gut-commensal flora enhances the severity of autoimmune disease in a T cell transfer model of arthritis.
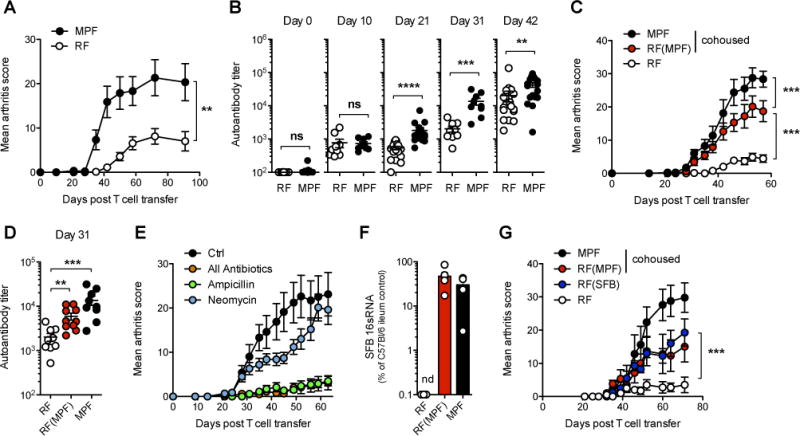
(A) Weekly arthritis score and (B) serum autoantibody titers in RF- or MPF-housed mPCC, Cd3e−/− hosts at indicated time points after naïve 5C.C7 T cell transfer. Data (mean±SEM) pooled from two (A) and four (B) independent experiments (n=9 and 18 mice per group respectively).
(C–D) One group of RF-housed mPCC, Cd3e−/− mice was cohoused with MPF-housed hosts for 3 weeks prior to T cell transfer (RF(MPF)). (C) Bi-weekly arthritis scores and (D) autoantibody titers at day 31. Data (mean±SEM) pooled from two independent experiments (n=9 RF, n=10 RF(MPF) and n=8 MPF).
(E) Various antibiotic formulations were given in the drinking water of MPF-housed mPCC, Cd3e−/− mice starting 3 weeks prior to T cell transfer and maintained for the length of the experiment. “All antibiotics” formulation contains ampicillin, neomycin, vancomycin and metronidazole. Data (mean±SEM) pooled from two independent experiments (n=10 mice per group).
(F) Amount of SFB 16sRNA in cecum contents of indicated mPCC, Cd3e−/− hosts 8 weeks post T cell transfer (n=5 mice per group, mean±SEM).
(G) Same as (C) with an additional group of RF-housed mPCC, Cd3e−/− mice cohoused with SFB-monocolonized GF mice 3 weeks prior to T cell transfer (RF(SFB)). (n=4 RF and RF(SFB), n=6 RF(MPF) and n=5 MPF, mean±SEM). See also Figure S1.
To better characterize the nature of such a factor, we next treated MPF-housed hosts with a cocktail of four antibiotics (ampicillin, neomycin, vancomycin and metronidazole), previously described to ablate most of the gut commensal bacterial species (Ivanov et al., 2008). This appeared sufficient to almost fully prevent the appearance of any sign of autoimmune arthritis (Figure 1E). Ampicillin alone, but not neomycin sulfate, led to a similar outcome. This suggested a key role for specific, ampicillin-sensitive, members of the MPF-gut commensal flora.
A similar impact of an ampicillin-sensitive member of the intestinal microbiota, segmented filamentous bacteria (SFB), has recently been reported in the K/BxN spontaneous model of rheumatoid arthritis (Wu et al., 2010). In light of this observation, we tested for and found SFB 16sRNA by quantitative PCR in both ileum and cecum of 6 week old MPF-housed mPCC, Cd3e−/− hosts, but not RF-housed hosts, at a level similar to that of control MPF-housed C57BL/6 mice from Taconic Farms (Ivanov et al., 2009) (Figure S1A). This commensal organism was easily transferred to cohoused-RF(MPF) hosts (Figure 1F), and absent from mice undergoing ampicillin, but not neomycin sulfate, treatment (Figure S1B). Finally, cohousing of RF-housed hosts with SFB-monocolonized Germ-free mice (cohoused-RF(SFB)) demonstrated the critical impact that the addition of only SFB to the host microbiota can have on the outcome of the autoimmune arthritis in this model (Figure 1G).
SFB promote the differentiation of arthritis-inducing Th1 cells
The impact of SFB on autoimmune responses has been previously linked to its ability to promote IL-17A-producing CD4+ T helper cells (Th17), as well as follicular helper T cells (Tfh), in gut-associated lymphoid tissues and spleen (Wu et al., 2010; Lee et al., 2011). In our model, however, no significant difference could be detected for the early differentiation of autoreactive IL-17A+ (Figure 2A–B), or PD-1+Bcl-6+ Tfh 5C.C7 T cells (Figure S2A) and germinal center B cells (Figure S2B–C), in both mLN and spleen of mPCC, Cd3e−/− hosts, housed under RF or MPF conditions. There were also no notable changes in Foxp3+ T cells, which were close to undetectable in both hosts by day 4 in any organ we looked at (Figure S2D and data not shown), as previously reported (Singh et al., 2006). IL-10 production by CD4+ T cells appeared also similar in mLN of RF- and MPF-housed hosts at day 4 and actually increased in the spleen of MPF-housed hosts (Figures S2E–F), supporting the idea that the RF commensal flora did not simply regulate the balance between pro- and anti-inflammatory T cell responses.
Figure 2. SFB-containing flora promotes the differentiation of arthritis-inducing Th1 cells.
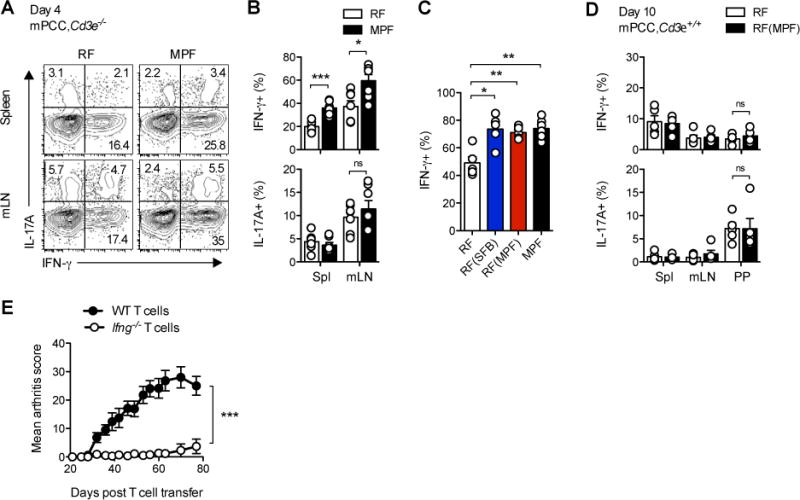
(A–C) 4 days post transfer of naïve 5C.C7 T cells into mPCC, Cd3e−/− hosts, the cells were stained for intracellular IL-17A and IFN-γ following a 3h stimulation with PMA and ionomycin. (A) Representative IFN-γ and IL-17A expression profiles, gated on live, CD4+Vβ3+ T cells and (B) frequencies of IFN-γ+ or IL17A+ CD4+Vβ3+ T cells in the indicated organs. (C) Frequencies of IFN-γ+ in CD4+Vβ3+ T cells in mLN of indicated host. Data (mean±SEM) pooled from 3 independent experiments each (n=7 (A–B) and n=5–6 (C) mice per group).
