Abstract
Background
This study aimed to evaluate the prognostic significance of co-polsomy of chromosome 1q and 19p in 1p/19q codeleted oligodendroglial tumors (ODGs).
Methods
In a series of 148 ODGs with 1p/19q deletion, co-polysomy of 1q and 19p was detected by fluorescence in situ hybridization (FISH). Log-rank analysis and Cox regression methods were used to compare Kaplan–Meier plots and identify factors associated with worse prognosis.
Results
There were 104 (70.3%) low-grade ODGs and 44 (29.7%) high-grade ODGs. Co-polysomy was independently associated with shorter progression-free survival and overall survival in 1p/19q codeleted ODGs, irrespective of tumor grades. The odds ratio of without and with co-polysomy was 0.263 (95% confidence interval [CI], 0.089–0.771; P = .015) for progression-free survival and 0.213 (95% CI, 0.060–0.756; P = .017) for overall survival. Subgroup analysis confirmed this trend in both low-grade and high-grade ODGs, although the P value for high-grade ODGs was marginally significant.
Conclusions
Co-polysomy of 1q and 19p could be used as a marker to independently predict worse prognoses and guide individual therapy in 1p/19q codeleted ODGs.
Keywords: 1p/19q co-deletion, co-polysomy, oligodendroglial tumors, prognosis, survival
Gliomas are the most common intracranial tumors, constituting 80% of primary malignant intracranial tumors.1 Although combined-modality therapy and individualized treatment of gliomas have been widely used, therapeutic effect and prognosis are still poor.2 Recently, molecular genetics revealed that a t(1;19)(q10;p10) mediated codeletion of chromosome 1p and 19q and predicted better prognosis in patients with oligodendroglioma (ODG).3 In the detection of 1p/19q codeletion by fluorescence in situ hybridization (FISH) method, polysomy of chromosome 1q and 19p was frequently encountered, which was defined as more than two 1q and 19p signals in gliomas. However, rare studies have reported the prognostic significance of polysomy of 1q and 19p in the context of 1p/19q codeletion in ODGs.4,5 To clarify the above pending questions, we counted the frequency of co-polysomy of chromosome 1q and 19p and then analyzed the relationship between polysomy and prognoses in a series of 148 1p/19q codeleted ODGs.
Materials and Methods
Ethics Statement
A series of 148 patients with ODG with 1p/19q codeletion were surgically treated and pathologically confirmed at Beijing Tiantan Hospital from May 2009 through June 2011. All patients provided written informed consent for the current study, and the clinical study was approved by the Medical Ethics Committee of Capital Medical University.
Pathological Examination
Fresh paraffin-embedded tumor tissue was made into 5-μm slides and stained with hematoxylin and eosin. All the pathological slides were examined morphologically and graded by 3 independent neuropathologists according to the 2007 World Health Organization Classification of Tumors of the Central Nervous System.6 The diagnosis was decided if all 3 pathologists agreed on it. If they could not agree on the final diagnosis, another ≥1 independent neuropathlogist would decide on a diagnosis. The slides were reviewed, and corresponding immunohistochemical staining would be performed if necessary. The final decision was decided by the majority, and at least 3 of them agreed on it.
Detection of 1p/19q Codeletion and Co-polysomy of 1q/19p by FISH Method
The 1p/19q fluorescent probe kit (Vysis) was used for the FISH test as was described previously.7 The assessment and interpretation of FISH results were performed according to guidelines defined by the International Society of Paediatric Oncology Europe Neuroblastoma Pathology and Biology and Bone Marrow Group.8 Tumors with >30% of nuclei showing DNA loss were defined as a tumor with chromosomal loss. The tumor was considered to have co-polysomy of 1q and 19p if 30% of nuclei showed >2 1q and 19p.4
Quality Control for FISH Detection of 1p/19q Codeletion
For each case, a paraffin-embedded tumor block was selected on the basis of tumor content, including the highest-grade component and representation of the predominant morphology of the individual case. Several sections were prepared for FISH. The first and last sections were hematoxylin and eosin stained, regions representing tumor were delineated, and the first section was examined to ensure that it met the standards by which the block was selected. For FISH analysis, the section immediately adjacent to the first hematoxylin and eosin-stained slide was used to minimize the effects of tumor heterogeneity. In the corresponding region rich of representing tumor cells, >100 nonoverlapping nuclei were enumerated per hybridization for each probe.
