Abstract
Isocitrate dehydrogenase (IDH) enzymes have recently become a focal point for research aimed at understanding the biology of glioma. IDH1 and IDH2 are mutated in 50%–80% of astrocytomas, oligodendrogliomas, oligoastrocytomas, and secondary glioblastomas but are seldom mutated in primary glioblastomas. Gliomas with IDH1/2 mutations always harbor other molecular aberrations, such as TP53 mutation or 1p/19q loss. IDH1 and IDH2 mutations may serve as prognostic factors because patients with an IDH-mutated glioma survive significantly longer than those with an IDH–wild-type tumor. However, the molecular pathogenic role of IDH1/2 mutations in the development of gliomas is unclear. The production of 2-hydroxyglutarate and enhanced NADP+ levels in tumor cells with mutant IDH1/2 suggest mechanisms through which these mutations contribute to tumorigenesis. Elucidating the pathogenesis of IDH mutations will improve understanding of the molecular mechanisms of gliomagenesis and may lead to development of a new molecular classification system and novel therapies.
Keywords: glioma, IDH1/2 mutation, 2-hydroxyglutarate, 2-HG, metabolism
Deregulation of cellular energetics is one of the hallmarks of cancer described by Hanahan and Weinberg.1 Malignant tumor cells exhibit fundamentally altered cellular energetics, such as increased aerobic glycolysis,2 which may contribute to tumorigenesis and malignancy.3 The inclusion of deregulated cellular energetics as a hallmark of cancer reflects the increasing recognition of this fundamental cellular process in malignant transformation.4
In recent years, researchers have found that genomic mutations often target key enzymes that regulate important metabolic pathways; they also have shown that aberrant expression of oncogenes and tumor suppressor genes alters the expression and activity of nutrient transporters and metabolic enzymes.5,6 The earliest known examples of mutations in genes coding for mitochondrial enzymes involved in metabolism include alterations in fumarate hydratase and succinate dehydrogenase in hereditary leiomyomatosis, renal cell carcinoma, and paragangliomas.7,8 The first mutations discovered in the genes encoding isocitrate dehydrogenases (IDHs; including IDH1 and IDH2) were identified in metastatic colon cancer,9 and this discovery represents one of the highlights of cancer biology research in the era of high-throughput sequencing.
In 2008, the genes encoding IDH1, and to a lesser extent IDH2, were found to be mutated in low-grade gliomas and a subset of secondary glioblastomas.10 IDH1/2 mutations have also been identified in other malignant tumors, including acute myeloid leukemia, oral squamous cell carcinoma, prostate cancer, non–small-cell lung cancer, enchondroma, cholangiocarcinoma, and chronic myelogenous leukemia.11–16 However, the highest frequencies of IDH mutations are found in glioma and acute myeloid leukemia.17,18
Gliomas are the most common cancers of the nervous system. They are classified as grade I to grade IV on the basis of histopathologic and clinical criteria established by the World Health Organization (WHO).19 Gliomas comprise 3 common histologic subtypes: astrocytomas, oligodendrogliomas, and ependymomas, which are based on their morphological similarities to their normal cellular counterparts. WHO grade I gliomas are often curable by surgical resection, while WHO grade II or III gliomas have a worse prognosis. Median survival for WHO grade IV tumors (glioblastomas) is only about 16 months, even after treatment with a multimodality regimen comprising surgery, radiation therapy, and adjuvant chemotherapy.20
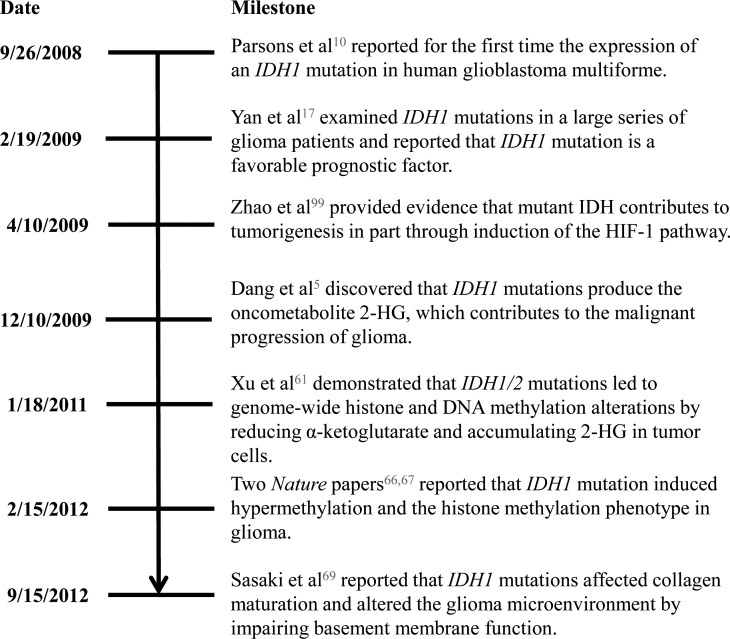
Glioblastoma most frequently arises de novo (“primary” glioblastoma), but approximately 10% of patients have a clinical history of a lower-grade astrocytoma. The tumors that progress from a lower grade are termed “secondary” glioblastoma.19 While IDH1 mutations are present in 70% of lower-grade gliomas and secondary glioblastomas, they are present in only 5% of primary glioblastomas.17 Even though the histopathologic features of primary and secondary glioblastomas are similar, a constellation of characteristics distinguishes glioblastomas expressing an IDH mutation from those without an IDH mutation, such as frontal location, weaker contrast enhancement, and necrosis.21 Furthermore, Lai et al21 found that glioblastomas with IDH mutation may originate from lineage-committed neural cells, whereas glioblastomas without IDH mutation may arise from neural stem cells. As Fig. 1 shows, research on IDH mutations in glioma has made considerable progress, from enhancing methods of detecting IDH mutations to improving understanding of the mechanism of tumorigenesis in IDH-mutant gliomas. In this review, we summarize the current understanding of the roles of IDH1 and IDH2 in glioma and the potential of IDH mutations as diagnostic and prognostic markers and therapeutic targets.
Fig. 1.
Significant milestones in research on IDH mutations in glioma.5,10,17,66,67,69,99
Biochemical Functions of Wild-type and Mutant IDH Enzymes
Physiological Structure and Function of Wild-type IDH
The human genome has 5 IDH genes, coding for 3 distinct IDH enzymes whose activities are dependent on either NADP (NADP+-dependent IDH1 and IDH2) or NAD (NAD+-dependent IDH3). Both IDH2 and IDH3 are localized in the mitochondria and participate in the citric acid cycle for energy production, whereas IDH1 is localized in the cytoplasm and peroxisomes.22 The IDH1 gene is located on 2q33.3 and encodes a 414-amino-acid protein; IDH2 is located on 15q26.1 and encodes a 419-amino-acid protein.22
IDH1 and IDH2 are homodimeric enzymes with 69% amino acid identity. Crystallographic studies have shown that the overall quaternary structures of the 2 proteins are nearly identical.22 IDH3 is a heteroctamer (α4β2γ2) formed by 3 gene products: IDH3A (15q25.1-2), IDH3B (20p13), and IDH3G (Xq28). It is known for its role in catalyzing a critical step of the citric acid cycle.23 Whereas IDH3 is critically important in cellular metabolism, it has not been shown to express cancer-associated mutations. Krell et al.24 speculated that mutations in IDH3 do not occur at an appreciable frequency in glioma because monoallelic IDH1 and IDH2 mutations occur more frequently by chance than the biallelic mutations expected for IDH3.