(D) 10 days post transfer of naïve CD45.1+ 5C.C7 T cells into 8–12 wk. old mPCC, Cd3e+/+ hosts, the cells were stained for intracellular IL-17A and IFN-γ following a 5.5h stimulation with PMA and ionomycin. Frequencies of IFN-γ+ or IL17A+ in CD45.1+ CD4+Vβ3+ T cells in indicated organs. Data (mean±SEM) pooled from 5 independent experiments, each representing one pool of 6 RF-housed or 2 cohoused-RF(MPF) mice. RF(MPF) hosts were cohoused for 2 weeks with MPF-housed mPCC, Cd3e−/− mice prior to T cell transfer.
(E) Bi-weekly arthritis scores of MPF-housed mPCC, Cd3e−/− mice post transfer of naïve 5C.C7 WT or Ifng−/− 5C.C7, Rag2−/− T cells. Data (mean±SEM) pooled from two independent experiments (n=10 mice per group). See also Figure S2.
Instead, a reproducible increase in the frequency of IFN-γ+ 5C.C7 T cells (Th1) was detectable in both spleen and mLN of MPF-housed, as well as cohoused-RF(SFB) and -RF(MPF), hosts (Figures 2A–C). Interestingly, a large majority of the IL-10-producing T cells co-expressed IFN-γ (Figure S2E), reminiscent of the IL-10-producing Th1 cells described in other models of Th1-driven chronic infections and autoimmunity (Anderson et al., 2007; Jankovic et al., 2007; Gabrysová et al., 2009). Confirming our observations in T cell-deficient hosts, SFB-independent differentiation of IL-17A+ 5C.C7 T cells could also be observed following transfer into RF-housed T cell-replete mPCC, Cd3e+/+ hosts, albeit slightly delayed and limited to Peyer’s patches (PPs) (Figure 2D and data not shown). The differentiation of IFN-γ+ 5C.C7 T cells appeared, however, fully blunted, as previously described and in line with the absence of autoimmune pathologies (Singh et al., 2006).
The importance of the Th1 cell component of the T cell response in driving the appearance of autoimmune pathologies was further investigated using Ifng−/− 5C.C7 T cells. Transfer of these cells into MPF-housed mPCC, Cd3e−/− hosts resulted in greatly reduced arthritis compared to that observed with WT 5C.C7 T cells (Figure 2E). This result confirmed an essential role for this proinflammatory cytokine in our T cell transfer model of autoimmune arthritis. More importantly, by demonstrating a key role for SFB in enhancing a Th1 cell-driven autoimmune disease, these results suggested that SFB has a much broader impact on autoimmune responses than the sole control of Th17 T cell differentiation.
SFB sustain chronic proliferation of self-reactive CD4+ T cells in gut-associated lymphoid tissues
SFB induces the full maturation of gut-associated helper T cell responses (Gaboriau-Routhiau et al., 2009; Chung et al., 2012). Analyzing polyclonal CD4+ T cell populations in RF- and MPF-housed mPCC, Cd3e+/+ hosts, we also found a clear accumulation of polyclonal Th17 and Th1 T cells in lamina propria (LP) and Peyer’s patches (PPs) of the small intestine in MPF-housed hosts (Figure S2G), in line with the colonization of the MPF gut-commensal flora by SFB. This was further associated with a marked increase in TCRβ+CD4+ T cell number (Figure S2H), correlating with an enhanced proliferation of such cells, as revealed by Ki-67 expression and EdU incorporation (Figures S2I–J). These results thus prompted us to investigate a potential impact of SFB-derived signals on autoreactive T cell proliferative responses in gut-associated lymphoid tissues.
Echoing observations made under steady-state homeostasis, we observed a sustained lymphadenopathy of MPF-housed mPCC, Cd3e−/− host mesenteric lymph nodes (mLN), as well as a slight splenomegaly following 5C.C7 T cell transfer (data not shown). In line with the overall size of the mLN, a 12 to 50 fold increased accumulation of 5C.C7 T cells could be detected as early as day 14 in mLN and lamina propria of the small intestine (LP) of MPF-housed hosts, as compared to that of RF-housed hosts (Figure 3A). Detailed kinetic analysis did not reveal any difference in the primary proliferation phase occurring in both hosts up to day 4 (Figure S3A). The secondary, chronic, expansion phase, however, was dramatically curtailed in RF-housed mPCC, Cd3e−/− hosts in both spleen and mLN. No significant differences in absolute T cell numbers were observed between the two hosts in peripheral lymph nodes (pLN) up to day 57 (data not shown).
Figure 3. SFB-containing flora sustains chronic proliferation of self-reactive CD4+ T cells.
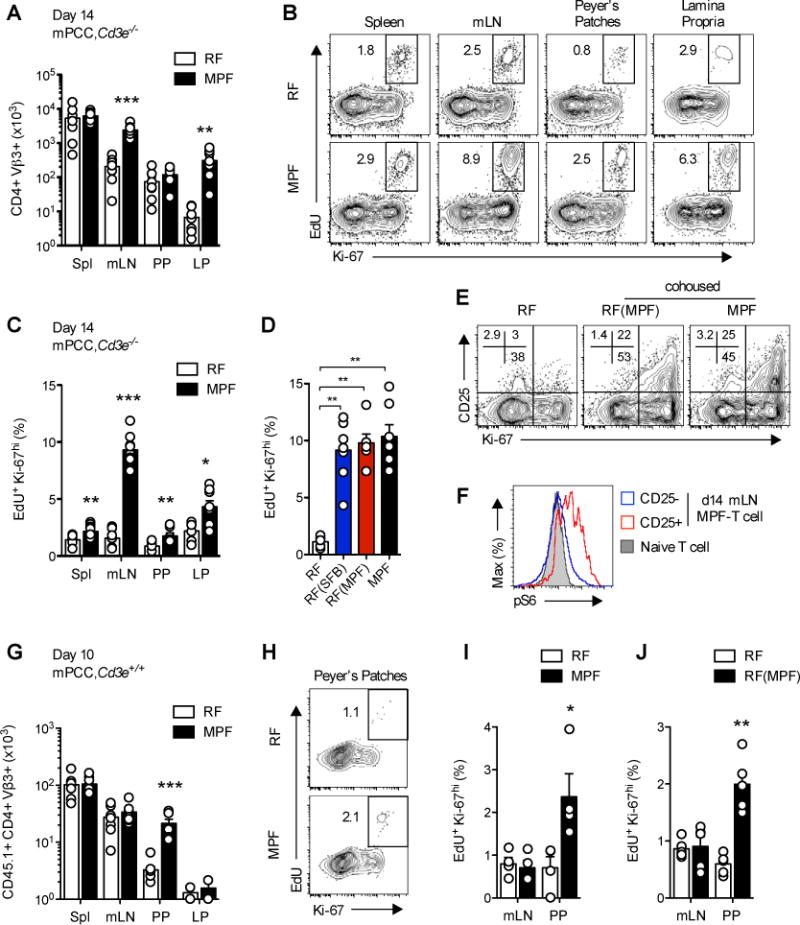
(A–D) 14 days post naïve 5C.C7 T cell transfer, mPCC, Cd3e−/− hosts were injected with EdU and sacrificed 1hr. later. (A) Absolute number of CD4+Vβ3+ T cells in spleen, mLN, Peyer’s patches (PP) and lamina propria (LP) of the small intestine. (B) Representative EdU and Ki-67 expression profiles and (C–D) Frequency of EdU+Ki-67hi cells in live, CD4+Vβ3+ T cells isolated from indicated organs of RF- or MPF-housed hosts (B–C) or mLN of indicated hosts (D). Data (mean±SEM) pooled from 3 independent experiments each ((A–C) n=8 and (D) n=6–7 mice per group).