Some parameters were used for quality control. The percentage of nuclei, in which the number of control signal is less than the number of target signal, is used for quality control to avoid the methodological error. Statistically, the probability of signal loss due to tissue section was the same for both control and target signals if there is no 1p/19q deletion. If the percentage for quality control is >10%, the section is of poor quality and another section will be evaluated for use.
Statistical Analysis
Survival as a function of time was plotted using the Kaplan–Meier method. Log-rank analysis was used to compare Kaplan–Meier plots and identify factors associated with prognosis. The multivariate proportional hazards regression analysis was used to identify independent factors associated with prognosis. This was done after controlling for clinical, operative, and pathological factors that have been shown to be associated with prognosis, including age, Karnofsky performance status (KPS), tumor removal degree, pathology, and adjuvant therapy9 by independent t test or χ2 test between groups. SPSS, version 13.0 (SPSS Inc., Chicago, IL) was used for statistical analysis. Probability value was obtained from 2-sided tests with a statistical significance of P < .05.
Results
Overall Characteristics of Study Population
Table 1 summarized the general information of 148 patients with 1p/19q codeleted ODG who were tested for co-polysomy of 1q/19p. There were 96 male and 52 female patients with a mean age of 42.6 ± 8.8 years (range, 19–67 years). The KPS ranged from 10 to 100; the median KPS was 90.
Table 1.
General information for 148 patients with ODG with 1p/19q codeletion
| Parameter | Values (%) |
|---|---|
| Age (years) | |
| Mean | 42.6 ± 8.8 |
| Range | 19–67 |
| Sex | |
| M | 96 (64.9) |
| F | 52 (35.1) |
| KPS | |
| Median | 90 |
| Range | 10–100 |
| Tumor removal degree | |
| GTR | 76 (51.4) |
| STR | 58 (39.2) |
| PTR | 13 (8.8) |
| N/A | 1 (0.7) |
| Pathology | |
| O | 61 (41.2) |
| OA | 43 (29.1) |
| AO | 19 (12.8) |
| AOA | 25 (16.9) |
| WHO Grade | |
| II | 104 (70.3) |
| III | 44 (29.7) |
| Co-polysomy | |
| Yes | 32 (21.6) |
| No | 107 (72.3) |
| N/A | 9 (6.1) |
| Postoperative chemotherapy | |
| Yes | 125 (84.5) |
| No | 14 (9.5) |
| N/A | 9 (6.1) |
| Postoperative radiotherapy | |
| Yes | 77 (52.0) |
| No | 60 (40.5) |
| N/A | 11 (7.4) |
| Follow-up (months) | |
| Median | 15.5 |
| Range | 0.5–29.5 |
Abbreviations: ODG, oligodendroglial tumor; KPS, Karnofsky performance scale; GTR, gross-total resection; STR, subtotal resection; PTR, partial resection; N/A, not available; O, oligodendroglioma; OA, oligoastrocytoma; AO, anaplastic oligodendroglioma; AOA, anaplastic oligoastrocytoma.
Gross total removal was achieved in 76 (51.4%) patients, subtotal removal in 58 (39.2%) patients, and partial removal in 13 (8.8%) patients.
There were 104 (70.3%) low-grade ODGs, including 61 oligodendrogliomas and 43 oligoastrocytomas. There were 44 (29.7%) high-grade ODGs, including 19 anaplastic oligodendrogliomas and 25 anaplastic oligoastrocytomas.
The frequencies of co-polysomy of 1q and 19p were 21.6% (32/148); 125 (84.5%) patients with ODG received postoperative chemotherapy, and 77 (52.0%) received postoperative radiotherapy.
All patients were followed up for 0.5–29.5 months after surgery, and the median follow-up was 15.5 months.