IDH enzymes catalyze the oxidative decarboxylation of isocitrate to produce α-ketoglutarate (also known as 2-oxoglutarate) and concomitantly produce NADPH from NADP+. IDH enzymes also catalyze the reductive carboxylation of α-ketoglutarate to form isocitrate and concomitantly produce NADP+ from NADPH.25 IDH1 contains a C-terminal peroxisome-targeting sequence and is thought to be the sole producer of NADPH in peroxisomes.22 Peroxisomal enzymes are involved in a broad spectrum of metabolic pathways, including conversion of lipids, amino acids, hydroxyacids, purines, and reactive oxygen species (ROS).26 NADPH in peroxisomes is used for biosynthetic reactions and for thiol-based antioxidant systems.27 IDH1 forms an asymmetric homodimeric complex with a hydrophilic active site cleft consisting of an NADP-binding site and an isocitrate metal ion–binding site.22 At the active site, the R132 residue forms hydrophilic interactions with the α-carboxylate of isocitrate. The R132 residue is strictly conserved in all bacterial and eukaryotic NADP+-dependent IDH1 enzymes.22 IDH2 serves to maintain redox balance in the mitochondria and to respond to oxidative damage.28 The R172 residue of IDH2 is located in the active site of the enzyme and is analogous to the R132 residue in IDH1.17 These 2 IDH enzymes also support growth in cells by the glutamine-dependent reductive carboxylation pathway.29
The common functions of IDH1 and IDH2 are production of NADPH and α-ketoglutarate. NADPH is a required cofactor for many cellular functions, including lipid metabolism,30 glucose metabolism,31 and defense against oxidative stress.32 NADPH is an important element in cellular defense against oxidative damage through its maintenance of a pool of reduced glutathione (GSH).33 α-Ketoglutarate also has roles in diverse cellular functions, serving as an essential cofactor for a family of oxygenases that includes prolyl hydroxylase, jumonji-C domain–containing histone demethylases (JHDMs), TETs (ten-eleven translocation enzymes), collagen prolyl-4-hydroxylases 1, 2, and 3 (C-P4H1–3), and procollagen-lysine 2-oxoglutarate 5-dioxygenases 1, 2, and 3 (PLOD1–3).34 Therefore, the by-products of the IDH1/2 enzymatic reactions can affect many different cellular processes.
Structure and Function of IDH Mutants
Somatic mutations in IDH1 and IDH2 were initially identified in low-grade gliomas and secondary glioblastomas. These mutations are always hemizygous, and almost all are missense mutations that result in an amino acid substitution of an arginine residue in the active site.17 Moreover, mutations of IDH1 and IDH2 are mutually exclusive in gliomas. A recent publication showed that IDH1 and IDH2 mutations resulted in differential elevation of 2-hydroxyglutarate (2-HG) expression.35 Over 90% of IDH mutations affect IDH1,17 and almost all IDH1 mutations affect the arginine at R132, within the enzyme's substrate-binding site.23 Approximately 90% of these missense mutations result in substitution of arginine with histidine (R132H).36 Other mutations of IDH1 are far less common but also affect R132: substitution of arginine by cysteine (R132C; frequency 3.6%–4.6%), serine (R132S; 0.8%–2.5%), glycine (R132G; 0.6%–3.8%), or leucine (R132L; 0.5%–4.3%).23 There are 2 additional conserved arginine residues within the substrate-binding site of the IDH1 enzyme. A single report identified mutations involving one of these (R100Q) in 2 WHO grade III anaplastic oligodendrogliomas and a WHO grade II astrocytoma.37 Other IDH1 mutations have been reported, including G97 and Y139.37
Like those in IDH1, mutations in IDH2 affect a conserved arginine residue (R172, synonymous with IDH1 R132) in the substrate-binding site of the IDH2 enzyme. Point mutations result in the substitution of arginine by lysine (R172K), methionine (R172M), glycine (R172G), or tryptophan (R172W).38 Mutation of IDH1/2 in the substrate-binding site results in markedly reduced affinity for its substrate isocitrate. These mutations therefore confer a loss of enzyme function. Crystal structure analysis has indicated that IDH1 R132H substitution causes alterations in the structure of the active site.5 In vitro studies demonstrated that mutant IDH1/2 may dominantly inhibit the enzyme activity of wild-type IDH1/2 through heterodimer formation, leading to reduced generation of NADPH and α-ketoglutarate.17,39 Moreover, IDH1/2 mutations partly impair reductive carboxylation of α-ketoglutarate.39 Besides causing this loss of function, IDH1/2 mutation confers a surprising but important new catalytic activity, production of 2-HG by reduction of α-ketoglutarate, which is accompanied by consumption of NADPH.5 Tumor samples harboring IDH1/2 mutations exhibit 2-HG levels as high as 100-fold greater than tumors with wild-type IDH1/2.5 However, not all IDH1/2 mutants gain this neomorphic activity. Some IDH1/2 mutants maintain their primary enzymatic function, while others exhibit loss of function without gaining additional functions (Table 1).
Table 1.
| IDH Function | IDH1 Mutation | IDH2 Mutation |
|---|---|---|
| 2-HG production | IDH1 R132_ | IDH2 R172 |
| IDH1 R100_ | IDH2 R140 | |
| IDH1 Y139D | ||
| Wild-type activity | IDH1 V71I | IDH2 V249M |
| IDH1 V178I | ||
| IDH1 I99M | ||
| IDH1 G123R | ||
| IDH1 I130M | ||
| IDH1 H133Q | ||
| Loss of function | IDH1 G70D | IDH2 F394_ |
| IDH1 A134D | ||
| IDH1 R49C |
IDH Mutations in Gliomas
Mutations in IDH1 were initially identified in a small number of glioblastomas that had been subjected to exome-wide deep sequencing.10 It was noted at the time that the glioblastomas harboring IDH1 mutations were mostly secondary glioblastomas that had arisen through progression from a lower-grade tumor, occurred mostly in younger patients, and were associated with better survival rates.10 In several subsequent analyses, IDH1 mutations were identified in 70%–80% of grades II and III astrocytomas, oligodendrogliomas, oligoastrocytomas, and secondary glioblastomas (Table 2). IDH1 mutations were also found in some cases of gliomatosis cerebri40 as well as approximately 5% of primary glioblastomas. A small group of gliomas (3%–5%) was found to harbor IDH2 mutations.17 In contrast to the diffuse gliomas, other WHO grades I and II central nervous system tumors such as pilocytic astrocytoma, subependymal giant cell tumor, typical ganglioglioma, ependymoma, and pleomorphic xanthoastrocytoma expressed few or no IDH mutations.41–44 Small numbers of relatively rare neuronal and glioneuronal tumors such as central neurocytoma, dysembryoplastic neuroepithelial tumor, rosetting glioneuronal tumor, and embryonal tumors such as medulloblastoma have also been examined and been found to lack IDH mutations.42 High-grade childhood gliomas are similar to adult primary glioblastoma in that they rarely carry IDH mutations.45
Table 2.
