Abstract
Carriers of BRCA2 germline mutations are at high risk to develop early-onset breast cancer. The underlying mechanisms of how BRCA2 inactivation predisposes to malignant transformation have not been established. Here, we provide direct functional evidence that human BRCA2 promotes homologous recombination (HR), which comprises one major pathway of DNA double-strand break repair. We found that up-regulated HR after transfection of wild-type (wt) BRCA2 into a human tumor line with mutant BRCA2 was linked to increased radioresistance. In addition, BRCA2-mediated enhancement of HR depended on the interaction with Rad51. In contrast to the tumor suppressor BRCA1, which is involved in multiple DNA repair pathways, BRCA2 status had no impact on the other principal double-strand break repair pathway, nonhomologous end joining. Thus, there exists a specific regulation of HR by BRCA2, which may function to maintain genomic integrity and suppress tumor development in proliferating cells.
DNA double-strand breaks (DSBs) can arise spontaneously during normal DNA metabolism or may be induced by exogenous DNA-damaging agents. Unrepaired or misrepaired DSBs are either lethal or mutagenic. Homologous recombination (HR) and nonhomologous end joining (NHEJ) are the two major mechanisms by which DSBs are repaired in mammalian cells (1, 2). NHEJ requires little or no homologous template and may be considered mutagenic because there is often a change of sequence at the site of the break. Defects in NHEJ result in hypersensitivity to ionizing radiation, severely impaired V(D)J recombination, and chromosome instability (3–5). In contrast to NHEJ, HR uses a homologous DNA template and may repair DSBs with high fidelity. However, HR also can give rise to loss of heterozygosity, a mechanism for unmasking a recessive gene mutation such as in a tumor suppressor gene (6); and chromosome translocation, which can give rise to gene mutations. Proteins involved in executing HR include Rad51, Rad52, Rad54, and other homologs (7, 8). Cells with a mutation in any of these genes display defective HR, extensive spontaneous chromosomal aberrations, impaired cell proliferation, and/or increased radiosensitivity (9, 10). Proteins involved in regulating the levels of HR include p53 and ataxia-telangiectasia mutated (ATM). Cells with nonfunctional p53 or ATM exhibit elevated HR frequencies (11–14) and show an increased frequency of loss of heterozygosity and decreased chromosomal stability (15). Therefore, the balanced regulation of chromosomal exchanges by HR and the precise execution of homology-directed DSB repair are essential for maintaining genomic integrity in mammalian cells.
Mutations in the hereditary breast cancer genes BRCA1 and BRCA2 predispose carriers to breast and ovarian cancer (reviewed in refs. 16 and 17). BRCA2 also has been found to be mutated in 10% of pancreatic cancers studied (18). Although stable interaction between BRCA1 and BRCA2 in mitotic and meiotic cells suggested their functional association (19), the mechanisms by which BRCA1 or BRCA2 inactivation gives rise to cancer have not been established. A function of BRCA1 in DNA repair has been established by its rapid hyperphosphorylation after DNA damage (20) by ATM (21) and CDS1 (22), as well as its involvement in regulation of multiple DNA repair pathways, including transcription-coupled repair (23), homologous recombinational repair (24), and probably nonhomologous-mediated chromosomal break rejoining (25). Little is known, however, about the function of BRCA2. A role of BRCA2 in DNA repair was implied by recent evidence (reviewed in ref. 26). The BRCA2 protein directly associates with the Rad51, a protein essential for HR and HR-mediated DNA repair (27). BRCA2 binds to Rad51 through its BRC repeats (27, 28), and each repeat has a different binding ability. The less conserved BRC5 and BRC6 are not required for binding. The BRC4 region, in which a familial breast cancer mutation, G1529R, has been found, showed threefold stronger binding to Rad51 than BRC1, which itself is a critical Rad51-binding repeat. It is reported that interaction of the BRC4 repeat with Rad51 inhibits the ability of Rad51 binding to DNA and nucleoprotein filament formation (29). In addition, the phenotypic changes seen in BRCA2 knockout mouse cells are similar to those observed in Rad51-deficient cells, including radiosensitivity, chromosomal aberrations, and inhibition of proliferation (30–32). In the present study, we provide direct functional evidence that BRCA2 promotes HR and has no effect on break-induced NHEJ. By specifically promoting homology-mediated high fidelity repair of DSBs generated endogenously during DNA metabolism, or exogenously by ionizing radiation, BRCA2 ensures genomic integrity in proliferating cells.
Methods
Cell Culture, Plasmids, and Transfection.
Capan-1 (human pancreatic cancer) was obtained from the American Type Culture Collection. Cells were grown in monolayer, cultured in Iscove's medium supplemented with 20% FBS, 2 mM l-glutamine, 100 units/liter penicillin, and 1 mg/ml streptomycin (all purchased from Sigma), and maintained at 37°C and 7.5% CO2.