(E) Representative CD25 and Ki-67 expression profiles, gated on CD4+Vβ3+ T cells and (F) representative phosphorylated S6 ribosomal protein (pS6) expression profile in CD25− (open, blue line) and CD25+ (open, red line) CD4+Vβ3+ T cells isolated from mLN of indicated hosts at day 14 and compared to naive 5C.C7 T cells (closed, grey). (G–I) 10 days post transfer of naïve CD45.1+ 5C.C7 T cells into 4–5 wk. old mPCC, Cd3e+/+ hosts, the recipient mice were injected with EdU and sacrificed 1 hr. later. (G) Absolute number of CD45.1+CD4+Vβ3+ T cells recovered at day 10 from indicated organs. (H) Representative EdU and Ki-67 expression profiles and (I) frequency of EdU+Ki-67hi cells in live, CD45.1+CD4+Vβ3+ T cells isolated from mLN and PP. Data (mean±SEM) pooled from 6 (G) and 4 (H–I) independent experiments. (n=6 pools of 1 to 3 mice per group and 4 pools of 3 mice per group respectively). (J) Same as (I) in 8–12 wk. old RF-housed mPCC, Cd3e+/+ mice cohoused for 2 weeks with MPF-housed mPCC, Cd3e−/− mice prior to T cell transfer. Data (mean±SEM) pooled from 5 independent experiments, each representing one pool of 6 RF-housed or 2 cohoused-RF(MPF) mice. (G–J) All EdU and Ki-67 stainings were performed on purified CD4+ T cells. See also Figure S3.
In favor of a second wave of active in vivo proliferation of chronically stimulated 5C.C7 T cells, a secondary peak of Ki-67 expression, correlating with strong EdU incorporation, could be detected around day 14 in all gut-associated lymphoid tissues (Figures 3B–C and S3B–C). In line with the incidence of arthritis in these hosts, the enhanced proliferation and accumulation of 5C.C7 T cells could be recapitulated by the sole addition of SFB to the RF flora (Figure 3D and data not shown). This secondary phase of T cell proliferation was additionally associated with signs of sustained T cell activation, as evidenced by the late re-expression of the high affinity IL-2R by a subpopulation of mLN T cells recovered from MPF-housed mPCC, Cd3e−/− hosts (Figure S3B–D). Expression of CD25 correlated with high expression of Ki-67 (Figure 3E) and an increased level of endogenous mTOR activation, as revealed by ex vivo analysis of S6 ribosomal protein phosphorylation (Figure 3F).
Of interest, the impact of the gut commensal flora on autoreactive T cell proliferative responses appeared to extend beyond the sole context of the lymphopenic host. Although cell numbers recovered from mPCC, Cd3e+/+ hosts were dramatically reduced as compared to mPCC, Cd3e−/− hosts, as previously described (Singh et al., 2006), and close to the limit of detection in LP of both hosts, a clear accumulation of transferred CD45.1+ 5C.C7 T cells could be observed in PPs of MPF-housed mPCC, Cd3e+/+ hosts (Figure 3G and S3E). This also correlated with enhanced proliferation (Figure 3H–I). Under these additional T cell extrinsic constraints, however, no difference in cell numbers recovered from either spleen or mLN could be observed up to day 10. Similar results were obtained in adult RF-hosts cohoused for 2 weeks with MPF-housed mPCC, Cd3e−/− hosts prior to T cell transfer (Figure 3J). Altogether, our results demonstrated a non-redundant role for specific members of the MPF-commensal flora, such as SFB, in promoting chronic activation and proliferation of auto-reactive T cells in gut-associated lymphoid tissues.
Gut microbiota modulates T cell tuning to self-antigen in gut-associated lymphoid tissues
Previous data from this experimental model indicates that the main constraint on chronic T cell activation is the result of a T cell-intrinsic adaptation of its activation threshold to the ambient level of antigen presentation in the host (Singh and Schwartz, 2003; Singh et al., 2006). The uncontrolled chronic T cell activation observed here could therefore reflect an inadequate and/or impaired tuning of 5C.C7 T cell sensitivity to the ambient level of PCC presentation in the mLN of MPF-housed hosts.
An inadequate tuning could result from a local, commensal flora-mediated, increase in TCR stimulation, e.g. through cross-reactivity to bacterial antigens or increased self-antigen presentation. However, transfer of carboxyfluorescein succinimidyl ester (CFSE)-labeled naïve 5C.C7 T cells into MPF- or RF-housed Cd3e−/− hosts didn’t reveal any differences in homeostatic expansion in the absence of PCC antigen, making it unlikely that 5C.C7 TCRs cross-react to bacterial antigens specifically present in the MPF microflora (Figure S4A). Similarly, ex vivo analysis of the antigen presentation capacity of various APCs isolated from RF- or MPF-housed mPCC, Cd3e−/− hosts didn’t indicate any significant changes in endogenous PCC presentation levels. Seven days after the initial T cell transfer, the ability to efficiently present endogenous PCC peptide to CD4+ T cells appeared mostly restricted to CD11c+ dendritic cells (DCs) in both hosts, and all DC populations tested demonstrated an equal potential to stimulate the in vitro proliferation of a population of pre-activated 5C.C7 T cells (Figure S4B) or the overnight activation of chronically stimulated 5C.C7 T cells purified from mLN (mLN T cells) of RF- or MPF-housed hosts at day 7 (Figure S4C–D).
In contrast, the response of MPF-mLN T cells to both sources of DCs appeared significantly enhanced as compared to the response of RF-mLN T cells (Figures S4C–D). Consistent with an impaired tuning of T cells residing in mLN of MPF-housed hosts, in vitro activation of MPF-mLN T cells with MCC peptide-pulsed naïve Cd3e−/− splenocytes revealed a selectively increased sensitivity of these cells in both their ability to upregulate CD69 (Figure 4A–B) and to proliferate (Figure 4C–D). Similar results were obtained using plate-bound anti-CD3 stimulation in the absence of any added APCs (Figure 4E). Sensitivity of MPF-mLN T cells was never restored fully to the level of naïve T cells, yet these cells displayed a distinctively lower activation threshold to MCC peptide as compared to RF-mLN, RF-spleen or MPF-spleen T cells.
Figure 4. SFB-containing flora locally modulates T cell intrinsic tuning of TCR activation threshold to endogenous antigen.