1q/19p Co-polysomy Independently Predicts Worse Prognoses in 1p/19q Co-deleted ODGs, Irrespective of Tumor Grade (Fig. 1)
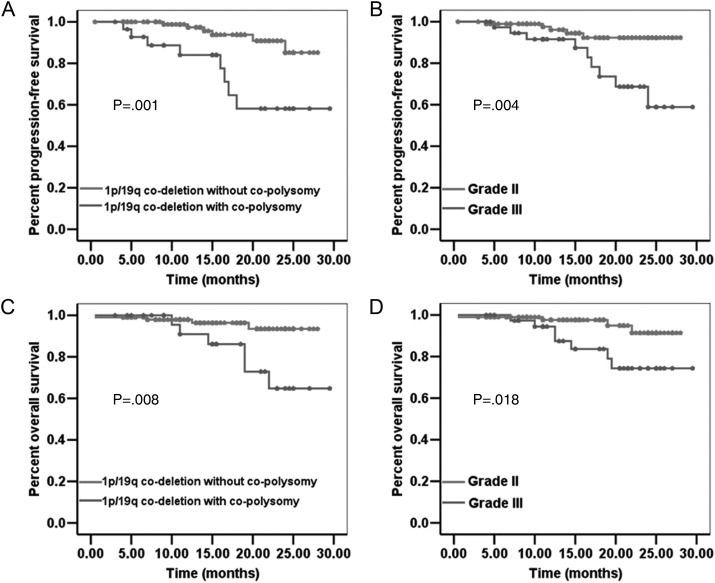
Fig. 1.
Patients with 1p/19q codeleted ODGs with co-polysomy have significantly shorter PFS (A, P = .001) and OS (C, P = .008) than do those without co-polysomy. Patients with 1p/19q codeleted ODGs with grade III have shorter PFS (B, P = .004) and OS (D, P = .018) than do those with grade II.
Patients with 1p/19q co-deleted ODG with co-polysomy had shorter progression-free survival (PFS) than did those without co-polysomy (Fig. 1A; P = .001). The median PFS rates for 1p/19q codeleted ODGs with and without co-polysomy were both unavailable. The 1-year and 2-year PFS rates for 1p/19q codeleted ODG with co-polysomy were 84.0% and 58.2%, respectively. The 1-year and 2-year PFS rates for 1p/19q codeleted ODG without co-polysomy were 97.3% and 85.2%, respectively.
Patients with 1p/19q codeleted ODG with co-polysomy had shorter overall survival (OS) than did those without co-polysomy (Fig. 1C; P = .008). The median OS for 1p/19q codeleted ODGs with and without co-polysomy were unavailable. The 1-year and 2-year OS rates among patients with 1p/19q codeleted ODG with co-polysomy were 90.9% and 64.8%, respectively. The 1-year and 2-year OS rates for 1p/19q co-deleted ODG without co-polysomy were 97.9% and 93.5%, respectively.
Log-rank analysis revealed that both co-polysomy (yes/no) and grade (World Health Organization II/III) were factors significantly associated with PFS and OS (Fig. 1). Thus, these 2 factors were considered in multivariate analysis. Cox regression analysis confirmed that both co-polysomy and higher grade were independent factors for shorter PFS, whereas only co-polysomy was an independent factor for shorter OS in 1p/19q co-deleted ODGs (Table 3). The odds ratio of without and with co-polysomy was 0.263 (95% confidence interval [CI], 0.089–0.771; P = .015) for PFS and 0.213 (95% CI, 0.060–0.756; P = .017) for OS.
Table 3.
Multivariate associations with survival for patients with 1p/19q codeleted ODG
| Multivariate associations with survival | ||
|---|---|---|
| Factors | OR (95% CI) | P value |
| Factors associated with prolonged PFS in 1p/19q co-deleted ODGs | ||
| Without co-polysomy | 0.263 (0.089–0.771) | .015 |
| Grade II | 0.281 (0.092–0.855) | .025 |
| Factors associated with prolonged OS in 1p/19q co-deleted ODGs | ||
| Without co-polysomy | 0.213 (0.060–0.756) | .017 |
| Factors associated with prolonged PFS in low-grade 1p/19q co-deleted ODGs | ||
| Without co-polysomy | 0.138 (0.023–0.824) | .030 |
| Factors associated with prolonged OS in low-grade 1p/19q co-deleted ODGs | ||
| Without co-polysomy | 0.09 (0.009–0.872) | .038 |
Abbreviations: ODG, oligodendroglial tumor; PFS, progression-free survival; OS, overall survival; OR, odds ratio; CI, confidence interval.