The pcDNA3 vector that expresses hemagglutinin (HA)-tagged human BRCA2 was provided by D. Livingston (Dana–Farber Cancer Institute, Boston). Vectors expressing the BRC4 repeat or mutant BRC4 allele were obtained from W. Lee (University of Texas Health Science Center, San Antonio, TX). Construction of homologous-targeting vectors, pΔ2 and pΔ3, was reported by Hamilton and Thacker (33). A puromycin resistance (puroR) gene for selection of stable chromosomal integrants was inserted into the EcoRI site of pΔ2. Plasmid DNA was transfected into Capan-1 cells by using Lipofectamine-Plus (Life Technology) following the manufacturer's instructions.
Immunoprecipitation and Western Blotting.
The HA-tagged BRCA2 protein (HA-BRCA2) expressed in transfected Capan-1 cells were immunoprecipitated from the nuclear extracts with anti-HA antibody (12CA5). The immunocomplexes were eluted from Protein-A Sepharose beads by boiling in reduced SDS-loading buffer. Proteins in immunocomplexes from Capan-1 cells, or in nuclear extracts from human breast cancer cell line MCF7, a control for BRCA2 expression, were separated by SDS/PAGE on 5% gel. The fractionated proteins were transferred to Immobilon-P polyvinylidene fluoride membrane (Millipore) and immunoblotted with anti-BRCA2 antibody (Ab-2, Oncogene Science), which recognizes a C-terminal epitope of BRCA2.
Homologous Targeting Assay.
To measure homology-directed chromosomal-targeting frequencies, Capan-1 cells were initially transfected with linearized pΔ2-puro and selected for stable chromosomal integration (Capan/pΔ2-puro) by their resistance to puromycin (0.5 μg/ml) but sensitivity to XHATM (1 μg/ml xanthine, 1 μg/ml mycophenolic acid, 1.36 μg/ml hypoxanthine, 0.017 μg/ml aminopterin, and 0.387 μg/ml thymidine). Next, Capan/pΔ2-puro cells were cotransfected with a wt BRCA2 expression vector or a control vector and the circular donor plasmid pΔ3 vector at a molar ratio of 1:5 with Lipofectamine, and subsequently allowed to grow under XHATM selection. Cells with random chromosomal integration of pΔ3 maintained their sensitivity to XHATM; whereas cells in which HR between the integrated pΔ2-puro and the episomal pΔ3 donor had led to restoration of the xgprt gene survived XHATM selection and formed viable colonies. HR frequencies were determined by the number of XHATM-resistant colonies per total number of viable cells seeded.
Clonogenic Survival Assays.
Cells were seeded onto 100-mm tissue culture dishes for 16 h. Ionizing radiation was applied to cells by using a Siemens Stabilipan 2 x-ray generator at 250 kVp, 12 mA, and dose rate of 2.08 Gy/min. For UV-C treatment, adherent cells were washed with PBS and directly exposed to UV-C light with no liquid on the top of the monolayers. Viable colonies containing at least 50 cells were counted after 3 weeks incubation time, and survival fractions were calculated as the plating efficiency of treated cells relative to the plating efficiency of untreated control cells.
DSB-Rejoining Assays by Pulse-Field Gel Electorphoresis (PFGE).
The DSB-rejoining assay using PFGE has been described elsewhere (34). In brief, equal numbers of exponentially growing cells labeled with [14C]thymidine were embedded in agarose and exposed to IR at the indicated doses at 4°C. For determination of break induction, cells were lysed immediately in the plug and subjected to PFGE. For determination of rejoining kinetics, cells were incubated at 37°C for various periods of time after expose to ionizing radiation to allow for fragment rejoining. Fragmented DNA migrated into the gel and appeared as a smear when visualized by UV light after ethidium bromide staining. The level of DNA breakage was estimated by the fraction of activity released from the plug into the gel. Fraction of activity released was quantified with PhosphorImager (Molecular Dynamics) and plotted as a function of radiation dose or repair time.
Results
BRCA2 Status of Capan-1 Cells.
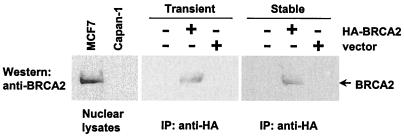
The pancreatic carcinoma line Capan-1 is the only known established human cell line with nonfunctional BRCA2 status (18). Whereas one wt allele is lost, the second allele carries a cancer-associated 6174delT mutation which gives rise to a BRCA2 product that is truncated at codon 1,982 with loss of ≈40% of the C terminus of the protein (19, 27). There are discrepancies about the function of this truncated BRCA2 protein, reported by different studies. Although this truncated form of BRCA2 protein still retains the Rad51 binding sites and interacts with Rad51 as determined by coimmunoprecipitation (27, 35), loss of two nuclear import signals results in its exclusion from the nucleus (36).It is possible that mutant BRCA2 in Capan-1 cells still retains residual nuclear functions. To restore wt function in various experimental settings, cells were transfected with a pcDNA3-based expression vector containing an HA-tagged wt BRCA2 (pBRCA2-HA) or an empty control vector (pcDNA3). Transient or stable BRCA2 expression in transfected Capan-1 cells were verified by immunoprecipitating with anti-HA mAb (12CA5) and immunoblotting by using a C-terminal BRCA2 antibody (Ab2) (Fig. 1). Endogenous mutant BRCA2 protein in parental Capan-1 or Capan-1/vector cells were therefore not visualized. An HA-tagged full-length BRCA2 protein from Capan-1/HA-BRCA2 cells comigrated with endogenous wt BRCA2 from MCF7 cells at ≈400 kDa. Expression of BRCA2 was detected in polyclonal populations, in an effort to avoid any bias resulting from clonal selection. No apparent changes on cell death, growth rate, or plating efficiency were observed upon reconstitution of wt BRCA2 in Capan-1 cells.