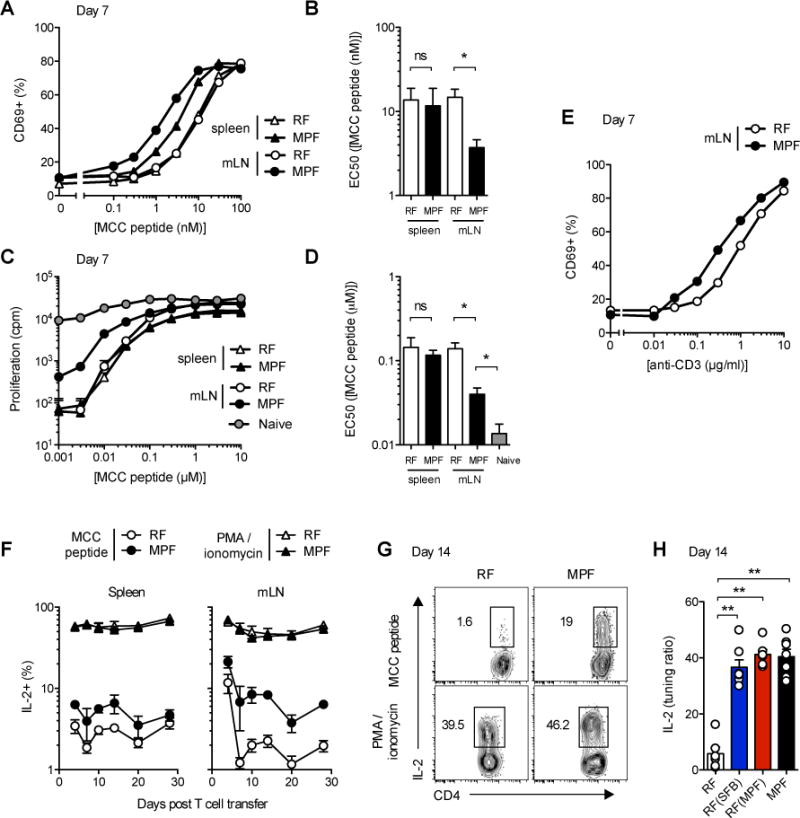
(A and E) CD69 expression and (C) 3H-Thymidine incorporation in CD4+Vβ3+ T cells purified from spleen or mLN of mPCC, Cd3e−/− hosts 7 days post naïve 5C.C7 T cell transfer and cultured for 16h (A and E) or 84h (B) with fresh Cd3e−/− splenocytes and various concentrations of MCC peptide (A and C) or plate-bound anti-CD3 (2C11) and soluble anti-CD28 in the absence of added APC (E). 3H-Thymidine was added to the culture for the last 24h. One representative experiment out of 4 (pooled triplicates) or 3, respectively. (B and D) Summary of EC50 for each independent T cell population tested. Data (mean±SEM) pooled from (A–B) 5 and (C–D) 3 independent experiments, * P<0.05, student t test.
(F) Frequencies of IL-2+ cells in live, CD4+Vβ3+ T cells isolated from spleen or mLN of mPCC, Cd3e−/− hosts at the indicated time points post T cell transfer and restimulated for 3h with PMA and ionomycin or 3μM MCC peptide. (G) Representative expression profiles and (H) tuning ratios, as described in experimental procedures, for IL-2 in live, CD4+Vβ3+ T cells isolated at day 14 in mLN of indicated host. Data (mean±SEM) pooled from 5 independent experiments (n=2 to 7 mice per time point) (F) and 3 independent experiments (n=6–7 mice per group) (H). See also Figure S4.
By primarily affecting the TCR proximal signaling machinery, and in particular Zap70-mediated phosphorylation of LAT (Choi and Schwartz, 2007; 2011), T cell tuning results in the downregulation of all functions downstream of TCR signaling, and in particular IL-2 production (Tanchot et al., 2001). Accordingly, and as early as day 7 after initial T cell transfer, less than 3 percent of 5C.C7 T cells recovered from either spleen or mLN of RF-housed mPCC, Cd3e−/− hosts displayed IL-2 production capacity following 3h of in vitro activation with a high dose of MCC peptide (3μM) (Figure 4F). In comparison, such potential appeared clearly greater in both spleen and mLN T cells isolated from MPF-housed hosts. This differential IL-2 production appeared maximal around day 14 (Figures 4F–G), thus mimicking the kinetics observed for CD25 and Ki-67 expression. More importantly, it persisted up to day 28, arguing for a stable state of impaired tuning of such cells rather than a delayed induction of T cell intrinsic tolerance. As a control, stimulation of T cells with PMA and ionomycin, which can bypass the block in proximal TCR signaling normally associated with T cell tuning (Chiodetti et al., 2006), gave comparable IL-2 responses from all the populations examined (Figures 4F–G). In this type of assay, the potential of tuned T cells to produce IL-2 is thus best represented as a ratio between the frequency of IL-2 producers obtained under MCC peptide- over PMA and ionomycin-mediated stimulation, referred to subsequently as the tuning ratio. Such analysis revealed that, at any time point studied after day 7, less than 5% of the IL-2 competent RF-mLN T cells appeared still capable of making IL-2 following TCR-mediated activation (Figure 4H and data not shown). In contrast, up to 20–30% of the MPF- or cohoused-RF(SFB) or RF(MPF) mLN T cells demonstrated such potential by day 14. Overall, these data demonstrated that specific members of the gut-commensal flora can oppose the ability of self-reactive T cells to fully tune their activation threshold to the level of self-antigen presented in gut-associated lymphoid tissues.
Modulation of T cell tuning by SFB dynamically affects all self-reactive CD4+ T helper subsets
Similar to IL-2, both IL-17A and IFN-γ production upon MCC peptide restimulation were increased in MPF-mLN T cells, as early as day 7 and up to day 28, as compared to RF-mLN T cells (Figure 5A–B). This was partially explained by differences in Th1 cell differentiation and Th17 cell maintenance, as measured upon PMA and ionomycin-mediated stimulation (Figure 5A–B). Comparing the tuning ratio for each individual cytokine, however, a respective 2.8 and 2.4 fold increase in the frequency of differentiated IFN-γ+Th1 or IL-17A+Th17 cells capable of producing their respective cytokines could be observed (Figure 5C). These results confirmed that regulation of T cell tuning acts as a second layer of regulation on self-reactive T cell effector functions to complement lineage differentiation, a layer partially removed in mLN T cells of SFB-harboring hosts. Of interest, T cell tuning appeared further reduced in T cells undergoing active in vivo proliferation in these organs (Figure 5D–E), suggesting heterogeneity of T cell tuning at the single cell level.
Figure 5. Modulation of T cell tuning by commensal flora broadly affects self-reactive CD4+ Th1 and Th17 subsets.
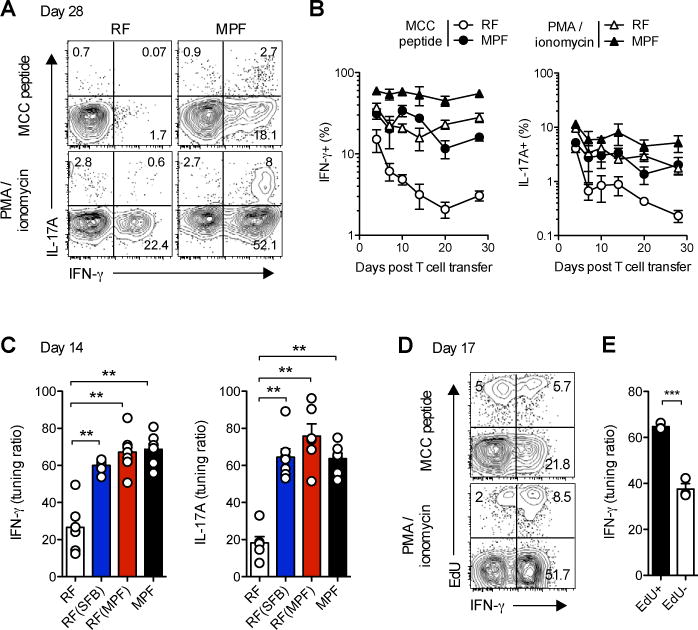
(A) Representative expression profiles at day 28 and (B) Frequencies of IFN-γ+ and IL17A+cells in live, CD4+Vβ3+ T cells isolated from mLN of mPCC, Cd3e−/− hosts at the indicated time points post naïve 5C.C7 T cell transfer and restimulated for 3h with PMA and ionomycin or 3μM MCC peptide. (C) Tuning ratios, as described in experimental procedures, for IFN-γ and IL17A in live, CD4+Vβ3+ T cells isolated from mLN of indicated hosts 14 days post T cell transfer and restimulated for 3h with PMA and ionomycin or 3μM MCC peptide. Data (mean±SEM) pooled from 3 independent experiments (n=6–7 mice per group).
