1-Introduction
Proton magnetic resonance spectroscopy (MRS) appeals to many clinicians and scientists as the application in the clinical setting can increase the specificity of magnetic resonance imaging (MRI) when implemented with appropriate questions for MRS to answer. However, enthusiasm for application of this technique is often diminished due to the complexities of data acquisition, processing, and interpretation. The original expectation of MRS included a concept of "disease specificity", meaning MRS would be a technique having distinct patterns for specific disease processes. Unfortunately, only a few disease processes have distinct MRS profiles unique to a particular disorder. Instead, MRS typically reveals the metabolic status of the region sampled. In the brain, MRS provides information content on neuronal/axonal viability, energetics of the cellular structures, and status of the cellular membranes. Yet, by examining the composite magnetic resonance spectrum, a pattern of disease involvement at a molecular level can compliment an imaging examination.
The objective of this article is to provide descriptive concepts of the technique and its application in vivo for a variety of patient populations. Hopefully, when appropriately incorporating MRS into the neuroradiological evaluation, this technique will produce relevant information to radiologists and clinicians for their understanding of adult and pediatric neurologically based disease processes.
2-Technical Concerns
For the reader previously unexposed to the MRS technique, many articles within the literature and textbooks describe the origin, mathematical approaches and physics of MRS acquisition in the brain.1–4 Conventional MRI systems with 1.5 and 3.0 Tesla (T) superconducting magnets offer suitable magnetic field strengths for performing proton MRS of the brain clinically. For the practicing radiologist familiar with MRI, there are several technical properties which must be especially considered for MRS. These include the concepts of chemical shift, spectral dispersion associated with magnetic field strength, shimming, signal suppression, combination schemes for signal processing from phased array coils, sequence approach and localization sequences.
2.1-Chemical Shift
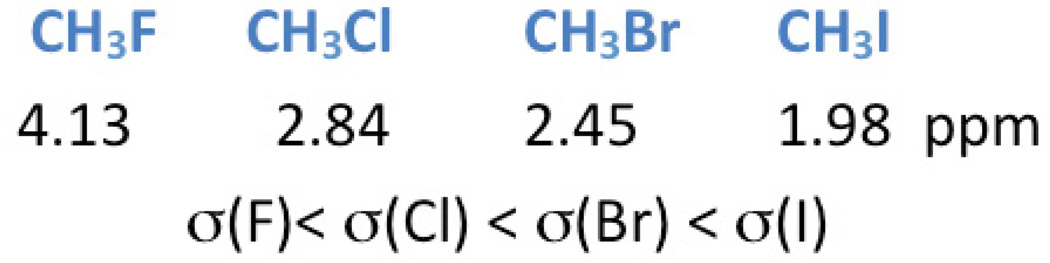
Spectroscopy uses signal intensity, line width and position to display information from molecules of interest. It remains important to note that multiple chemical regions of the same molecule may contribute distinct signals to the spectrum. Typically, these “molecules of interest” or “neurochemicals” from the human brain are referred to as “metabolites”. Proton MRS plots hydrogen atom (proton) metabolite signal intensity versus an observation frequency. Recalling from organic chemistry, the protons from the backbone components of the molecule (primarily carbon atoms) can produce so called “peaks” or “resonances” when in the MR environment with signal intensity proportional to the relative number of protons. The “position” of the metabolite peaks on the x-axis reflects the local chemical and magnetic environment of the molecule. Shielding factors influence the position of the peak (Fig.1). The position is described by employing the parts-per-million (ppm) scale. As locations in Hertz (Hz) would change as field strengths vary, a dimensionless unit was necessary to normalize field strengths. The chemical shift for a given peak location is calculated by dividing the difference in frequency of two peaks (with one peak being defined as the reference) by the operating frequency of the MR scanner. This scale refers to the fact that the magnetic field strength is on the order of 106 or a million Hz. This allows comparison of a peak location found on the spectrum obtained on a 1.5T scanner comparable to that found at that location on a different field strength scanner. Examples of this property include the methyl (CH3) resonance on the acetyl group of N-acetyl aspartate that appears at 2.0 ppm and the creatine N-methyl resonance that always appears at 3.0 ppm, regardless if measured on a 1.0 T, 1.5T, 3T or a 4T scanner. In general, the locations of the metabolites are stable, as the brain pH does not change sufficiently to result in a change in peak assignment.