Figure 1.
Expression of BRCA2 in Capan-1 cell. Nuclear extracts from untransfected Capan-1 cells and Capan-1 cells transfected (transiently or stable integration) with a pcDNA3-based expression vector coding for HA-tagged wt BRCA2 or an empty control vector were immunoprecipitated with anti-HA antibody, followed by Western blotting with anti-BRCA2 antibody. The nuclear extracts from MCF7 were used as control for endogenous expression of wt BRCA2.
BRCA2 Promotes Spontaneous HR.
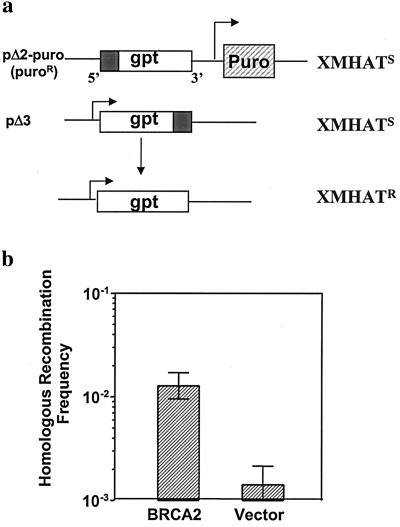
HR has been commonly studied by means of various plasmid-based assays (11, 14, 24, 37). For example, Essers and coworkers (8) measured HR in Rad54-deficient murine cells and their normal counterparts by using a gene-targeting technique. They found that reduced homologous plasmid-targeting frequencies in the absence of Rad54 protein were associated with cellular radiosensitivity, thus implying a defect in homology-directed DSB repair. With a similar approach to determine the effect of BRCA2 on HR in a chromosomal context, we used a chromosomal homologous-targeting system by using the plasmid substrates pΔ2-puro and pΔ3 (Fig. 2a). Each plasmid carries a copy of the bacterial xgprt gene, which has been inactivated either by a 5′ or 3′ deletion (pΔ2-puro or pΔ3, respectively). Neither of the xgprt mutations are revertible, and only HR, via either gene conversion or gene crossover between these two mutant copies can reconstitute a functional xgprt gene, thereby conferring resistance to XHATM drug selection in cell culture (14, 33). For cells without wt BRCA2, a mean homology-mediated chromosomal-targeting frequency of 14.2 × 10−4 was observed (Fig. 2b). After reconstitution of wt BRCA2 status, the spontaneous HR frequencies were elevated by 10-fold.
Figure 2.
Human BRCA2 promotes HR. (a) Schematic map of the chromosomal homologous-targeting substrates. Donation of wt xgprt sequence from the episomal pΔ3 to the chromosomally integrated pΔ2-puro through either gene conversion or crossover events restores xgprt function and confers resistance to XHATM. (b) Reconstitution of wt BRCA2 function in BRCA2-deficient Capan-1 cells enhances HR frequencies. Capan-1 cells with stable pΔ2-puro integration (Capan/pΔ2-puro) were transfected with donor plasmid pΔ3 and pcDNA3 based HA-BRCA2 expression plasmid (19) or control plasmid. The plating efficiency of cells was used to calculate the HR frequencies, i.e., HR frequency equals the number of XHATM-resistant colonies per total number of viable cells seeded. Logarithmic means of HR frequencies +/− SEM are based on four independent experiments.
Because the level of HR-mediated targeting can be influenced by both plasmid uptake capacity and NHEJ-mediated random integration efficiency of the cell, we next examined whether BRCA2 affects these two processes. To measure plasmid uptake, pCMVGFP, which express green fluorescent protein, were cotransfected with BRCA2 expression vector pBRCA2-HA or empty control vector pcDNA3 into capan-1 cells by lipofection. Expression of green fluorescent protein was visualized under fluorescent microscopy 48 h after transfection. Green fluorescent protein-positive frequencies were consistently at ≈10% of transfected cells regardless BRCA2 status (data not shown). To evaluate the NHEJ-mediated random integration, we cotransfected pΔ2-puro with pBRCA2-HA or pcDNA3 into Capan-1 cells. Puromycin was used for selecting random integration events. As seen in Fig. 5a, BRCA2 has no effect on random integration frequencies, which were 1.1–2.2 × 10−4. It should be noted that the gene-targeting frequency is much higher than the random integration frequency, implying that HR targeting can be the dominant integration event in this assay. Therefore, the data demonstrate that wt BRCA2 promotes homology-directed recombination in human cells. In agreement with our data, low levels of HR were reported in Capan-1 cells with a single integrated copy of intrachromosomal HR substrate, which detect gene conversion event only (38). The same study also reported that the levels of HR targeting in Brca2lex1/lex2 mouse ES cells were reduced compared to normal ES cells (38), supporting that the effect of BRCA2 on HR targeting is not unique to Capan-1 cells. A comparable role for BRCA1 also has been suggested in a recent study of HR in mouse embryo stem cells, either null or heterozygous for BRCA1 (24). The effect of BRCA2 on HR depends on its interaction with Rad51.