1q/19p Co-polysomy in 1p/19q Codeleted Low-Grade ODGs (Fig. 2A and 2B)
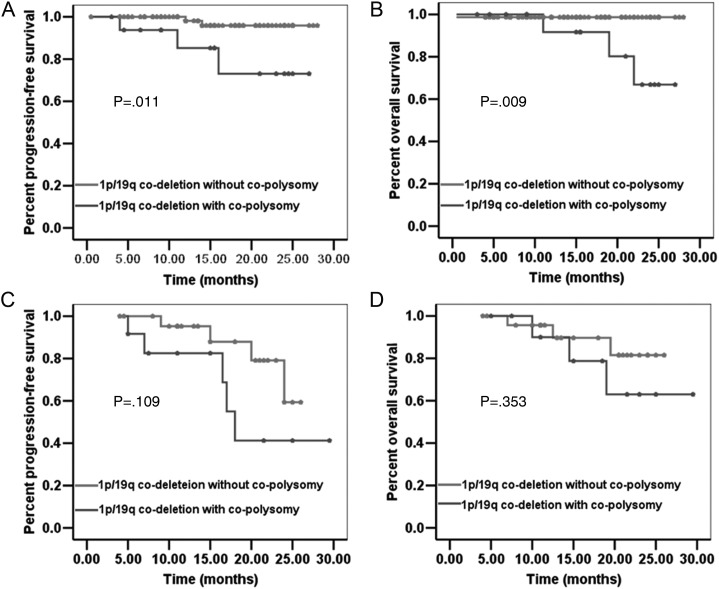
Fig. 2.
The PFS and OS of 1p/19q codeleted low-grade ODGs with co-polysomy are significantly shorter than those of 1p/19q codeleted low-grade ODGs without co-polysomy (A, P = .011 and B, P = .009). The PFS and OS of 1p/19q codeleted high-grade ODGs with co-polysomy show a trend to be shorter than those of 1p/19q codeleted high-grade ODGs without co-polysomy, although the difference is marginally significant (C, P = .109 and D, P = .353).
Patients with low-grade ODGs with concurrent 1p/19q codeletion and co-polysomy had significantly shorter PFS and OS than did those with 1p/19q codeletion but without co-polysomy (P = .011 and P = .009, respectively). There were no significant differences between the 2 groups with regard to the following parameters: age, KPS, pathology, tumor removal degree, postoperative chemotherapy, and postoperative radiotherapy (Table 2).
Table 2.
Comparison of characteristics of patients with ODG with 1p/19q codeletion with and without co-polysomy*
| Factor | Total | 1p/19q codeletion in low-grade ODGs |
P value | Total | 1p/19q codeletion in high-grade ODGs |
P value | ||
|---|---|---|---|---|---|---|---|---|
| With co-polysomy (%) | Without co-polysomy (%) | With co-polysomy (%) | Without co-polysomy (%) | |||||
| Number of tumors | 98 | 18 (18.4) | 80 (81.6) | 41 | 14 (34.1) | 27 (65.9) | ||
| Age (years) | ||||||||
| Mean | 98 | 45.3 ± 10.1 | 42.3 ± 7.9 | .168 | 41 | 40.7 ± 11.9 | 43.5 ± 8.3 | .383 |
| Range | 30–67 | 24–67 | 19–67 | 30–64 | ||||
| KPS | ||||||||
| Median | 98 | 90 | 90 | .188 | 41 | 90 | 85 | .893 |
| Range | 10–100 | 50–100 | 10–90 | 20–100 | ||||
| Tumor removal degree | ||||||||
| GTR | 45 | 6 (33.3) | 39 (48.8) | .448 | 25 | 9 (64.3) | 16 (59.3) | >.05 |
| STR | 42 | 9 (50.0) | 33 (41.3) | 14 | 4 (28.6) | 10 (37.0) | ||
| PTR | 11 | 3 (16.7) | 8 (10.0) | 1 | 0 (0.0) | 1 (3.7) | ||
| N/A | 1 | 1 (7.1) | 0 (0.0) | |||||
| Pathology | ||||||||
| O (AO) | 58 | 11 (19.0) | 47 (81.0) | .854 | 18 | 6 (33.3) | 12 (66.7) | .923 |
| OA (AOA) | 40 | 7 (17.5) | 33 (72.5) | 23 | 8 (34.8) | 15 (65.2) | ||
| Postoperative chemotherapy | ||||||||
| Yes | 84 | 16 (88.9) | 68 (85.0) | .695 | 33 | 12 (85.7) | 21 (77.8) | .522 |
| No | 11 | 1 (5.6) | 10 (12.5) | 3 | 0 (0.0) | 3 (11.1) | ||
| N/A | 3 | 1 (5.6) | 2 (2.5) | 5 | 2 (14.3) | 3 (11.1) | ||
| Postoperative radiotherapy | ||||||||
| Yes | 41 | 7 (38.9) | 34 (42.5) | .823 | 34 | 11 (78.6) | 23 (85.2) | 1.000 |