(D–E) 17 or 26 days post T cell transfer, MPF-housed mPCC, Cd3e−/− hosts were injected with EdU and sacrificed 1hr later. (D) Representative EdU and IFN-γ expression profiles in live, CD4+Vβ3+ T cells isolated from mLN at day 17. (E) Tuning ratio for IFN-γ in live, EdU+ versus EdU− CD4+Vβ3+ T cells. Data (mean±SEM) pooled from 2 independent experiments (n=3 mice per group).
On a molecular basis, the phenomena we have observed could be the consequence of active signaling in T cells occurring either at the early stages of T cell priming, preventing later tuning to chronic self antigen, or late during the chronic phase of the autoimmune response, bypassing and/or negatively regulating already established tuning in chronically stimulated T cells. To assay the full extent of T cell tuning plasticity in vivo, we purified T cells from mLN of RF-housed mPCC, Cd3e−/− hosts 7 days after the initial transfer, retransferred them into an MPF-housed host, itself pre-transferred with congenic CD45.1+ 5C.C7 T cells, and then analyzed their “new” responsiveness 7 days later (Figure 6A). Ex vivo re-stimulation revealed similarly high responsiveness of both T cell populations as measured by IL-2 (Figure 6B). This was further associated with restored proliferation and effector functions as measured by Ki-67 expression and IFN-γ production. A more complete panel of T cell transfer experiments confirmed this to be true regardless of the T cell’s organ of origin (Figure 6B). Furthermore, the opposite experiment, with T cells of MPF origin transferred into RF-housed hosts, gave all low responsiveness, i.e. transferred MPF-T cells showed restored tuning 7 days later, to the same extent as endogenous RF-T cells (Figure 6B). Overall these results demonstrated that tuned T cells can dynamically adapt to a change in the local microenvironment by either down- or up-regulating their TCR activation threshold. Furthermore, they demonstrated that the local microenvironment of an MPF-housed host’s mLN can actively induce the local reactivation of previously fully tuned self-reactive T cells, leading to restored proliferative and effector functions.
Figure 6. Host microbiota dynamically regulates self-reactive T cell tuning.
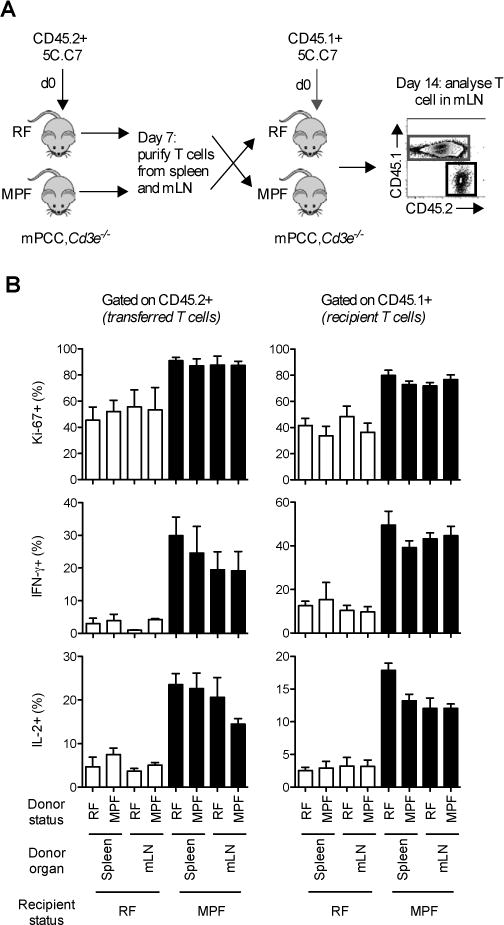
(A) Two cohorts of mPCC, Cd3e−/− mice were injected simultaneously with either naïve CD45.2+ 5C.C7 T cells or CD45.1+ 5C.C7 T cells. 7 days later, spleen and mLN were harvested from hosts injected with CD45.2+ T cells. Purified T cells (106) were further reinjected into the hosts previously transferred with CD45.1+ T cells. mLN cells were stained at day 14 for Ki-67 expression or for intracellular IL-2 and IFN-γ following 3h stimulation with 3μM MCC peptide (MCC). (B) Frequency of Ki-67+ (top), IFN-γ+ (middle), or IL-2+ (bottom) in live, CD4+Vβ3+ T cells in the CD45.2+ donor (left) or the CD45.1+ recipient (right) fractions. Data (mean±SEM) pooled from 3 independent experiments. See also Figure S5.
Inflammatory gut microbiota-derived signals impact self-reactive T cell chronic activation
Changes in the local microenvironment of MPF-housed gut-associated tissues could result from T cell dependent signals linked to the enhanced IFN-γ production (Shalapour et al., 2010) and/or the appearance of autoimmune inflammatory symptoms. IFN-γ production by T cells, however, didn’t seem to play a critical role as Ifng−/− 5C.C7 T cells responded identically to wild type T cells in the MPF-housed host’s mLN microenvironment (Figure S5A–B). Similarly, the impact of the gut microbiota on chronically activated 5C.C7 T cells appeared unaffected by the absence of B cells and overt autoimmune manifestations (Singh et al., 2006), as observed upon transfer into mPCC, Rag2−/− hosts (Figure S5C–G). Subsequent transfer of naïve B cells further demonstrated the enhanced B cell helper and arthritis-inducing potential of chronically stimulated 5C.C7 T cells in MPF-housed T and B-cell deficient Rag2−/− hosts (Figure S5H). These results thus prompted us to look for an impact of the gut microbiota on the mLN microenvironment during the chronic phase of T cell activation.
Direct modulation of antigen presentation in mLN didn’t seem to play a critical role in our model (Figure S4). Therefore, we investigated whether these effects could be the result of direct signaling of chronically activated T cells by one or more commensal florainduced molecule(s). Analysis of the expression pattern of over 30 cytokines in the serum of MPF- versus RF-housed hosts, enabled us to identify four (G-CSF, IL-1Ra, IL-6 and IL-12p40) as significantly increased in host sera during the course of an MPF-induced chronic autoimmune T cell response (Figure 7A). Both IL-6 and IL-23-mediated signaling can affect T cell long-term proliferative responses, as evidenced in murine colitis models (Tajima et al., 2008; Ahern et al., 2010). To test their role in our model, we treated MPF-housed hosts with neutralizing mAb between day 7 and 14. In vivo neutralization of IL-12p40, but not IL-6, in an MPF-housed host was sufficient to reduce ongoing proliferation of, and prevent the secondary CD25 expression normally observed on MPF-mLN T cells (Figures 7B–C and S6A–B). Similarly, analysis of the IL-2, IL-17A and IFN-γ expression profiles and related tuning ratios confirmed a non-redundant role for IL-12p40, but not IL-6, in maintaining TCR responsiveness of chronically stimulated T cells, in addition to their Th1 and Th17 differentiation programs (Figure 7D–E and data not shown).
Figure 7. In vivo IL-12p40 blockade restores optimal T cell tuning in MPF-housed hosts.