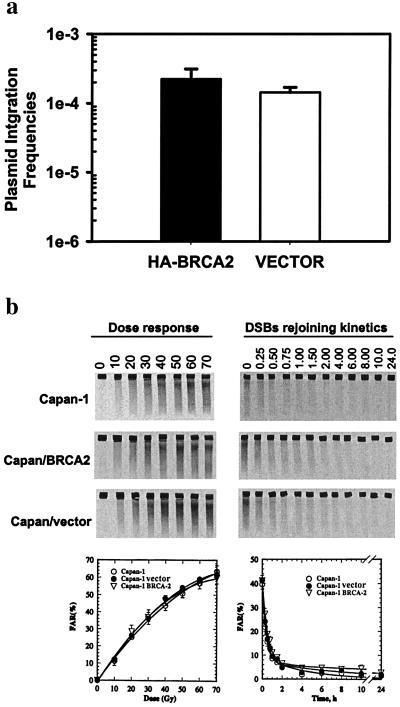
Figure 5.
BRCA2 is not involved in NHEJ. (a) Random integration of plasmid pΔ2-puro with or without BRCA2 expression vector was assessed via selection by puromycin in Capan-1 cells. Integration frequencies were calculated from the number of puroR colonies per number of viable cells seeded. Logarithmic means +/− SEM are based on four independent experiments. (b) Human BRCA2 deficiency does not compromise rejoining of IR-induced DSBs. Induction of DSB (Left) and DSB-rejoining kinetics (Right) by pulse-field gel electrophoresis in Capan-1 cells with or without constitutive wt BRCA2 expression. (Top Left and Right) Typical gels. (Bottom Left and Right) Quantification of the results obtained in three independent experiments. Plotted is the fraction of activity released (FAR) as a function of radiation dose or as a function of time after exposure to 40-Gy x-rays.
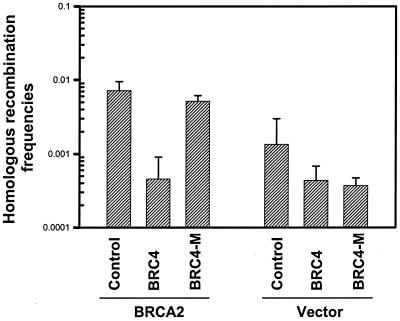
To further explore the molecular mechanism of the function of BRCA2 in regulation of HR, we asked whether the binding of BRCA2 to Rad51 (27) is critical because Rad51 is a highly conserved from bacterial to human cell and essential for HR. It has been reported that expression of wt BRC4 effectively disrupted the interaction of BRCA2 with Rad51 (39) and thus exhibiting a dominant-negative effect leading to cellular hypersensitivity to ionizing radiation (39). In contrast, the T to A mutation in BRC4, namely BRC4–M5, eliminated its binding to Rad51 in vitro and had no dominant-negative effect on the BRCA2–Rad51 interaction in vivo. We cotransfected an expression vector carrying the wt BRC4 repeat together with BRCA2 and pΔ3 into Capan/pΔ2-puro cells. The enhanced HR frequency in cells with wt BRCA2 (71.5 × 10−4), which compared to 13.5 × 10−4 in cells without wt BRCA2, was inhibited 15-fold upon BRC4 coexpression (Fig. 3). However, BRCA2-mediated up-regulation of HR was not significantly compromised when a mutant BRC4 allele, BRC4-M5, was coexpressed. This result suggests that the interaction of BRCA2 with Rad51 is critical for the function of BRCA2 in regulation of HR. The basal levels of HR frequency in BRCA2-deficient cells were not affected by either wt or mutant BRC4 coexpression.
Figure 3.
The function of BRCA2 on HR enhancement depends on its interaction with Rad51. HR frequencies were assessed with the same assay as described in Fig. 2b. Donor plasmid pΔ3 and BRCA2-HA expression vector or control vector were cotransfected with wt BRC4 or mutant BRC4-M5 repeats into Capan/pΔ2-puro cells. The wt BRC4, but not mutant BRC4-M5 repeat, dominant-negatively inhibited the enhancement effect of BRCA2 on HR. Logarithmic means +/− SEM are based on four independent experiments.
BRCA2-Enhanced HR Is Associated with Radioresistance in Capan-1 Cells.