| Glioma Subtype | Number of Patients | IDH1 Mutation (%) | IDH2 Mutation (%) | IDH1/2* Mutation (%) |
|---|---|---|---|---|
| Diffuse astrocytoma | 342 | 248 (72.51) | 4 (1.17) | 252 (73.68) |
| Oligodendroglioma | 311 | 242 (77.81) | 8 (2.57) | 250 (80.39) |
| Oligoastrocytoma | 140 | 114 (81.43) | 1 (0.71) | 115 (82.14) |
| Anaplastic astrocytoma | 232 | 113 (48.71) | 5 (2.16) | 118 (50.86) |
| Anaplastic oligoastrocytoma | 253 | 168 (66.40) | 11 (4.35) | 179 (70.75) |
| Anaplastic oligodendroglioma | 308 | 101 (32.79) | 12 (3.90) | 113 (36.69) |
| Primary glioblastoma | 577 | 25 (4.43) | 0 (0) | 25 (4.43) |
| Secondary glioblastoma | 54 | 37 (68.52) | 0 (0) | 37 (68.52) |
| Ganglioglioma | 8 | 3 (37.50) | 0 (0) | 3 (37.5) |
| Anaplastic ganglioglioma | 5 | 3 (60.00) | 0 (0) | 3 (60.00) |
| Pilocytic astrocytoma | 21 | 0 (0) | 0 (0) | 0 (0) |
| Subependymal giant cell astrocytoma | 2 | 0 (0) | 0 (0) | 0 (0) |
| Pleomorphic xanthoastrocytoma | 7 | 1 (14.29) | 0 (0) | 1 (14.29) |
| Primary pediatric glioblastoma | 15 | 0 (0) | 0 (0) | 0 (0) |
*The number of patients and frequency of IDH1/2 mutation are combined from 6 published reports that measure IDH1/2 mutation frequency by direct sequencing.
Table 2 shows the reported rates of IDH1 and IDH2 mutation in the different subtypes of glioma. This table also shows that the reported rates of IDH1 mutation were significantly greater than those of IDH2 mutation in glioma. This is in contrast to acute myeloid leukemia, where the reported rates of IDH1 (6.6%) and IDH2 (10.8%) mutation were similar.18 This suggests that the rates of IDH1 and IDH2 mutations may vary with tumor type. Additionally, IDH1 and IDH2 appear to function in different cellular compartments.24 Therefore, mutation of these 2 genes may impact different cellular pathways and exert tumorigenic function differently. Further studies are needed for a more in-depth understanding.
IDH Mutations and Other Genetic and Epigenetic Alterations in Glioma
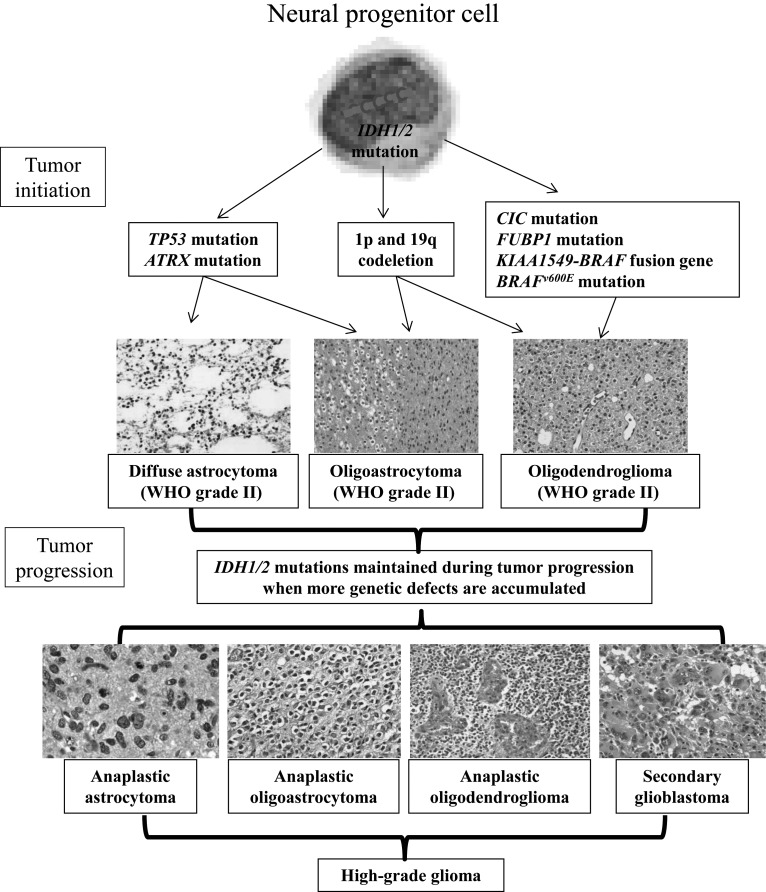
IDH1/2 mutations are associated with other genetic alterations that occur frequently in glioma.46 For example, TP53 mutations and IDH1/2 mutations frequently occur together in tumors of astrocytic origin, while IDH1/2 mutations and 1p/19q deletions frequently occur together in tumors of oligodendroglial origin (Fig. 2).47–49 These co-occurrences specific to histologic type indicate that IDH1 and IDH2 mutations are frequent and early genetic alterations in the development of astrocytomas and oligodendrogliomas.50 Moreover, IDH mutation status is stable during the progression of low-grade gliomas to secondary high-grade gliomas.50 An analysis of initial and recurrent tumors from 51 patients showed that most carried an IDH mutation together with either mutant TP53 or 1p/19q codeletion at the time of initial resection. In no case did an IDH mutation occur after acquisition of the other alteration, while a few showed an isolated IDH mutation in the initial specimen and acquisition of either the TP53 mutation or 1p/19q codeletion in the recurrent tumor.48 Moreover, Figarella-Branger et al51 analyzed IDH1 mutation, TP53 mutation, and 1p/19q codeletion in 88 low-grade glioma patient samples. The patients were divided into 4 groups: group 1, IDHR132H+/p53–/1p19q–; group 2, IDHR132H+/p53–/1p19q+; group 3, IDHR132H+/p53+/1p19q–; and group 4, triple negative gliomas. In the 88 patients studied, p53 mutation and 1p/19q codeletion were always associated with IDH mutation.51 This finding led to the hypothesis that IDH mutation occurs prior to other genetic events in gliomagenesis.
Fig. 2.
Relationship between IDH mutations and other genetic alterations in glioma. The timeline represents glioma development from earlier events (top) to later events (bottom). Acquisition of an IDH1 mutation in a common tumor progenitor cell leads to gliomagenesis in the presence of another concurrent molecular feature such as TP53 mutation or 1p/19q codeletion. Tumors may then acquire additional molecular events, such as citrate carrier (CIC) mutation, which cooperate to transform a previously low-grade glioma into a high-grade glioma, such as anaplastic astrocytoma, anaplastic oligodendroglioma, anaplastic oligoastrocytoma, or secondary glioblastoma.
IDH mutation is also found to frequently co-occur with mutation of alpha-thalassemia/mental retardation syndrome X-linked (ATRX), citrate carrier, far upstream element binding protein 1 (FUBP1), or BRAFv600E or presence of the KIAA1549-BRAF fusion gene (Fig. 2).47–50,52,53 IDH mutation may occur in a precursor cell that can give rise to both oligodendrocytes and astrocytes.48 Following IDH mutation, additional mutations may specify tumor development along an astrocytic or oligodendrocytic pathway. These additional genetic events may also play important roles in the progression of low-grade gliomas to high-grade gliomas (Fig. 2).