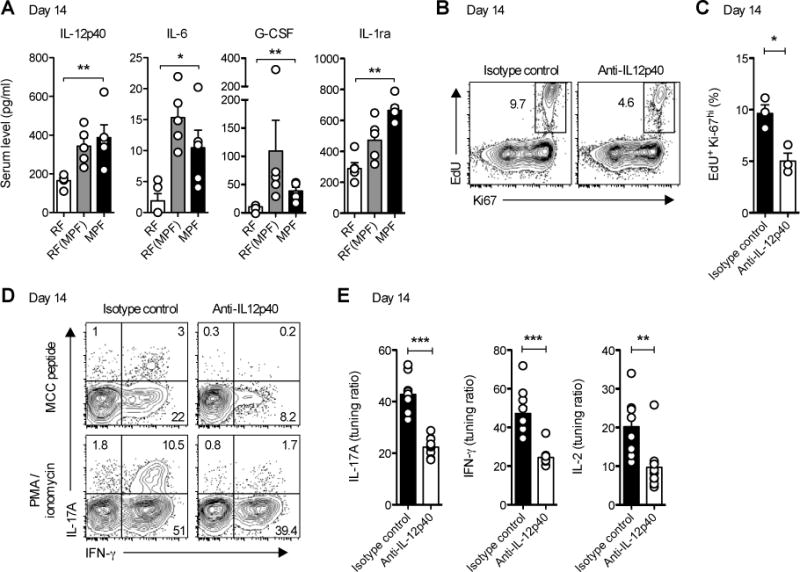
(A) IL-12p40, IL-6, G-CSF and IL-1Ra levels in sera of mPCC, Cd3e−/− hosts 14 days post naïve 5C.C7 T cell transfer. Data (mean±SEM) pooled from 2 independent experiments (n=5 mice per group).
(B–E) Starting at day 7, MPF-housed mPCC, Cd3e−/− mice were further injected twice daily i.p. with anti-IL12p40 or a corresponding isotype control mAb. At day 14, mice were injected with EdU and sacrificed 1hr later. (B) Representative EdU and Ki67 expression profiles and (C) frequency of EdU+Ki-67hi cells in live, CD4+Vβ3+ T cells isolated from mLN (n=3 mice per group, mean±SEM). * P<0.05, student t test. (D) Representative IL-17A and IFN-γ expression profiles and (E) tuning ratio for IL-17A (left), IFN-γ (middle) and IL-2 (right) production in live, CD4+Vβ3+ T cells isolated from mLN at day 14 and restimulated for 3h with PMA and ionomycin or 3μM MCC peptide. Data (mean±SEM) pooled from 4 independent experiments (n=9–10 mice per group). See also Figure S6.
IL-12p40 is an integral part of IL-12p70 as well as IL-23 heterodimers. Interestingly, IL-12p70 directly induces CD25 expression in a STAT4 dependent manner and acts as a secondary signal for proliferation of CD4+ Th1 cell clones (Yanagida et al., 1994; Nishikomori et al., 2002; O’Sullivan et al., 2004). Consistent with this, both RF- and MPF-mLN T cells showed responsiveness to IL-12p70 stimulation in vitro, as measured by CD25 upregulation (Figure S6C). The frequency of responding cells, however, appeared greatly increased in MPF-mLN T cells, in line with the increased Th1-driven response in MPF-housed hosts. As expected, IL-12p70 had no direct impact on naïve T cells, and IL-23 did not induce any CD25 expression above that of the medium control in any of the T cell populations tested (Figure S6C). More importantly, IL-12p70, but not IL-23, was able to enhance pS6 activation in MPF-mLN T cells in response to low levels of MCC peptide in vitro (Figure S6D). As observed for CD25 up-regulation, IL-12p70 also impacted RF-mLN T cell pS6 activation, albeit at much lower levels (data not shown). These results thus suggested IL-12p70 signaling as a potential key promoter of chronic self-reactive T cell responses in this murine model of autoimmune arthritis.
DISCUSSION
Taking advantage of the commensal flora-driven nature of a T cell transfer model of autoimmune arthritis, we revisited in this study the impact of commensal flora-derived signals on the responses of self-reactive CD4+ T cells to chronic self-presentation in vivo. As expected from recent studies in other autoimmune models (Wu et al., 2010; Lee et al., 2011), effects on the maintenance, albeit not the early induction of, Th17 cells could be observed. In vivo neutralization experiments with an anti-IL-17A monoclonal Ab, however, did not reveal any role for such cells in our model (unpublished data). Instead, transfer of Ifng−/− T cells clearly demonstrated the key pathogenic role played by IFN-γ-producing Th cells in this arthritis model. This observation further extends the scope of autoimmune pathologies that can be influenced by colonization of host gut-associated bacterial niches with specific member(s) of the gut flora, such as segmented filamentous bacteria, and all subsequent changes that it might induce for other commensal bacteria or viruses (Kane et al., 2011; Kuss et al., 2011).
How does SFB regulate IFN-γ production by chronically stimulated 5C.C7 T cells in mPCC, Cd3e−/− hosts? SFB reproducibly enhanced Th1 differentiation of autoreactive T cells, reminiscent of observations made in other models of autoimmunity (Wu et al., 2010; Lee et al., 2011). Observed as early as day 4 following T cell transfer and quite stably up to day 57 in both mLN and spleen, this effect of the gut flora, however, only accounted for a 1.6 (day 4) to 2-fold (day 14 and up) increase in the frequency of IFN-γ-producing T cells in MPF-housed hosts. Indeed the major impact of the gut flora in this model system was observed during the chronic phase of the autoimmune T cell response, a phase during which the SFB-induced cytokine milieu locally favored the maintenance of both the sensitivity and the proliferative response of autoimmune CD4+ T cells, a process normally controlled by the T cell intrinsic down-regulation of TCR activation thresholds naturally occurring under chronic TCR activation, which we refer to as T cell tuning (Tanchot et al., 2001; Singh and Schwartz, 2003). Taken all together, these effects led to close to a 50 fold increase in the absolute number of recovered 5C.C7 T cells demonstrating IFN-γ-producing capacity following TCR restimulation on day 14 in mLN of SFB-harboring hosts.
It had been previously suggested that gut-draining lymphoid organs serve as the starting place of systemic T cell responses (Wu et al., 2010; Lee et al., 2011). In mPCC, Cd3e−/− hosts, however, effects on T cell activation appeared mostly limited to the mLN, although small differences in T cell proliferation and sensitivity could be detected as far away as the spleen later in the response (unpublished data). These effects, however, preceded a similarly localized enhanced B cell response, with a ten fold increase in absolute number of GC B cells by day 14 and plasma cells by day 21 in mLN (unpublished data). Although an input from the later splenic responses cannot be excluded (Maccioni et al., 2002), it is highly probable that these locally enhanced T and B cell responses drive the detectably higher levels of high affinity autoantibodies in the serum of MPF-housed hosts as early as day 21, a prerequisite for the induction of the arthritis pathologies (Singh et al., 2006).
The most interesting finding from this study is the demonstration that SFB colonization of host microbiota can locally enhance T cell responsiveness to endogenous antigens, as recently suggested for macrophage responsiveness to IFN (Abt et al., 2012). T cell sensitivity has been previously described to be dynamically tunable to the level of antigen in the host (Tanchot et al., 2001; Singh and Schwartz, 2003; Han et al., 2010) or the T cell density (Tanchot et al., 2001). Detailed ex vivo analysis, secondary T cell transfer experiments and in vivo IL-12p40 blockade experiments clearly demonstrated here that T cell sensitivity could be further modulated by flora-derived inflammatory signals. The ability of the inflammatory milieu, and in particular IL-12p70, to imprint the sensitivity of individual T cell clones at the level of proximal TCR signaling pathways, has recently been shown for CD8 T cells (Raué et al., 2013; Richer et al., 2013). Our data further extend this role to autoreactive CD4+ T cells in the context of gut-associated lymphoid organs. Although we cannot fully exclude other independent regulatory mechanisms, it is tempting to speculate that chronic proliferation is an expected consequence of the maintenance of 5C.C7 T cell responsiveness to the endogenous level of PCC by the floraspecific inflammatory milieu. This would provide a conceptual framework to explain how chronic infections can sustain an ongoing immune response while autoantigens normally do not.