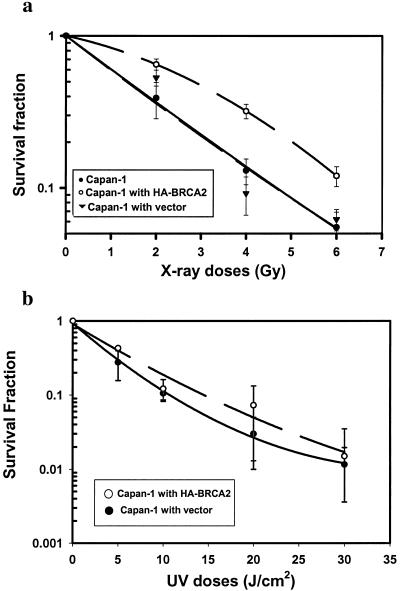
Several lines of evidence indicate that plasmid-based spontaneous HR, which may reflect repair of endogenous DNA damage, are closely correlated with exogenous DSB-induced HR (24, 37). For example, BRCA1-null murine embryonic stem cells showed reduced levels of both spontaneous HR- and restriction endonuclease (I-Sce-I)-induced HR repair of DSBs, when compared to BRCA1 heterozygotes (24). Overexpression of Rad51 in CHO cells resulted in increased spontaneous homologous exchange frequencies in a chromosomal plasmid substrate, which was correlated with an increase in cellular radioresistance, thereby linking plasmid-based HR to homology-mediated repair of exogenous DSBs (40). Conversely, Rad54-null embryonic stem cells exhibited reduced levels of homologous gene targeting and reduced survival upon exposure to IR (8). Recently, a 5- to 6-fold reduction in I-Sce-I induced DSB repair was reported in ES cells from BRCA2lex1/lex2 mouse, compared to BRCA2+/+ mouse (38). However, DSB ends generated by the endonuclease I-Sce-I or by ionizing radiation may require different repair processes. Therefore, we sought to determine whether increased radiosensitivity in BRCA2-deficient cells, as reported by others, is linked to impaired HR. Specifically, we tested whether BRCA2-mediated enhancement of HR was associated with increased resistance to IR-induced cell killing. By using a colony formation assay to measure cellular radiosensitivity, we found that exogenous expression of wt BRCA2 in Capan-1 cells resulted in a significant increase in cellular radioresistance as compared to control vector transfection or to parental Capan-1 cells (Fig. 4a). The fraction of cells surviving at a dose of 2 Gy (SF2), were 0.59, 0.35, and 0.34 in Capan-1 cells transfected with wt BRCA2, a control vector, and with no vector, respectively. A difference of 0.2 between SF2 values is considered significant for clinical radiotherapy (41). In comparison, reintroduction of wt BRCA2 into Capan-1 cells did not affect cell sensitivity to UV irradiation (Fig. 4b), which is mainly determined by the function of nucleotide excision repair. Our findings from human Capan-1 cell, together with the observation from the BRCA2lex1/lex2 mouse (38), indicate that the effect of BRCA2 is relatively specific for IR and not a general effect in abrogating cell death or regulating nucleotide excision repair. These data suggest that BRCA2-mutant tumor cells are radiosensitive because of, at least in part, an impaired ability to carry out HR-mediated DSB repair.
Figure 4.
Restoration of wt BRCA2 enhances clonogenic survival of Capan-1 cells after exposure to ionizing radiation, but not to UV light. (a) Surviving fractions after irradiation were determined for untransfected Capan-1 cells and in derivatives with or without constitutive wt BRCA2 expression. (b) Clonogenic survival following exposure to UV-C was assessed as for x-ray exposure. Error bars are SEM.
BRCA2 Status Has No Impact on Break-Induced NHEJ.
As both HR and NHEJ pathways are important determinants of radiosensitivity in mammalian cells, we wanted to clarify whether the dependence of cellular radiosensitivity on BRCA2 status also can be attributed to functions in the NHEJ pathway. For this, we used two different approaches. In the first approach, we evaluated the random chromosomal integration frequencies of transfected plasmids, a process which involves NHEJ and is impaired in Ku-deficient cells (42). Cotransfection of linearized pΔ2-puro plasmid either with a BRCA2 expression vector or with a control vector into Capan-1 cells and subsequent selection for chromosomal integration by cell resistance to puromycin yielded similar mean integration frequencies: 2.2 ± (SEM) 0.889 × 10−4 with a BRCA2 expression vector and 1.1 ± (SEM) 0.268 × 10-4 with control vector, (t test, P = 0.886) (Fig. 5a).
Secondly, we measured radiation-induced DNA fragmentation and break-rejoining kinetics in Capan-1 cells by PFGE. There was no difference either in the yield of radiation-induced DSBs, or in the rejoining kinetics of DSBs in Capan-1 cells, compared to cells that are NHEJ proficient (data not shown). Furthermore, reconstitution of wt BRCA2 function in Capan-1 did not change the DSB-rejoining capacity (Fig. 5b). Together, these observations strongly suggest that BRCA2 is not involved in break-induced repair by NHEJ. In line with other HR-deficient cells, resulting from mutations in either XRCC2, XRCC3 (43–44), Rad51, Rad52, or Rad54 (G.I., unpublished data), no deficiency in the rejoining of fragmented DNA (PFGE or other methods) was seen. Thus, the increased radiosensitivity in BRCA2-mutated Capan-1 cells is likely because of deficient homology-mediated repair of exogenously induced DSBs.