| No | 53 | 10 (55.6) | 43 (53.8) | 2 | 1 (7.1) | 1 (3.7) | ||
| N/A | 4 | 1 (5.6) | 3 (3.8) | 5 | 2 (14.3) | 3 (11.1) | ||
| Progression | 5 | 3 (16.7) | 2 (2.5) | 9 | 5 (35.7) | 4 (14.8) | ||
| Median PFS (months) | N/A | N/A | .011* | 18.0 | N/A | .109 | ||
| 1-year PFS rate (%) | 85.2 | 98.1 | 82.5 | 95.2 | ||||
| 2-year PFS rate (%) | 73.1 | 95.9 | 41.3 | 59.3 | ||||
| Death | 4 | 3 (16.7) | 1 (1.3) | 6 | 3 (21.4) | 3 (11.1) | ||
| Median OS (months) | N/A | N/A | .009* | N/A | N/A | .353 | ||
| 1-year PFS rate (%) | 91.7 | 98.7 | 90.0 | 95.7 | ||||
| 2-year PFS rate (%) | 66.8 | 98.7 | 63.0 | 81.5 | ||||
*Co-polysomy was not available for 6 low-grade ODGs and 3 high-grade ODGs.
For low-grade 1p/19q codeleted ODGs, Cox regression analysis included patient sex, age (>40 or ≤40 years), pathology (oligodendroglioma/oligoastrocytoma), removal degree (gross total resection or non-gross total resection), co-polysomy status (yes/no), postoperative chemotherapy (yes/no), and postoperative radiotherapy (yes/no). Co-polysomy was confirmed as an independent factor for shorter PFS and OS (Table 3). The odds ratio of without and with co-polysomy was 0.138 (95% CI, 0.023–0.824; P = .030) for PFS and 0.09 (95% CI, 0.009–0.872; P = .038) for OS.
1q/19p Co-polysomy in 1p/19q Codeleted High-Grade ODGs (Fig. 2C and 2D)
The PFS and OS among patients with high-grade ODG with concurrent 1p/19q codeletion and co-polysomy showed a trend to be shorter than those among patients with 1p/19q codeletion but without co-polysomy (Fig. 2C and D), although the difference was not significantly different (P = .109 and P = .353, respectively). There were no significant differences between the 2 groups with regard to the following parameters: age, KPS, pathology, tumor removal degree, postoperative chemotherapy, and postoperative radiotherapy (Table 2).
Discussion
High incidence of 1p and 19q deletion is observed in oligodendroglioma and oligoastrocytoma.10 It has been reported that codeletion of 1p and 19q is associated with longer PFS and longer median survival time, thus representing an independent prognostic factor in anaplastic oligodendroglial tumors (World Health Organization grade III).11–14 Similarly, 1p/19q codeletion also predicts a longer radiographic response to temozolomide and is associated with both superior OS and PFS in low-grade ODG tumors.3,15–17 It was speculated that 1p/19q codeletion was associated with more sensitivity to adjuvant therapies; however, the molecular mechanism was still unclear.