In CD8 T cells, IL-12p70 can sustain mTOR activation (Rao et al., 2010), a key step in preventing the induction of clonal anergy in vitro (Allen et al., 2004; Zheng et al., 2007). We report here that IL-12p70, but not IL-23, also actively signals in tuned 5C.C7 CD4+ T cells, leading to a similar enhancement of mTOR activation in response to low levels of MCC peptide stimulation. The enhanced in vitro responsiveness of MPF-mLN T cells to anti-CD3 stimulation suggests, however, a process that goes beyond simply providing a third signal during T cell activation. The T cell phenotype observed in this study could thus reflect a direct targeting by IL-12p70, through mTOR, of the expression and/or maintenance of the yet uncharacterized “anergic factors” associated with tuning. Alternatively, it could be an indirect consequence of an IL-12p70-induced, enhanced proliferation of the chronically stimulated T cells, itself responsible for diluting out these “anergic factors”, as once proposed for clonal anergy (Powell et al., 2001). A more detailed analysis of the molecular profile of tuned T cells in response to IL-12p70, or other cytokines, in combination with in vivo genetic experiments, are needed to fully decipher the molecular mechanisms in play here.
One interesting question that remains is whether similar mechanisms are at play in endogenous T cells residing in gut-associated lymphoid organs and could explain the broad impact that SFB has on the maturation of the murine gut-adaptive immune system (Talham et al., 1999; Gaboriau-Routhiau et al., 2009; Chung et al., 2012). Numerous tissue and APC-derived signals, such as pathogen-derived IL-12p70, IL-18 and IFN-I (Wang et al., 2012; Raué et al., 2013; Richer et al., 2013), gut-derived retinoic acid (Hall et al., 2011) and IL-23 (Ahern et al., 2010), or skin-derived IL-1 (Naik et al., 2012) can locally impact various aspect of CD8+ or CD4+ T cell fitness, although T cell sensitivity has never been carefully analyzed so far. Experiments done here in T cell-replete mPCC, Cd3e+/+ hosts revealed a striking similarity between the effect of the MPF gut flora on the proliferative response of chronically stimulated 5C.C7 and endogenous CD4+ T cells. T cell extrinsic mechanisms did, however, limit the strength and location of this proliferative response - differences could only be detected in Peyer’s Patches - as well as severely blunting autoreactive T cell differentiation toward the Th1 lineage. Understanding the functional redundancy between the extended family of inflammatory signals as well as whether such signals can locally impact the sensitivity of various self and non-self-specific T cells will clearly require further studies, but could be key in understanding the general impact of the gut flora on the adaptive immune system.
In the case of autoimmune disease, dynamic regulation of T cell sensitivity represents a built-in negative feedback mechanism designed to prevent overexuberant autoreactive T cell responses, a key tenant of the tunable activation threshold model (Grossman and Paul, 1992; 2001). Such negative regulation of T cell responsiveness over time has also been described in various chronic pathogen or tumor models (Carmichael et al., 1993; Rehermann et al., 1996; Oxenius et al., 1998; Staveley-O’Carroll et al., 1998; Zajac et al., 1998), two disease states in which it prevents full eradication of an unwanted target by the immune system. Unraveling the molecular mechanisms at play in bypassing this mechanism will be of key interest to help design clinical approaches aimed at locally manipulating chronic T cell responses.
EXPERIMENTAL PROCEDURES
Mice
Mice used in this study have all been described previously (Tanchot et al., 2001; Singh et al., 2006). All colonies used in this study were bred under the NIAID contract at Taconic Farms (Germantown, New York, USA), an AAALAC accredited facility, in a Restricted FloraTM run barrier unit (IBU37), except for one colony of B10.A mPCC, Cd3e−/− mice bred in a Murine Pathogen FreeTM run barrier unit (IBU40). In addition to the known murine pathogens monitoring occurring in Taconic MPFTM barrier unit, Taconic RFTM health standard excludes specified unacceptable bacteria, of which only Klebsiella oxytoca could be tested for in our MPFTM colonies (www.taconic.com/Health_Reporting). Mice were then housed in micro-isolator cages for the length of our experiments in an NIAID/NIH run AAALAC accredited facility. All experiments in this study were performed in accordance with the guidelines of the National Institutes of Health Animal Care and Use Committee.
Arthritis experiments, Cohousing and Antibiotic treatments
The arthritic score used in this study has been described previously (Singh et al., 2006). Detailed experimental procedures for autoantibodies titration are given in Supplemental Information.
For cohousing experiments, mice were mixed upon arrival at our NAID facility. For association of RF-housed mice with SFB, the mice were cohoused with SFB monoassociated germ free C57BL/6 mice for 3 weeks. At that time point, SFB reconstitution was confirmed by qPCR of fecal 16S rDNA and mice showing SFB titers similar to MPF-housed mice were injected with T cells and then further cohoused with mice displaying lower SFB titers. Detailed experimental procedures for SFB quantification and monoassociation of GF mice are given in Supplemental Information. For antibiotic treatments, freshly diluted combinations of 1mg/ml each of Ampicillin (sodium salt), Neomycin sulfate, Metronidazole and 0.5mg/L Vancomycin hydrochloride (all from Sigma) were added to the drinking water and changed weekly (Ivanov et al., 2008). In vivo neutralization of IL-12p40 was achieved by injecting 0.5mg of mAb C17.8 at day 7 and 0.25mg every other day until day 14 and compared to control Rat IgG2a (2A3) (all BioXCell). Cytokines in serum were analyzed using Ray Biotech Quantibody mouse array (Ray Biotech Inc.).
Cell isolation, adoptive transfer and ex vivo analysis of T cells
Naïve 5C.C7 T cells were isolated from B10.A 5C.C7 TCR transgenic, Rag2−/− mice and 0.5–1 × 106 cells were injected i.v. in indicated hosts. Tuned 5C.C7 T cells were isolated from B10.A mPCC, Cd3e−/− and B10.A mPCC, Cd3e+/+ hosts, restimulated in vitro and stained as previously described (Singh and Schwartz, 2003; Singh et al., 2006). The tuning ratio for individual cytokines was calculated by dividing the frequencies of individual cytokines producers obtained under moth cytochrome c (MCC) peptide- over PMA and ionomycin-mediated stimulation, and presented as a percent. Detailed experimental procedures are in supplemental information.
Statistics
All statistical analyses were performed using GraphPad Prism Software version 5 (GraphPad, San Diego, California, USA). The minimal level of confidence at which the result was considered significant was P<0.05. For all kinetic experiments, statistical significance between each group over time was determined using a two-way ANOVA test. For all other statistical analysis, Mann Whitney non-parametric tests were used unless otherwise noted. * P<0.05, ** P<0.01, *** P<0.001 and **** P<0.0001.