Figure 1.
The influence of the environment on the methyl (CH3−) group location due to the shielding factor (σ) of the adjacent atom (in this example, fluoride (F), chloride (Cl), bromide (Br) and iodide (I)) or groups of atoms. The adjacent atom influences the position (chemical shift value) on the x-axis in ppm.
Metabolites with a singlet peak, such as those with N-acetyl aspartate (NAA) and creatine (including phosphocreatine Cr), corresponding to a methyl group do not change chemical shift assignment. However, metabolites with adjacent methylene (CH2) and methine (CH) elements may produce a slightly different signal position and appearance at different field strengths; however, this does not occur from a pH change. The signal from methylene and methine groups may split peaks into multiplet patterns. The local chemical and magnetic environment influences the appearance of the peaks and reflects local coupling constants between protons within the molecule. Myo-inositol (mI), glutamate, glutamine, glucose, and aspartate are a few of the molecules with coupled methylene and methine spin systems visible on clinical proton MRS. For the purpose of this discussion, the only routinely reported metabolite with distinct alterations in its assignment and spectral appearance due to field strength is myo-inositol. For MRS performed at 1.5 T, the mI peak normally is distinct with four of the molecule’s methine protons magnetically indistinguishable, thereby co-resonating at the same location (3.57 ppm). However, increased spectral dispersion (discussed further below) inherent at higher field strengths now produces two distinct peaks (at 3T, 3.55 and 3.61 ppm) for the four protons, effectively reducing the signal intensity by half. Normal mI levels visually appear lower at higher field strengths such as 3T in comparison to 1.5T. While some reports have found improved detection of mI at high field strength arising from increased signal to noise ratio, it may be problematic depending on the acquisition conditions.5 It is important to also note that a short echo approach (i.e. echo time (TE) ≤35 milliseconds (msec)) must be employed due to the relatively fast relaxation rate in order to detect mI.
2.2-Benefits and Challenges at Fields of 3T and Higher: Relaxation times, signal to noise ratio, spectral dispersion
The transition from 1.5 T to 3T or higher field strengths illustrates some of the fundamental principals of MRS, by perturbing properties such as relaxation rate, signal-to-noise ratio (SNR), and spectral dispersion. Just as with MRI, the relaxation properties influence the metabolite signal appearance. T1 relaxation reflects the time it takes for the perturbed magnetization to return to equilibrium. T2 relaxation represents the time it takes for dephasing or loss of coherence within the applied field. For the metabolites, T1 relaxation rates lengthen slightly while T2 shortens upon moving to fields of 3T or higher compared with 1.5T. For metabolites (N-acetyl aspartate (NAA), creatine (Cr) and cholines (Cho)) the longitudinal relaxation T1 times approximate 1.2–1.4 seconds at 1.5T and increase slightly to 1.2–1.6 seconds at 4T, with Cho the shortest.6–9 These values impact the repetition time (TR) necessary to conduct the MRS experiment. At least five times the longest T1 value is needed to obtain a fully relaxed experimental condition. For practical purposes, most clinical MRS sites employ TR times on the order 2000 msec to balance the signal afforded from decaying metabolites and beginning a new excitation. For transverse relaxation, T2, several common metabolites have values on the order of 270–480 milliseconds (msec) at 1.5T and 150–210 msec at 3T, 140–185 msec at 4T, with Cr values as the shortest.7, 9 These three primary metabolites will be observed at long echo times greater than 135 msec. Other metabolites generally have much shorter T2 values, thus requiring echo times (TE) on the order of 35 msec or less. (These will be discussed later in the section on the specific metabolites.) It remains important to note that the protocols employed must consider the relaxation properties of the metabolites and how the field strength influences them.