IDH1 and IDH2 mutations rarely co-occur in the same tumor. IDH1/2 mutations have been shown to be inversely related to or even mutually exclusive of EGFR and PTEN abnormalities.50 IDH1 mutations are associated with specific gene-expression clustering patterns in glioblastoma and are almost always associated with the proneural subtype.54
When gliomas are classified according to the level of epigenetic methylation, tumors with IDH1 mutations are classified as methylation rich.55 Analysis of DNA promoter alterations in 272 tumors from The Cancer Genome Atlas revealed a distinct set of samples with concerted hypermethylation at a large number of loci. These same samples were also highly associated with IDH1 mutation.55 These and other data provide evidence suggesting the existence of a glioma–cytosine-phosphate-guanine island methylator phenotype (G-CIMP). This phenotype is tightly correlated with both the proneural subgroup and IDH1 mutations.53 Furthermore, and of particular prognostic relevance in glioma, IDH1 mutations are associated with methylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter.55 SongTao et al56 reported that patients whose glioblastoma had MGMT promoter methylation survived significantly longer than those whose glioblastoma did not have MGMT promoter methylation after receiving temozolomide, demonstrating that IDH1 mutation is a prognostic factor in glioma.56
Possible Mechanisms Through Which IDH1/2 Mutations Contribute to Tumorigenesis
Differences in metabolism between cancer and normal cells have long been noted by cancer researchers. The aberrant cellular metabolism of cancer cells may alter cellular gene expression. On the other hand, the genetic alterations observed in tumors may lead to deregulation of cellular metabolism.57 Aberrant cellular metabolism and aberrant gene expression affect each other and together drive cancer pathogenesis. The mechanisms by which IDH mutation contributes to the development of cancer are still not completely understood, but some studies indicate that aberrant levels of 2-HG competitively inhibit α-ketoglutarate–dependent dioxygenase enzymes to affect tumorigenesis.34 Losman et al58 reported that R-2-HG is sufficient to promote leukemogenesis. It is important to note that IDH1/2 mutation induces the R-enantiomer of 2-HG, whereas it is L-2-HG that increased in patients with L-2-hydroxyglutaric aciduria, a rare autosomal recessive inherited encephalopathy.59 In addition to 2-HG, aberrant cellular metabolism of nutrients and metabolites also plays a role in driving gliomagenesis, especially metabolism of glucose, glutamine, and NADPH.
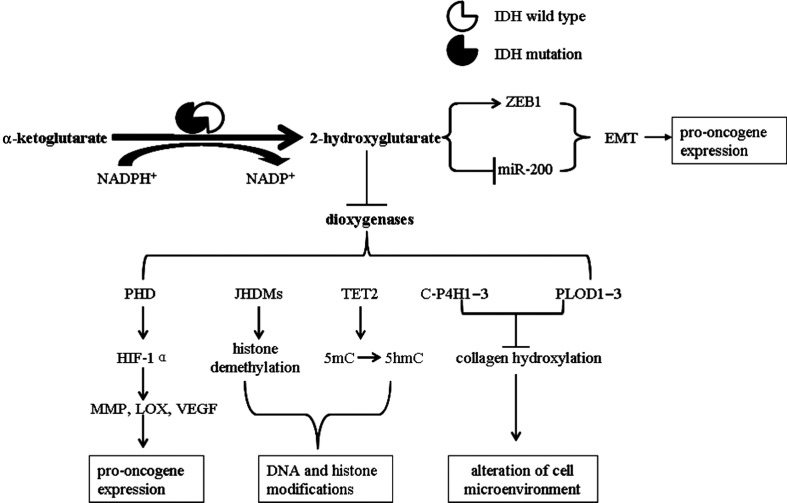
More than 60 different dioxygenase enzymes are present in cells, and α-ketoglutarate is an important cofactor for many dioxygenases involved in a variety of cellular processes. Dang et al5 demonstrated that IDH1/2 mutations produce 2-HG, which functions as an oncometabolite and contributes to the formation and malignant progression of gliomas. In structure, 2-HG is similar to α-ketoglutarate and competitively inhibits the enzymatic activity of dioxygenases.60 Thus, inhibition of dioxygenases by 2-HG is believed to be one mechanism through which IDH1/2 mutations contribute to the pathogenesis of gliomas (Fig. 3). The lines of evidence supporting this suggestion are summarized in the following paragraphs.
Fig. 3.
Proposed mechanism by which IDH mutations produce 2-HG and promote gliomagenesis. IDH mutations produce 2-HG, which inhibits α-ketoglutarate–dependent dioxygenases. Inhibition of C-P4H1–3 and PLOD1–3 prevents proper prolyl and lysyl hydroxylation of collagen, affecting basement membrane integrity and altering the cellular microenvironment. Inhibition of TET enzymes blocks 5-hydroxy methylcytosine modification, which leads to cytosine demethylation. Inhibition of JHDMs leads to altered histone methylation on multiple histone H3 lysine residues. Inhibition of prolyl hydroxylase domain–containing proteins (PHDs) stabilizes HIF-1α and promotes epithelial-to-mesenchymal transition (EMT). Together with 2-HG–induced DNA modifications, these alterations promote oncogene expression and promote oncometabolism. IDH mutations also alter glucose metabolism and increase glycolysis. miR-200, microRNA-200.