Supplementary Material
Acknowledgments
We thank N.J. Singh for helpful discussion and comments on the design of the experiments; N.J. Singh, Y. Belkaid, J. Davoust and D.A. Gross for helpful comments on the manuscript; N. Shulzenko and A. Morgun for help on the design of the antibiotic treatment experiment and the analysis of SFB colonization; E.J. Chuang for technical assistance and D. Trageser-celser, C. Acevedo and the NIAID germ free facility for maintaining the mice used in these experiments and technical assistance. This work was supported by the Intramural Research Program of the National Institutes of Allergy and Infectious Diseases at the National Institutes of Health, Bethesda, MD, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A, Zheng Y, Gardner L, Safford M, Horton MR, Powell JD. The novel cyclophilin binding compound, sanglifehrin A, disassociates G1 cell cycle arrest from tolerance induction. J Immunol. 2004;172:4797–4803. doi: 10.4049/jimmunol.172.8.4797. [DOI] [PubMed] [Google Scholar]
- Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4(+)CD25(−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204:285–297. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DG, Teyton L, Oldstone MBA, McGavern DB. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. Journal of Virology. 2005;79:10514–10527. doi: 10.1128/JVI.79.16.10514-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael A, Jin X, Sissons P, Borysiewicz L. Quantitative analysis of the human immunodeficiency virus type 1 (HIV-1)-specific cytotoxic T lymphocyte (CTL) response at different stages of HIV-1 infection: differential CTL responses to HIV-1 and Epstein-Barr virus in late disease. J Exp Med. 1993;177:249–256. doi: 10.1084/jem.177.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- Chiodetti L, Choi S, Barber DL, Schwartz RH. Adaptive tolerance and clonal anergy are distinct biochemical states. J Immunol. 2006;176:2279–2291. doi: 10.4049/jimmunol.176.4.2279. [DOI] [PubMed] [Google Scholar]
- Choi S, Schwartz RH. Molecular mechanisms for adaptive tolerance and other T cell anergy models. Seminars in Immunology. 2007;19:140–152. doi: 10.1016/j.smim.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Schwartz RH. Impairment of immunological synapse formation in adaptively tolerant T cells. J Immunol. 2011;187:805–816. doi: 10.4049/jimmunol.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Gabrysová L, Nicolson KS, Streeter HB, Verhagen J, Sabatos-Peyton CA, Morgan DJ, Wraith DC. Negative feedback control of the autoimmune response through antigen-induced differentiation of IL-10-secreting Th1 cells. J Exp Med. 2009;206:1755–1767. doi: 10.1084/jem.20082118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman Z, Paul WE. Adaptive cellular interactions in the immune system: the tunable activation threshold and the significance of subthreshold responses. Proc Natl Acad Sci USA. 1992;89:10365–10369. doi: 10.1073/pnas.89.21.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman Z, Paul WE. Autoreactivity, dynamic tuning and selectivity. Curr Opin Immunol. 2001;13:687–698. doi: 10.1016/s0952-7915(01)00280-1. [DOI] [PubMed] [Google Scholar]
- Hall JA, Cannons JL, Grainger JR, Santos Dos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, et al. Essential Role for Retinoic Acid in the Promotion of CD4(+) T Cell Effector Responses via Retinoic Acid Receptor Alpha. Immunity. 2011;34:435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Asoyan A, Rabenstein H, Nakano N, Obst R. Role of antigen persistence and dose for CD4+ T-cell exhaustion and recovery. Proc Natl Acad Sci USA. 2010;107:20453–20458. doi: 10.1073/pnas.1008437107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos R de L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M, Wynn TA, Kamanaka M, Flavell RA, Sher A. Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J Exp Med. 2007;204:273–283. doi: 10.1084/jem.20062175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccioni M, Zeder-Lutz G, Huang H, Ebel C, Gerber P, Hergueux J, Marchal P, Duchatelle V, Degott C, van Regenmortel M, et al. Arthritogenic monoclonal antibodies from K/BxN mice. J Exp Med. 2002;195:1071–1077. doi: 10.1084/jem.20011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikomori R, Usui T, Wu CY, Morinobu A, O’Shea JJ, Strober W. Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL-12R beta 2 chain expression and signaling. J Immunol. 2002;169:4388–4398. doi: 10.4049/jimmunol.169.8.4388. [DOI] [PubMed] [Google Scholar]
- O’Sullivan A, Chang HC, Yu Q, Kaplan MH. STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J Biol Chem. 2004;279:7339–7345. doi: 10.1074/jbc.M309979200. [DOI] [PubMed] [Google Scholar]
- Oxenius A, Zinkernagel RM, Hengartner H. Comparison of activation versus induction of unresponsiveness of virus-specific CD4+ and CD8+ T cells upon acute versus persistent viral infection. Immunity. 1998;9:449–457. doi: 10.1016/s1074-7613(00)80628-7. [DOI] [PubMed] [Google Scholar]
- Powell JD, Bruniquel D, Schwartz RH. TCR engagement in the absence of cell cycle progression leads to T cell anergy independent of p27(Kip1) Eur J Immunol. 2001;31:3737–3746. doi: 10.1002/1521-4141(200112)31:12<3737::aid-immu3737>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué HP, Beadling C, Haun J, Slifka MK. Cytokine-mediated programmed proliferation of virus-specific CD8(+) memory T cells. Immunity. 2013;38:131–139. doi: 10.1016/j.immuni.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B, Chang KM, McHutchinson J, Kokka R, Houghton M, Rice CM, Chisari FV. Differential cytotoxic T-lymphocyte responsiveness to the hepatitis B and C viruses in chronically infected patients. Journal of Virology. 1996;70:7092–7102. doi: 10.1128/jvi.70.10.7092-7102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer MJ, Nolz JC, Harty JT. Pathogen-specific inflammatory milieux tune the antigen sensitivity of CD8(+) T cells by enhancing T cell receptor signaling. Immunity. 2013;38:140–152. doi: 10.1016/j.immuni.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- Shalapour S, Deiser K, Sercan O, Tuckermann J, Minnich K, Willimsky G, Blankenstein T, Hämmerling GJ, Arnold B, Schüler T. Commensal microflora and interferon-gamma promote steady-state interleukin-7 production in vivo. Eur J Immunol. 2010;40:2391–2400. doi: 10.1002/eji.201040441. [DOI] [PubMed] [Google Scholar]
- Singh NJ, Schwartz RH. The strength of persistent antigenic stimulation modulates adaptive tolerance in peripheral CD4+ T cells. J Exp Med. 2003;198:1107–1117. doi: 10.1084/jem.20030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NJ, Chen C, Schwartz RH. The impact of T cell intrinsic antigen adaptation on peripheral immune tolerance. PLoS Biol. 2006;4:e340. doi: 10.1371/journal.pbio.0040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staveley-O’Carroll K, Sotomayor E, Montgomery J, Borrello I, Hwang L, Fein S, Pardoll D, Levitsky H. Induction of antigen-specific T cell anergy: An early event in the course of tumor progression. Proc Natl Acad Sci USA. 1998;95:1178–1183. doi: 10.1073/pnas.95.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima M, Wakita D, Noguchi D, Chamoto K, Yue Z, Fugo K, Ishigame H, Iwakura Y, Kitamura H, Nishimura T. IL-6-dependent spontaneous proliferation is required for the induction of colitogenic IL-17-producing CD8+ T cells. J Exp Med. 2008;205:1019–1027. doi: 10.1084/jem.20071133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanchot C, Barber DL, Chiodetti L, Schwartz RH. Adaptive tolerance of CD4+ T cells in vivo: multiple thresholds in response to a constant level of antigen presentation. J Immunol. 2001;167:2030–2039. doi: 10.4049/jimmunol.167.4.2030. [DOI] [PubMed] [Google Scholar]
- Wang Y, Swiecki M, Cella M, Alber G, Schreiber RD, Gilfillan S, Colonna M. Timing and Magnitude of Type I Interferon Responses by Distinct Sensors Impact CD8 T Cell Exhaustion and Chronic Viral Infection. Cell Host Microbe. 2012;11:631–642. doi: 10.1016/j.chom.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida T, Kato T, Igarashi O, Inoue T, Nariuchi H. Second signal activity of IL-12 on the proliferation and IL-2R expression of T helper cell-1 clone. J Immunol. 1994;152:4919–4928. [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Collins SL, Lutz MA, Allen AN, Kole TP, Zarek PE, Powell JD. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. J Immunol. 2007;178:2163–2170. doi: 10.4049/jimmunol.178.4.2163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