Discussion
BRCA2 Is a Positive Regulator of HR.
We have obtained direct functional evidence demonstrating that human BRCA2 promotes HR (Fig. 2b). In recent studies, BRCA2 protein was found to interact with Rad51, a central HR effector protein (45), as well as with BRCA1, a newly discovered regulator of HR (24). Although a low level of HR was recently reported in Capan-1 cells (38), its mechanism remains unknown. By using plasmid-based isogenic-targeting assay, which is considered to be one of the most efficient measure of HR in mammalian cells (46, 47), we found an average of 10-fold increase of HR-mediated targeting frequency when wt BRCA2 was reintroduced into BRCA2-deficient Capan-1 cells. Unlike Rad51-deficient cells however (10, 48), basal level HR still can occur even in the absence of functional BRCA2. These data argue that BRCA2 is more likely to be a regulator of overall HR levels, similar to BRCA1, p53, and ATM (11–14) rather than an essential effector of HR, such as Rad51.
Mechanisms for Function of BRCA2 in HR Regulation.
It has been shown that expression of wt, but not mutant BRC4 repeat (39), diminished the interaction of wt BRCA2 with Rad51. This dominant-negative effect of BRC4 on the interaction of BRCA2 and Rad51 resulted in increased radiosensitivity, and disabled the formation of Rad51 foci after DNA damage. In an in vitro biochemical assay, it was shown that the interaction of BRC4 repeat with Rad51 prevents the binding of Rad51 to DNA and the formation of nucleoprotein filament (29). We found that the enhancing effects of BRCA2 on HR-mediated gene targeting in Capan-1 was suppressed significantly on expression of the wt, but not mutant, BRC4 repeat. Together, the data indicate that one important mechanism underlying the regulation of HR by BRCA2 is through its interaction with Rad51.
In this experiment, we also noticed that the basal level of HR targeting in Capan-1 cells transfected with wt BRC4 repeat tended to be lower than that in control vector transfection. Although the reduction in not significant statistically, it suggests a residual interaction of rad51 with the endogenous mutant BRCA2 protein in Capan-1, as reported by others (27, 35). However, a reduction in HR also was seen with mutant BRC4 transfection, raising the possibility that the effect is not specific to the mutant BRCA2–Rad51 interaction. Because the levels of Rad51 expression were not affected by BRCA2 status in our study (data not shown), it is less likely that an alteration of Rad51 expression would be a significant determinant of BRCA2-dependent HR. In a recent study (38), I-Sce-I-induced DSB repair was found to be 5–6 times lower in ES cells from the BRCA2lex1/lex2 mouse than from the BRCA2+/+ mouse. However, the interaction of BRCA2 with Rad51 and their nuclear location are not disrupted by the C-terminal deletion of BRCA2 in BRCA2lex1/lex2 mouse. The explanation for the decreased HR in BRCA2lex1/lex2 is unclear. Other mechanisms involving BRCA1, Bub1, or p53, which physically or functionally associate with BRCA2 (19, 35, 49), also may be relevant in this respect.
BRCA2 Is Not Involved in NHEJ Pathway.
Previously, Patel et al. (32) reported normal level of expression of V(D)J recombination product, μ heavy-chain, in developing B cells of BRCA2 knockout mouse. The V(D)J recombination uses the same obligatory repair components, namely DNA-PKcs (the catalytic subunit of the DNA-dependent protein kinase) (50), Ku70 and Ku80 (51), ligase IV (52), and XRCC4 (53). Therefore, a normal level of V(D)J recombination in mammalian cells correlates with the proficient NHEJ in mouse BRCA2TRC/TRC lymphoid cells. In the present study, we further investigated the role of human BRCA2 in NHEJ-mediated DSB repair in Capan-1 cells by using two different in vivo assays: random plasmid chromosomal integration and rejoining of chromosome fragments as assessed by PFGE. Both the frequency of plasmid integration and the efficiency of DNA fragments rejoining are commonly used measures of NHEJ. Cells deficient in NHEJ because of functional loss of Ku70/80, DNA-PKcs, or XRCC4 (54) displayed decreased plasmid integration frequency, as well as slowed and incomplete rejoining of DSBs (50). However, cells with impaired HR resulting from inactivation of Rad51, Rad52, Rad54 (G.I., unpublished data), XRCC2, or XRCC3 (55) showed efficient DSB rejoining in this assay. Therefore, our results from in vivo studies in human BRCA2-deficient cells are in accord with observations from V(D)J recombination in mouse B cells (32), and in vitro biochemical assays of linear plasmid rejoining in Capan-1 cells (56). These observations are also in line with results demonstrating that Capan-1 cells use the DNA-PK-dependent component of NHEJ to the same degree as cells with wt BRCA2 (57). It is noted that a recent study in Capan-1 cells reported nearly absent DSB rejoining (58). However, such an absence of DSB rejoining has not been previously observed in either HR-deficient or NHEJ-deficient cells, raising the concern that this observation may be incorrect. In another study using BRCA2-knockout mouse cells (31), impaired DNA repair after exposure to IR was reported by using the comet assay. However, the mechanisms that affect the formation of a comet tail in this assay are complicated, and include chromosomal breaks, alterations in chromosomal structure, and changes in histone proteins.