2-HG Inhibits Prolyl Hydroxylase to Stabilize Hypoxia-inducible Factor–1α
One of the dioxygenases inhibited by 2-HG is prolyl hydroxylase, which regulates the stability and activation of hypoxia-inducible factor (HIF)–1α.61,62 Under normal oxygen conditions, HIF-1α is modified by prolyl hydroxylase at specific proline residues, which triggers binding of the tumor suppressor von Hippel–Lindau protein and subsequent ubiquitination and proteasomal degradation.62 In hypoxic conditions, however, oxygen-dependent hydroxylation does not occur, leading to accumulation of HIF-1α and its subsequent translocation into the nucleus and induction of HIF-1α downstream gene targets. The reduced production of α-ketoglutarate and increased production of 2-HG resulting from mutant IDH1 in glioma cells lead to inhibition of prolyl hydroxylase and therefore stabilization of HIF-1α.63 Once stabilized, HIF-1α transcriptionally activates a number of genes that might promote tumor cell growth, invasion, angiogenesis, and metastasis. Examples include genes encoding growth factors such as transforming growth factor–β and platelet-derived growth factor–β, invasion-promoting genes such as those encoding matrix metalloproteinases (MMPs) and lysyl oxidase (LOX), and angiogenesis-promoting genes such as the gene encoding vascular endothelial growth factor (VEGF).62
2-HG Inhibits JHDMs to Induce Aberrant Histone Demethylation
Using biochemical, structural, and cellular assays, Chowdhury et al64 showed that 2-HG inhibits α-ketoglutarate–dependent oxygenases with varying potencies. Half-maximal inhibitory concentrations of 2-HG varied from approximately 25 μM for the histone demethylase jumonji-C domain–containing protein 2A (JMJD2A) to more than 5 mM for the HIF prolyl hydroxylase. This indicates that candidate oncogenic pathways in IDH-associated malignancies should also include those that are regulated by α-ketoglutarate oxygenases other than HIF hydroxylases, in particular those involving regulation of histone methylation.64 In humans, aberrant histone demethylation has been associated with cancer development and aggressiveness.65 Lu et al.66 reported that IDH mutants producing 2-HG can prevent the histone demethylation that is required for differentiation of lineage-specific progenitor cells into terminally differentiated cells. They found that the H3K9 demethylase JMJD2A, which can be inhibited by 2-HG, was induced during cell differentiation and that RNA-interference suppression of JMJD2A was sufficient to block differentiation. These findings demonstrate that 2-HG can inhibit histone demethylation and that inhibition of histone demethylation can be sufficient to block the differentiation of nontransformed cells.66
In addition to histone methylation, 2-HG may also impact DNA methylation. As mentioned, IDH1 mutations are closely linked to the epigenetic program.67 The CIMP phenotype is a powerful determinant of glioma pathogenicity. Examination of the epigenome of a large set of intermediate-grade gliomas demonstrated a distinct G-CIMP phenotype that is highly dependent on the presence of IDH mutation. Turcan et al.67 reported that mutation of IDH1 contributes to the establishment of G-CIMP by remodeling the methylome. Their findings demonstrated that IDH mutation is a molecular basis of CIMP in gliomas, providing a framework for understanding oncogenesis in these tumors. One potential mechanism by which IDH mutation establishes the glioma hypermethylator phenotype is by inhibition of the TET family of α-ketoglutarate–dependent dioxygenases. TET catalyzes the sequential oxidation of 5-methylcytosine to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine, and 5-carboxylcytosine, leading to eventual DNA demethylation.68 Production of 2-HG in IDH-mutant cells inhibits this process, maintaining DNA in a methylated state.34 Xu et al61 also demonstrated that ectopic expression of mutant IDH1/2 reduces TET-catalyzed 5hmC production to maintain DNA in a methylated state.
2-HG Inhibits PLOD1–3 and C-P4H1–3 to Impair Collagen Maturation
In a recent report, Sasaki et al established a brain-specific IDH1 R132H conditional knock-in mouse model and found that these mice did not develop glioma. Unexpectedly, brain-specific IDH1 R132 mutation resulted in perinatal lethality, and the mutant IDH1 protein induced massive hemorrhage within the cerebral hemispheres and cerebellum. They also found that 2-HG inhibited PLOD1–3 and C-P4H1–3 to impair collagen maturation, resulting in basement membrane aberrations.69 Basement membrane is a highly crosslinked and insoluble sheetlike structure that separates epithelial cells from the surrounding stroma. So far, there have been no reports of basement membrane aberrations inducing tumorigenesis, but an intact basement membrane prevents tumor invasion, and destruction of the basement membrane is the first step in cancer cell stromal invasion and metastasis.70 The 2 major components of the basement membrane are laminin and collagen type IV, lack of which was associated with poor outcome following curative resection of pancreatic head cancers.70,71
Besides inhibiting α-ketoglutarate–dependent dioxygenase enzymes, 2-HG also alters the gene expression profile of IDH-mutated cells, affecting signaling pathways. Grassian et al72 recently reported that high levels of 2-HG upregulated zinc-finger E-box binding homeobox 1 (ZEB1) and downregulated microRNA-200, activating the epithelial-mesenchymal transition to promote tumorigenesis.72 In the future, we need more comprehensive studies, employing microarray technology (mRNA, noncoding RNA) and bioinformational analysis methods to elucidate how a high level of 2-HG affects various signaling pathways.
Aberrant Metabolism in Cells With IDH Mutation
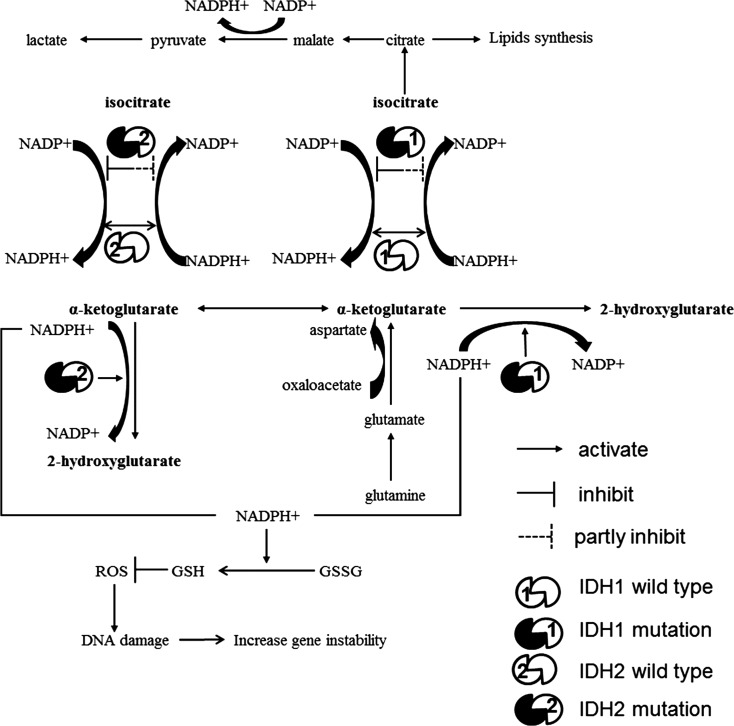
Tumor cells often take up nutrients in excess of their bioenergetic needs and shunt metabolites into pathways that support macromolecular biosynthesis.1,73 During cell proliferation, tumor cells depend on aerobic glycolysis to meet bioenergy needs and to generate intermediates for macromolecule biosynthesis, such as ribose sugars for nucleotides, glycerol and citrate for lipids, and nonessential amino acids. This is particularly important in cells with mitochondrial malfunction.73 One study demonstrated that glioblastoma cells utilize mitochondrial glucose oxidation during aggressive tumor growth in vivo,74 but almost all glioblastoma cells studied expressed wild-type IDH1/2 and maintained normal mitochondrial function.17 In IDH-mutant cells, glucose oxidation is weakened. To meet bioenergetic and biosynthesis needs, glioma cells with mutant IDH may maintain cell proliferation by the glutamate metabolism pathway. This is the reason that inhibition of the production of glutamate from glutamine slows cell growth selectively in IDH1-mutant cells.75 Mutant IDH is still capable of carrying out 50% of the cell’s capacity for NADPH-dependent reductive carboxylation, but the production of bioenergy and intermediates for biosynthesis is decreased, in contrast to that in cells with wild-type IDH.25 This may be one reason that cells with IDH mutations grow at a slower rate.76
The production of NADPH maintains cellular redox balance and regulates ROS.77 Decreased NADPH levels and increased ROS levels in gliomas with mutant IDH may contribute to tumorigenesis. GSH is a major antioxidant that is of crucial importance in defense against the oxidative stress caused by ROS.77 NADPH is required for the reduction of glutathione disulfide (GSSG) to GSH and maintenance of the antioxidant state of GSH in the cell. Mutation impairs the ability of IDH, the major producer of NADPH in the cell, to generate NADPH from NADP+ and also depletes NADPH by consuming it as a cofactor to convert α-ketoglutarate into 2-HG, producing NADP+.21,22 Thus, cells with reduced NADP-dependent IDH activities have increased oxidative DNA damage, a higher ratio of GSSG to total GSH, increased lipid peroxidation, and reduced survival on exposure to oxidative stress.78 It is possible that mutation of IDH renders cells vulnerable to oxidative DNA damage (Fig. 4). This may promote further genetic changes, such as TP53 mutation or t(1;19) translocation, leading to development of either astrocytoma or oligodendroglioma.