FISH method has been widely used for the deletion of chromosome 1p and 19q in gliomas especially in those with the ODG component. Compared with polymerase chain reaction (PCR), FISH is more sensitive when detecting deletions in specimens of mixed cellularity.18 FISH analysis can detect deletions in tumor cell populations making up as little as 15%–30% of all cells in the specimen compared with a requirement of 60%–90% tumor cell content for loss of heterozygosity PCR studies.19 In this study, detection of chromosome 1p and 19q deletion was routinely tested by FISH method in a series of ODGs. Meanwhile, polysomy of chromosome 1q and 19p was recorded as described in Materials and Methods. This study aimed to determine the significance of co-polysomy for ODGs in the context of 1p/19q codeletion, although polysomy was not an area as hot as 1p/19q codeletion. Thus far, to our knowledge, this is the third study on co-polysomy of 1q and 19p.4,5 In this study, we presented the largest series of 1p/19q codeleted ODGs with co-polysomy by FISH analysis. According to our analyses, we found the prognostic significance of co-polysomy in 1p/19q codeleted ODGs.
Co-polysomy Independently Predicted Shorter Survival in 1p/19q Codeleted ODGs
The most important finding in our study is that co-polysomy was an independent factor for shorter survival among patients with 1p/19q codeleted ODG. In 148 ODGs, log-rank analysis revealed that co-polysomy and higher grade were 2 factors associated with shorter PFS and OS. Cox regression further confirmed both of them as independent factors for shorter PFS and only co-polysomy as an independent factor for shorter OS. Thus, detection of polysomy of 1q and 19p together in 1p/19q codeleted ODGs has its merit in prognosis prediction and guiding individual treatment.
To our knowledge, this is the third study on polysomy of 1q and 19p with the largest series of patients. All the studies on the significance of polysomy of 1q and/or 19p in English literature are listed in Table 4. For the first time in 2009, Snuderl et al. analyzed 64 high-grade ODGs and found that polysomy showed shorter PFS than in those without polysomy among those with 1p/19q codeletion (P = .0048). The trend for OS was similar with PFS, but the difference was not significant because of the smaller cohort.4 For the second time in 2012, Wiens et al. analyzed 84 consecutive ODGs (including 68 low grade and 16 high grade) and found that polysomy of 1q and/or 19p is associated with less favorable clinical outcome (P = .06, P = .09, and P = .03, respectively), regardless of histological tumor grade.5 Our study found that co-polysomy was an independent factor for shorter PFS and OS, irrespective of tumor grade.
Table 4.
Studies on polysomy of chromosome 1q and 19p in ODGs reported in English literature
| Authors | Sample size and pathology | Findings |
|---|---|---|
| Snuderl et al in 2009 (4) | n = 64 (AO) | Polysomy showed shorter PFS than those without polysomy among those with 1p/19q co-deletion (P = .0048). The trend for OS was similar with PFS, but the difference was not significant due to the smaller cohort (P = .303). The Ki-67 labeling index was not associated with polysomy (P = 1.0). |
| Wiens et al in 2012 (5) | n = 84 (68 O and 16 AO) | Polysomy of chromosome 1 and/or 19 is associated with less favorable clinical outcome (P = .06 for 1, P = .09 for 19, and P = .03 for 1 and 19, respectively), regardless of histological tumor grade. |
| Present report | n = 148 (61 O, 19 AO, 43 OA, and 25 AOA) | Co-polysomy was independently associated with shorter PFS and OS in 1p/19q co-deleted ODGs, irrespective of tumor grades. The odds ratio of without and with co-polysomy was 0.263 (95% CI, 0.089–0.771; P = .015) for PFS and 0.213 (95% CI, 0.060–0.756; P = .017) for OS. Subgroup analysis confirmed this trend in both low-grade and high-grade ODGs, although the P-value for high-grade ODGs was marginally significant. |
Abbreviations: ODG, oligodendroglial tumor; O, oligodendrogliomas; AO, anaplastic oligodendrogliomas; OA, oligoastrocytomas; AOA, anaplastic oligoastrocytomas.
Co-polysomy Independently Predicted Shorter Survival in 1p/19q Codeleted Low-Grade ODGs
Co-polysomy was an independent factor associated with shorter PFS and OS among patients with 1p/19q codeleted low-grade ODG; 18.4% harbored co-polysomy of 1q and 19p. To analyze the role of co-polysomy in low-grade ODGs, log-rank analysis and Cox regression were performed in 104 low-grade ODGs with codeletion. Co-polysomy was confirmed as an independent factor for shorter PFS and OS. The odds ratio of without and with co-polysomy was 0.138 (95% CI, 0.023–0.824; P = .030) for PFS and 0.09 (95% CI, 0.009–0.872; P = .038) for OS.