BRCA2 Promotes HR-Mediated DSB Repair to Maintain Genomic Stability.
In prokaryotes, it has been shown that HR-mediated DNA repair plays a major role in repairing DSBs, generated either endogenously during DNA replication in S-phase or exogenously by genotoxic agents, thereby maintaining genomic integrity, and cell proliferation (6, 59). In mammalian cells, accumulating evidence shows that HR plays a important role in mitotic and meiotic recombinational repair of DSBs as well (61). High spontaneous chromosome instability, resulting from unrepaired endogenous DSBs, and increased radiosensitivity, linked to deficient repair of exogenous DSBs, were observed in mammalian cells that are HR deficient. This combination of features is thought to be because of mutations arising in genes that participate in the HR process, namely, Rad51, Rad54, XRCC2, and XRCC3 (7, 8, 55, 49, 62). Accordingly, it is possible that the reduced proliferative capacity, the increased levels of spontaneous chromosomal aberrations, and the radiosensitivity seen in BRCA2-mutated cells (30–32) might be because of compromised homology-mediated repair of chromosomal breaks arising from DNA replication and ionizing radiation. A plausible working hypothesis is that the function of BRCA2 is to be a regulator of HR, to ensure high-fidelity repair of replication intermediates, and thereby, maintaining genomic integrity in proliferating cells.
In summary, we present the first direct evidence that BRCA2 specifically promotes HR, a process which accurately repairs DSBs that arise during DNA replication in proliferating cells or after exposure to ionizing radiation. However, unlike BRCA1, which is involved in multiple DNA repair pathways including transcription-coupled DNA repair, HR repair (24), and also NHEJ (25), BRCA2 is restricted to the HR pathway through direct physical interaction with Rad51 and has no effect on NHEJ. We propose that compromised mitotic and meiotic homologous recombinational repair in BRCA2-deficient breast epithelial cells, together with their continued proliferation capacity, because of inactivation of cell cycle checkpoints, results in accumulation of genetic mutations at a faster rate in these cells than in their BRCA2 wt counterparts. The early onset of breast cancer in BRCA2 mutation carriers may arise through a similar process, but whether heterozygotes have detectable genetic instability and why cancer predisposition is largely restricted to breast cancer remains to be determined.
Acknowledgments
We thank Drs. D. M. Livingston, J. Thacker, W. Lee, and P. Chen for providing reagents, and Drs. D. Haber and D. Waver for critical reading of the manuscript. This work was supported in part by a Breast Cancer Research grant from the Massachusetts Department of Public Health to F.X. and a Dana–Farber/Partners Cancer Care grant to S.N.P.
Abbreviations
- NHEJ
nonhomologous end joining
- DSB
double-strand break
- HR
homologous recombination
- wt
wild-type
- ATM
ataxia-telangiectasia mutated
- PFGE
pulse field gel electorphoresis
- HA
hemagglutinin
- I-Sce-I
restriction endonuclease
References
- 1.Haber J E. Nature (London) 1999;398:665–667. doi: 10.1038/19423. [DOI] [PubMed] [Google Scholar]
- 2.Kanaar R, Hoeijmakers J H, van Gent D C. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 3.Difilippantonio M J, Zhu J, Chen H T, Meffre E, Nussenzweig M C, Max E E, Ried T, Nussenzweig A. Nature (London) 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Ferguson D O, Xie W, Manis J P, Sekiguchi J, Frank K M, Chaudhuri J, Horner J, DePinho R A, Alt F W. Nature (London) 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 5.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moynahan M E, Jasin M. Proc Natl Acad Sci USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinohara A, Ogawa T. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 8.Essers J, Hendriks R W, Swagemakers S M, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H, Kanaar R. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 9.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W, et al. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 10.Sonoda E, Sasaki M S, Buerstedde J M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mekeel K L, Tang W, Kachnic L A, Luo C M, DeFrank J S, Powell S N. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 12.Willers H, McCarthy E E, Wu B, Wunsch H, Tang W, Taghian D G, Xia F, Powell S N. Oncogene. 2000;19:632–639. doi: 10.1038/sj.onc.1203142. [DOI] [PubMed] [Google Scholar]
- 13.Meyn S M. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- 14.Luo C M, Tang W, Mekeel K L, DeFrank J S, Anne P R, Powell S N. J Biol Chem. 1996;271:4497–4503. doi: 10.1074/jbc.271.8.4497. [DOI] [PubMed] [Google Scholar]
- 15.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 16.Yang X H, Lippman M E. Breast Cancer Res Treat. 1999;54:1–10. doi: 10.1023/a:1006189906896. [DOI] [PubMed] [Google Scholar]
- 17.Welcsh P L, Schubert E L, King M C. Clin Genet. 1998;54:447–458. doi: 10.1111/j.1399-0004.1998.tb03764.x. [DOI] [PubMed] [Google Scholar]