Fig. 4.
Proposed mechanisms through which IDH mutations produce aberrant oncometabolites and promote gliomagenesis. (A) IDH mutation impairs the oxidative decarboxylation of isocitrate to produce α-ketoglutarate. IDH mutants convert α-ketoglutarate to 2-HG and maintain only 50% of the cell's capacity to carry out reductive carboxylation. The resultant decrease in tricarboxylic acid intermediates affects the rate of macromolecular biosynthesis and impairs cell proliferation. IDH mutations decrease levels of NADPH+ and increase levels of NADP+. NADPH+ maintains cellular redox balance. When level of NADPH+ decreases and level of NADP+ increases, GSSG does not produce enough GSH to inhibit ROS production. ROS-induced DNA damage may promote the incidence of malignancy.
Clinical Application of IDH Mutation
IDH Mutation as a Prognostic Marker
IDH1 mutation is strongly correlated with good prognosis in patients with glioma. Median overall survival of patients whose glioblastoma expressed an IDH mutation was significantly longer than that of patients whose glioblastoma expressed wild-type IDH1 and IDH2.79 Mutations of IDH also are associated with better prognosis in patients with anaplastic astrocytoma.80 Multivariate analysis confirmed that IDH1 mutation is an independent favorable prognostic marker in glioblastoma and anaplastic glioma after adjustment for other genomic profiles and treatment modality.81 IDH mutation in glioma patients can also distinguish diffuse infiltrating tumor cells from reactive astrocytosis, as well as astrocytic and oligodendroglial tumors from ependymomas and grade I pilocytic astrocytomas.23 Therefore, detection of IDH mutations is of clear clinical significance in terms of tumor classification and assessment of prognosis.
Methods for Detection of IDH Mutations
Initial studies identified IDH mutations using traditional Sanger sequencing, and this method is still commonly used to detect mutations other than IDH1R132H. While sequencing is considered the standard for detection of mutations, the sensitivity of this method is approximately 20% of mutant sequences in a wild-type background.44 Brain tumor biopsy specimens universally contain contaminating nontumor tissues that add noise in Sanger sequencing. Immunohistochemistry is currently the preferred method for detection of IDH mutations in glioma. Two monoclonal antibodies, H09 and IMab-1, have been developed against mutated IDH1.82 The H09 antibody is commercially available (Dianova) and works well in routine immunohistochemical analysis on formalin-fixed, paraffin-embedded tissue.83 The sensitivity and specificity of the H09 antibody for detecting IDH1 R132H reportedly approaches 100%,38 and IMab-1 seems to perform comparably.82 The current commercially available clone is highly specific for IDH1 R132H; it does not detect other rare substitutions in IDH1 (R132C, R132S, R132L, R132G).38 More recently, Kaneko et al84 established an antibody specific to anti–IDH2 mutation to detect IDH2 R172K and IDH2 R172M in tumors.
Alternatively, the tightly clustered character of the IDH mutations makes them ideal candidates for pyrosequencing analysis. The major advantages of pyrosequencing for somatic mutation detection are its quantitative nature and sensitivity: it reportedly is able to detect a minimum of 5% of IDH mutant alleles in a wild-type background.85 Pyrosequencing may thus be particularly useful for the analysis of diffusely infiltrating, low-grade gliomas.
Determination of IDH status requires tumor tissue for genetic analysis. A noninvasive method for assessing the presence of IDH mutation indirectly would be useful in the clinical setting. As many known mutations in either IDH1 or IDH2 have been shown to result in an increase of the 2-HG level, 2-HG may serve as a surrogate marker for such mutations.45,86,87 Sahm et al88 detected 2-HG levels by using gas chromatography/mass spectrometry, but this method requires tumor tissue. Blood samples contain measurable 2-HG, which can serve as a prognostic factor in acute myeloid leukemia.89 However, 2-HG concentration in serum from patients with glioma does not correlate with IDH1/2 mutation status or tumor size.90 Boisselier et al.91 recently reported using a combination of coamplification at lower denaturation temperature and digital polymerase chain reaction to detect IDH1 (R132H) mutation in the plasma of patients with glioma.91 This technique may provide the combination of accuracy and convenience required for clinical applications.
Brain MRI is the primary modality for clinical evaluation of glioma patients. The advent of MR spectroscopy (MRS) has provided a noninvasive diagnostic tool for biochemical characterization of pathophysiological processes in the brain. The ability to detect 2-HG in tumors by MRS could provide important diagnostic and prognostic information. Choi et al92 reported the noninvasive detection of 2-HG by proton MRS. They developed and optimized the pulse sequence with numerical and phantom analyses for 2-HG detection and estimated concentrations of 2-HG using spectral fitting in the tumors of 30 subjects. Detection of 2-HG correlated with mutations in IDH1 or IDH2 and with increased levels of 2-HG detected by mass spectrometry of the resected tumors.92 These results indicate that MRS may become a reliable, noninvasive tool for detection and quantification of 2-HG in patients with glioma. Pope et al93 also found that water-suppressed proton (1H) MRS provides a noninvasive measure of 2-HG in gliomas. Other studies have demonstrated the feasibility of using MRI- and MRS-based technology as specific and selective tools for noninvasive detection and quantification of 2-HG levels for the diagnosis and classification of IDH1/2 mutation–positive gliomas.94 These data indicate the value of metabolite screening approaches that specifically and sensitively identify IDH-mutant tumors in which the 2-HG level is elevated.
IDH Mutation as a Therapeutic Target
Beyond tumor classification and prognosis, there has been very little evidence of a potential role for IDH enzymes or 2-HG as a molecular target. Seltzer et al75 reported that as mutant IDH1-mediated production of α-ketoglutarate is reduced, there is potentially more reliance on alternative sources of α-ketoglutarate, such as glutamate conversion. Inhibition of the production of glutamate from glutamine through inhibition of glutaminase has been demonstrated to slow cell growth selectively in IDH1-mutant cells.75 Jin et al95 found that disruption of wild-type IDH1 suppressed d-2-hydroxyglutarate production in IDH1-mutated gliomas. This may be a strategy for treating these tumors. More recently, Rohle et al96 used a selective IDH1R132H inhibitor (AGI-5198) to impair the growth of IDH1-mutant containing, but not IDH1-wild-type containing, glioma cells without appreciable changes in genome-wide DNA methylation.96 At the same time, Wang et al97 used a selective IDH2R140Q inhibitor (AGI-6780) to induce differentiation of TF-1 erythroleukemia and primary human acute myelogenous leukemia cells in vitro.97 These studies suggest some potential for therapeutically targeting mutant IDH1, but further studies are needed.