Study Limitation
The primary limitation of this study is the short follow-up for ODGs, especially for high-grade ODGs. This may be the cause for marginal significance for some P values. In spite of these limitations, this study reported some important findings and convincible conclusions.
In conclusion, co-polysomy was independently associated with shorter PFS and OS in 1p/19q codeleted ODGs, irrespective of tumor grades. Subgroup analysis confirmed this trend in both low-grade and high-grade ODGs, although the P value for high-grade ODGs was marginally significant. Co-polysomy of 1q and 19p could be used as a marker to independently predict worse prognoses and guide individual treatment in 1p/19q codeleted ODGs.
Funding
This work was supported by Beijing Natural Science Foundation (7122061).
Conflict of interest statement. None declared.
Acknowledgments
We acknowledge financial support by Beijing Natural Science Foundation (7122061).
References
- 1.Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. 1–516. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19) (q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 4.Snuderl M, Eichler AF, Ligon KL, et al. Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res. 2009;15:6430–6437. doi: 10.1158/1078-0432.CCR-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiens AL, Cheng L, Bertsch EC, Johnson KA, Zhang S, Hattab EM. Polysomy of chromosomes 1 and/or 19 is common and associated with less favorable clinical outcome in oligodendrogliomas: fluorescent in situ hybridization analysis of 84 consecutive cases. J Neuropathol Exp Neurol. 2012;71:618–624. doi: 10.1097/NEN.0b013e31825b5f7a. [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren X, Cui X, Lin S, et al. Co-deletion of chromosome 1p/19q and IDH1/2 mutation in glioma subsets of brain tumors in Chinese patients. PLoS One. 2012;7:e32764. doi: 10.1371/journal.pone.0032764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros PF, Ambros IM. Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol. 2001;37:492–504. doi: 10.1002/mpo.1242. [DOI] [PubMed] [Google Scholar]
- 9.Chaichana KL, Chaichana KK, Olivi A, et al. Surgical outcomes for older patients with glioblastoma multiforme: preoperative factors associated with decreased survival. Clinical article. J Neurosurg. 2011;114:587–594. doi: 10.3171/2010.8.JNS1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraus JA, Koopmann J, Kaskel P, et al. Shared allelic losses on chromosomes 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol. 1995;54:91–95. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- 11.van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006;24:2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 12.Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24:2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 13.Kros JM, Gorlia T, Kouwenhoven MC, et al. Panel review of anaplastic oligodendroglioma from European Organization For Research and Treatment of Cancer Trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J Neuropathol Exp Neurol. 2007;66:545–551. doi: 10.1097/01.jnen.0000263869.84188.72. [DOI] [PubMed] [Google Scholar]
- 14.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 15.Kaloshi G, Benouaich-Amiel A, Diakite F, et al. Temozolomide for low-grade gliomas: predictive impact of 1p/19q loss on response and outcome. Neurology. 2007;68:1831–1836. doi: 10.1212/01.wnl.0000262034.26310.a2. [DOI] [PubMed] [Google Scholar]
- 16.Kujas M, Lejeune J, Benouaich-Amiel A, et al. Chromosome 1p loss: a favorable prognostic factor in low-grade gliomas. Ann Neurol. 2005;58:322–326. doi: 10.1002/ana.20543. [DOI] [PubMed] [Google Scholar]
- 17.Hoang-Xuan K, Capelle L, Kujas M, et al. Temozolomide as initial treatment for adults with low-grade oligodendrogliomas or oligoastrocytomas and correlation with chromosome 1p deletions. J Clin Oncol. 2004;22:3133–3138. doi: 10.1200/JCO.2004.10.169. [DOI] [PubMed] [Google Scholar]
- 18.Scheie D, Andresen PA, Cvancarova M, et al. Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am J Surg Pathol. 2006;30:828–837. doi: 10.1097/01.pas.0000213250.44822.2e. [DOI] [PubMed] [Google Scholar]
- 19.Fuller CE, Perry A. Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol. 2002;12:67–86. doi: 10.1111/j.1750-3639.2002.tb00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]