- 18.Goggins M, Schutte M, Lu J, Moskaluk C A, Weinstein C L, Petersen G M, Yeo C J, Jackson C E, Lynch H T, Hruban R H, et al. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 19.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 20.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 21.Cortez D, Wang Y, Qin J, Elledge S J. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 22.Jong J S, Collins K M, Brown A l, Lee C H, Chung J H. Nature (London) 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 23.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 24.Moynahan M E, Chiu J W, Koller B H, Jasin M. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 25.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston D M. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 26.Kote-Jarai Z, Eeles R A. Br J Cancer. 1999;81:1099–1102. doi: 10.1038/sj.bjc.6690814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen P L, Chen C F, Chen Y, Xiao J, Sharp Z D, Lee W H. Proc Natl Acad Sci USA. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong A K C, Pero R, Ormonde P A, Tavtigian S V, Bartel P L. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 29.Davies A A, Masson J, Mcllwraith M J, Stasiak A Z, Stasiak A, Venkitaraman A R, West S C. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 30.Sharan S K, Morimatsu M, Albrecht U, Lim D S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Nature (London) 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 31.Connor F, Bertwistle D, Mee P J, Ross G M, Swift S, Grigorieva E, Tybulewicz V L, Ashworth A. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 32.Patel K J, Vu V P, Lee H, Corcoran A, Thistlethwaite F C, Evans M J, Colledge W H, Friedman L S, Ponder B A, Venkitaraman A R. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton A A, Thacker J. Mol Cell Biol. 1987;7:1409–1414. doi: 10.1128/mcb.7.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dibiase S J, Guan J, Curran W J, Jr, Iliakis G. Int J Radiat Oncol Biol Phys. 1999;45:743–751. doi: 10.1016/s0360-3016(99)00229-1. [DOI] [PubMed] [Google Scholar]
- 35.Marmorstein L Y, Ouchi T, Aaronson S A. Proc Natl Acad Sci USA. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brian B H, Larson C J, Shihabuddin L S, Gage F H, Verma I M. Proc Natl Acad Sci USA. 1999;96:13920–13925. doi: 10.1073/pnas.96.24.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taghian D G, Nickoloff J A. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moynahan M E, Pierce A J, Jasin M. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 39.Chen C F, Chen P L, Zhong Q, Sharp Z D, Lee W H. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 40.Vispe S, Cazaux C, Lesca C, Defais M. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall E J. Radiobiology for the Radiologist. Philadelphia: Lippincott; 1988. [Google Scholar]
- 42.Finnie N J, Gottlieb T M, Blunt T, Jeggo P A, Jackson S P. Proc Natl Acad Sci USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thacker J. Biochimie. 1999;81:77–85. doi: 10.1016/s0300-9084(99)80041-8. [DOI] [PubMed] [Google Scholar]
- 44.Tebbs R S, Zhao Y, Tucker J D, Scheerer J B, Siciliano M J, Hwang M, Liu N, Legerski R J, Thompson L H. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baumann P, West S C. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 46.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 47.te Riele H, Maandag E B, Berns A. Proc Natl Acad Sci USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrini J H, Bressan D A, Yao M S. Semin Immunol. 1997;9:181–188. doi: 10.1006/smim.1997.0067. [DOI] [PubMed] [Google Scholar]
- 49.Lee H, Trainer A H, Friedman L S, Thistlethwaite F C, Evans M J, Ponder B A, Venkitaraman A R. Mol Cell. 1999;4:1–10. doi: 10.1016/s1097-2765(00)80182-3. [DOI] [PubMed] [Google Scholar]
- 50.Jackson S P, Jeggo P A. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 51.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 52.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 53.Riballo E, Critchlow S E, Teo S H, Doherty A J, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett C F, et al. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 54.Kabotyanski E B, Gomelsky L, Han J O, Stamato T D, Roth D B. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuller L F, Painter R B. Mutat Res. 1988;193:109–121. doi: 10.1016/0167-8817(88)90041-7. [DOI] [PubMed] [Google Scholar]
- 56.Yu V, Koehler M, Steinlein C, Schmid M, Hanakahi L A, van Gool A J, West S C, Venkitaraman A R. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Zeng Z, Bui T, DiBiase S, Qin W, Xia F, Powell S, Iliakis G. Cancer Res. 2001;61:270–277. [PubMed] [Google Scholar]
- 58.Abbott D W, Freeman M L, Holt J T. J Natl Cancer Inst. 1998;90:978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 59.Haber J E. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 60.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 61.Liang F, Han M, Romanienko P J, Jasin M. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caldecott K, Jeggo P. Mutat Res. 1991;255:111–121. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]