There are conflicting reports linking IDH mutations to treatment response. In some studies, gliomas with mutated IDH were identified as more sensitive to irradiation and chemotherapy than those with wild-type IDH.55 However, other reports indicate that these mutations are not linked to improved treatment response.98 Ultimately, the utility of the IDH enzymes or 2-HG as a molecular target requires further study. Comprehensive understanding of the roles of IDH mutations in gliomagenesis may help to answer these questions in the future.
Conclusions
Several mutations affecting metabolic enzymes are known to play a causal role in cancer, and aberrant cellular metabolism can affect oncogenic signaling pathways. Mutant IDH is an example of such a mutant metabolic enzyme. Despite the increasing number of publications examining IDH mutations in glioma, the roles of these mutations in tumorigenesis remain unclear. However, these studies have demonstrated that IDH mutation is an early event in gliomagenesis, indicating the clinical relevance of these mutations in tumor classification and prognosis. Whether IDH1 mutations are significant and sufficient driver mutations for glioma development remains an open question, since there is no direct in vivo evidence demonstrating the causal role of these mutations alone or in combination with other coincident mutations. Through future studies of both the genome and the metabolome, we hope to gain more information on the mechanisms by which IDH mutation affects gliomagenesis and glioma progression. This understanding will be critical in the design and implementation of novel glioma therapeutics that target this pathway.
Funding
This work was partially supported by NIH grants CA141432, CA09850305, and U24CA143835 to W.Z. and an MD Anderson Cancer Center core grant from the National Cancer Institute (CA16672).
Conflict of interest statement. None declared.
Acknowledgments
We thank Kathryn L. Hale of the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing this manuscript.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17:291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 4.Israël M, Schwartz L. The metabolic advantage of tumor cells. Mol Cancer. 2011;10:70. doi: 10.1186/1476-4598-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frezza C, Zheng L, Folger O, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 8.Gill AJ. Succinate dehydrogenase (SDH) and mitochondrial driven neoplasia. Pathology. 2012;44:285–292. doi: 10.1097/PAT.0b013e3283539932. [DOI] [PubMed] [Google Scholar]
- 9.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 10.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang JY, Chang CC, Lin PC, Chang JG. Isocitrate dehydrogenase mutation hot spots in acute lymphoblastic leukemia and oral cancer. Kaohsiung J Med Sci. 2012;28:138–144. doi: 10.1016/j.kjms.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghiam AF, Cairns RA, Thoms J, et al. IDH mutation status in prostate cancer. Oncogene. 2012;31:3826. doi: 10.1038/onc.2011.546. [DOI] [PubMed] [Google Scholar]
- 13.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22:2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 15.Borger DR, Tanabe KK, Fan KC, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makishima H, Jankowska AM, McDevitt MA, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117:e198–e206. doi: 10.1182/blood-2010-06-292433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y, Zhu YM, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 19.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma—are we there yet? Neuro Oncol. 2013;15:4–27. doi: 10.1093/neuonc/nos273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29:4482–4490. doi: 10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X, Zhao J, Xu Z, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 23.Weller M, Wick W, von Deimling A. Isocitrate dehydrogenase mutations: a challenge to traditional views on the genesis and malignant progression of gliomas. Glia. 2011;59:1200–1204. doi: 10.1002/glia.21130. [DOI] [PubMed] [Google Scholar]
- 24.Krell D, Assoku M, Galloway M, Mulholland P, Tomlinson I, Bardella C. Screen for IDH1, IDH2, IDH3, D2HGDH and L2HGDH mutations in glioblastoma. PLoS One. 2011;6:e19868. doi: 10.1371/journal.pone.0019868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonardi R, Subramanian C, Jackowski S, Rock CO. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem. 2012;287:14615–14620. doi: 10.1074/jbc.C112.353946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rokka A, Antonenkov VD, Soininen R, et al. Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLoS One. 2009;4:e5090. doi: 10.1371/journal.pone.0005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Q, Minard KI, McAlister-Henn L. Dual compartmental localization and function of mammalian NADP+-specific isocitrate dehydrogenase in yeast. Arch Biochem Biophys. 2008;472:17–25. doi: 10.1016/j.abb.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SY, Lee SM, Shin SW, Park JW. Inactivation of mitochondrial NADP+-dependent isocitrate dehydrogenase by hypochlorous acid. Free Radic Res. 2008;42:467–473. doi: 10.1080/10715760802098834. [DOI] [PubMed] [Google Scholar]
- 29.Mullen AR, Wheaton WW, Jin ES, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koh HJ, Lee SM, Son BG, et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J Biol Chem. 2004;279:39968–39974. doi: 10.1074/jbc.M402260200. [DOI] [PubMed] [Google Scholar]
- 31.Ronnebaum SM, Ilkayeva O, Burgess SC, et al. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J Biol Chem. 2006;281:30593–30602. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]
- 32.Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic Biol Med. 2011;51:1289–1301. doi: 10.1016/j.freeradbiomed.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Kim KY, Jang HS, et al. Role of cytosolic NADP+-dependent isocitrate dehydrogenase in ischemia-reperfusion injury in mouse kidney. Am J Physiol Renal Physiol. 2009;296:F622–F633. doi: 10.1152/ajprenal.90566.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih AH, Levine RL. IDH1 mutations disrupt blood, brain, and barriers. Cancer Cell. 2012;22:285–287. doi: 10.1016/j.ccr.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Ward PS, Lu C, Cross JR, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288:3804–3815. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 37.Ward PS, Cross JR, Lu C, et al. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 2012;31:2491–2498. doi: 10.1038/onc.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capper D, Weissert S, Balss J, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narasimhaiah D, Miquel C, Verhamme E, Desclée P, Cosnard G, Godfraind C. IDH1 mutation, a genetic alteration associated with adult gliomatosis cerebri. Neuropathology. 2012;32:30–37. doi: 10.1111/j.1440-1789.2011.01216.x. [DOI] [PubMed] [Google Scholar]
- 40.Ichimura K, Pearson DM, Kocialkowski S, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takano S, Tian W, Matsuda M, et al. Detection of IDH1 mutation in human gliomas: comparison of immunohistochemistry and sequencing. Brain Tumor Pathol. 2011;28:115–123. doi: 10.1007/s10014-011-0023-7. [DOI] [PubMed] [Google Scholar]
- 42.Horbinski C, Kofler J, Yeaney G, et al. Isocitrate dehydrogenase 1 analysis differentiates gangliogliomas from infiltrative gliomas. Brain Pathol. 2011;21:564–574. doi: 10.1111/j.1750-3639.2011.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solis OE, Mehta RI, Lai A, et al. Rosette-forming glioneuronal tumor: a pineal region case with IDH1 and IDH2 mutation analyses and literature review of 43 cases. J Neurooncol. 2011;102:477–484. doi: 10.1007/s11060-010-0335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byeon SJ, Myung JK, Kim SH, Kim SK, Phi JH, Park SH. Distinct genetic alterations in pediatric glioblastomas. Childs Nerv Syst. 2012;28:1025–1032. doi: 10.1007/s00381-012-1773-1. [DOI] [PubMed] [Google Scholar]
- 45.Juratli TA, Kirsch M, Robel K, et al. IDH mutations as an early and consistent marker in low-grade astrocytomas WHO grade II and their consecutive secondary high-grade gliomas. J Neurooncol. 2012;108:403–410. doi: 10.1007/s11060-012-0844-1. [DOI] [PubMed] [Google Scholar]
- 46.Gorovets D, Kannan K, Shen R, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18:2490–2501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 47.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226:7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lass U, Nümann A, von Eckardstein K, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1-mutation as common tumor-initiating event. PLoS One. 2012;7:e41298. doi: 10.1371/journal.pone.0041298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figarella-Branger D, Bouvier C, de Paula AM, et al. Molecular genetics of adult grade II gliomas: towards a comprehensive tumor classification system. J Neurooncol. 2012;110:205–213. doi: 10.1007/s11060-012-0953-x. [DOI] [PubMed] [Google Scholar]
- 52.Badiali M, Gleize V, Paris S, et al. KIAA1549-BRAF fusions and IDH mutations can coexist in diffuse gliomas of adults. Brain Pathol. 2012;22:841–847. doi: 10.1111/j.1750-3639.2012.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan W, Zhang W, You G, et al. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One. 2012;7:e30339. doi: 10.1371/journal.pone.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.SongTao Q, Lei Y, Si G, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 57.Reitman ZJ, Jin G, Karoly ED, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steenweg ME, Jakobs C, Errami A, et al. An overview of L-2-hydroxyglutarate dehydrogenase gene (L2HGDH) variants: a genotype-phenotype study. Hum Mutat. 2010;31:380–390. doi: 10.1002/humu.21197. [DOI] [PubMed] [Google Scholar]
- 60.Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate–dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pappalardi MB, McNulty DE, Martin JD, et al. Biochemical characterization of human HIF hydroxylases using HIF protein substrates that contain all three hydroxylation sites. Biochem J. 2011;436:363–369. doi: 10.1042/BJ20101201. [DOI] [PubMed] [Google Scholar]
- 63.Jin G, Reitman ZJ, Spasojevic I, et al. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS One. 2011;6:e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 66.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One. 2012;7:e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sasaki M, Knobbe CB, Itsumi M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Zee JA, van Eijck CH, Hop WC, et al. Tumour basement membrane laminin expression predicts outcome following curative resection of pancreatic head cancer. Br J Cancer. 2012;107:1153–1158. doi: 10.1038/bjc.2012.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirashima K, Iyama KI, Baba Y, et al. Differential expression of basement membrane type IV collagen α2 and α6 chains as a prognostic factor in patients with extrahepatic bile duct carcinoma. J Surg Oncol. 2013;107:402–407. doi: 10.1002/jso.23225. [DOI] [PubMed] [Google Scholar]
- 72.Grassian AR, Lin F, Barrett R, et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/mir-200–dependent epithelial-mesenchymal (EMT) transition. J Biol Chem. 2012;287:42180–42194. doi: 10.1074/jbc.M112.417832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benjamin DI, Cravatt BF, Nomura DK. Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab. 2012;16:565–577. doi: 10.1016/j.cmet.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marin-Valencia I, Yang C, Mashimo T, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seltzer MJ, Bennett BD, Joshi AD, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bralten LB, Kloosterhof NK, Balvers R, et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol. 2011;69:455–463. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 77.Bonner MY, Arbiser JL. Targeting NADPH oxidases for the treatment of cancer and inflammation. Cell Mol Life Sci. 2012;69:2435–2442. doi: 10.1007/s00018-012-1017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 79.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 80.Qi ST, Yu L, Lu YT, et al. IDH mutations occur frequently in Chinese glioma patients and predict longer survival but not response to concomitant chemoradiotherapy in anaplastic gliomas. Oncol Rep. 2011;26:1479–1485. doi: 10.3892/or.2011.1428. [DOI] [PubMed] [Google Scholar]
- 81.Kato Y, Jin G, Kuan CT, McLendon RE, Yan H, Bigner DD. A monoclonal antibody IMab-1 specifically recognizes IDH1R132H, the most common glioma-derived mutation. Biochem Biophys Res Commun. 2009;390:547–551. doi: 10.1016/j.bbrc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaneko MK, Tian W, Takano S, et al. Establishment of a novel monoclonal antibody SMab-1 specific for IDH1-R132S mutation. Biochem Biophys Res Commun. 2011;406:608–613. doi: 10.1016/j.bbrc.2011.02.102. [DOI] [PubMed] [Google Scholar]
- 83.Horbinski C, Kofler J, Kelly LM, Murdoch GH, Nikiforova MN. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68:1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 84.Kaneko MK, Morita S, Tsujimoto Y, et al. Establishment of novel monoclonal antibodies KMab-1 and MMab-1 specific for IDH2 mutations. Biochem Biophys Res Commun. 2013;432:40–45. doi: 10.1016/j.bbrc.2013.01.088. [DOI] [PubMed] [Google Scholar]
- 85.Setty P, Hammes J, Rothämel T, et al. A pyrosequencing-based assay for the rapid detection of IDH1 mutations in clinical samples. J Mol Diagn. 2010;12:750–756. doi: 10.2353/jmoldx.2010.090237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sonoda Y, Kumabe T, Nakamura T, et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 2009;100:1996–1998. doi: 10.1111/j.1349-7006.2009.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hartmann C, Hentschel B, Tatagiba M, et al. Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res. 2011;17:4588–4599. doi: 10.1158/1078-0432.CCR-10-3194. [DOI] [PubMed] [Google Scholar]
- 88.Sahm F, Capper D, Pusch S, et al. Detection of 2-hydroxyglutarate in formalin-fixed paraffin-embedded glioma specimens by gas chromatography/mass spectrometry. Brain Pathol. 2012;22:26–31. doi: 10.1111/j.1750-3639.2011.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Capper D, Simon M, Langhans CD, et al. 2-Hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. Int J Cancer. 2012;131:766–768. doi: 10.1002/ijc.26425. [DOI] [PubMed] [Google Scholar]
- 91.Boisselier B, Gállego Pérez-Larraya J, Rossetto M, et al. Detection of IDH1 mutation in the plasma of patients with glioma. Neurology. 2012;79:1693–1698. doi: 10.1212/WNL.0b013e31826e9b0a. [DOI] [PubMed] [Google Scholar]
- 92.Choi C, Ganji SK, DeBerardinis RJ, et al. 2-Hydroxyglutarate detection by magnetic resonance spectroscopy in subjects with IDH-mutated gliomas. Nat Med. 2012;18:624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pope WB, Prins RM, Albert Thomas M, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107:197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lazovic J, Soto H, Piccioni D, et al. Detection of 2-hydroxyglutaric acid in vivo by proton magnetic resonance spectroscopy in U87 glioma cells overexpressing isocitrate dehydrogenase-1 mutation. Neuro Oncol. 2012;14:1465–1472. doi: 10.1093/neuonc/nos258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jin G, Reitman ZJ, Duncan CG, et al. Disruption of wild type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang F, Travins J, Delabarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 98.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 99.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 101.Jha P, Suri V, Sharma V, et al. IDH1 mutations in gliomas: first series from a tertiary care centre in India with comprehensive review of literature. Exp Mol Pathol. 2011;91:385–393. doi: 10.1016/j.yexmp.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 102.Mellai M, Piazzi A, Caldera V, et al. IDH1 and IDH2 mutations, immunohistochemistry and associations in a series of brain tumors. J Neurooncol. 2011;105:345–357. doi: 10.1007/s11060-011-0596-3. [DOI] [PubMed] [Google Scholar]