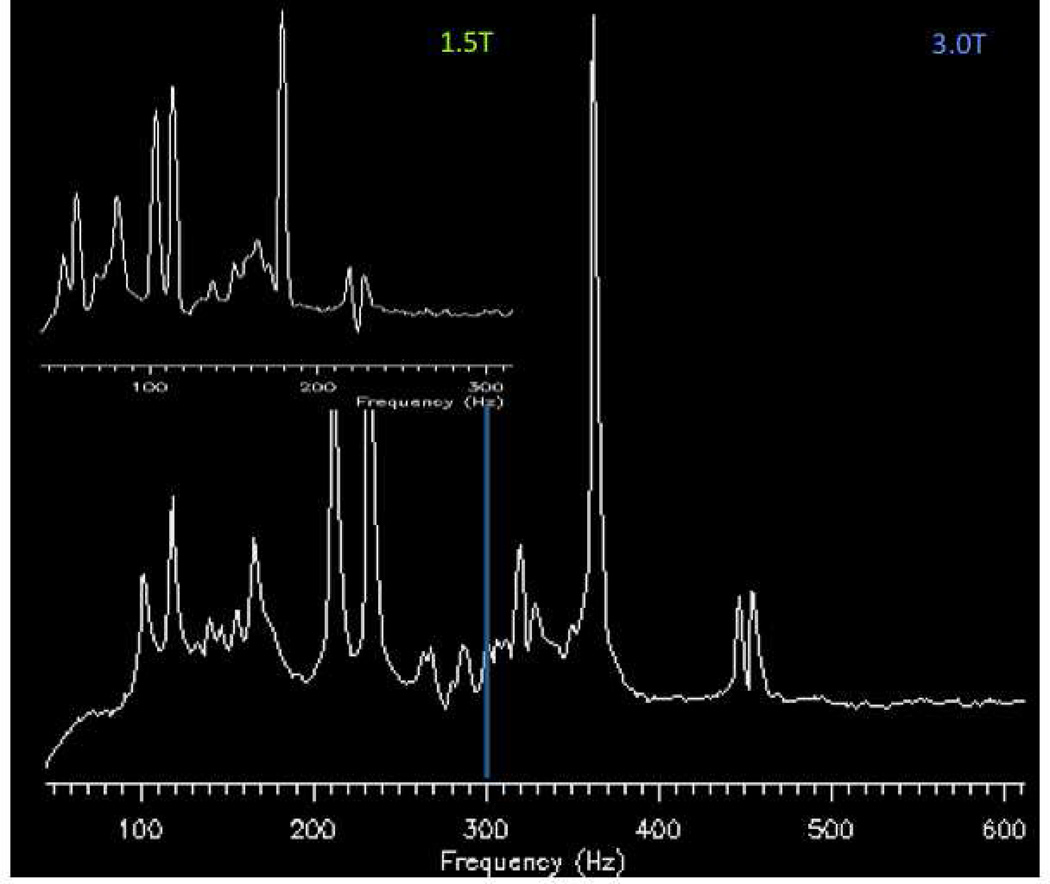
Acquiring proton MRS data at 3T compared with 1.5T should provide a significant benefit with an increase in signal to noise ratio (SNR), which can then be applied for increased speed of acquisition (temporal resolution), spatial resolution (smaller voxel sizes with equivalent SNR to lower field) or some hybrid of speed and voxel size. However, this increase has been actually found to be about 20% for short echo (TE 20 ms) and approaching 100% for long echo single voxel proton MRS.9 At short echo times, it appears that the peaks acquired at 3T are wider than those at 1.5T. With any increase in magnetic field strength, there is a decrease in metabolite T2* relaxation times resulting in increased metabolite line widths.10 It is important to mention that other post-processing factors also influence the overall appearance of the peaks (i.e. spectral apodization as known as line broadening) (Fig.2). The improved resolution afforded from increased field strength is accomplished with increased spectral dispersion. Simply stated, the spectrum is dispersed or spread apart as the frequency between peaks is greater at higher field strength (Fig.3). The ability to distinguish where one peak begins and ends is improved. For instance, the fat and water peaks are separated by 220 Hz at 1.5T but 440 Hz at 3T with the creatine and choline peaks appearing further apart at 3T than at 1.5T.
Figure 2.
Illustration of field strength effects on spectral dispersion. Both spectra were obtained using a water based phantom with lactate, NAA, glutamate (Glu), Cr, Cho, and mI included with the water location set at 0 Hertz. Insert, a spectrum obtained at 1.5T demonstrates the metabolites within the spectrum appear from 100 to 300 Hz. For the spectrum below obtained at 3T, metabolites within the spectrum appear from 100 to 600 Hz.
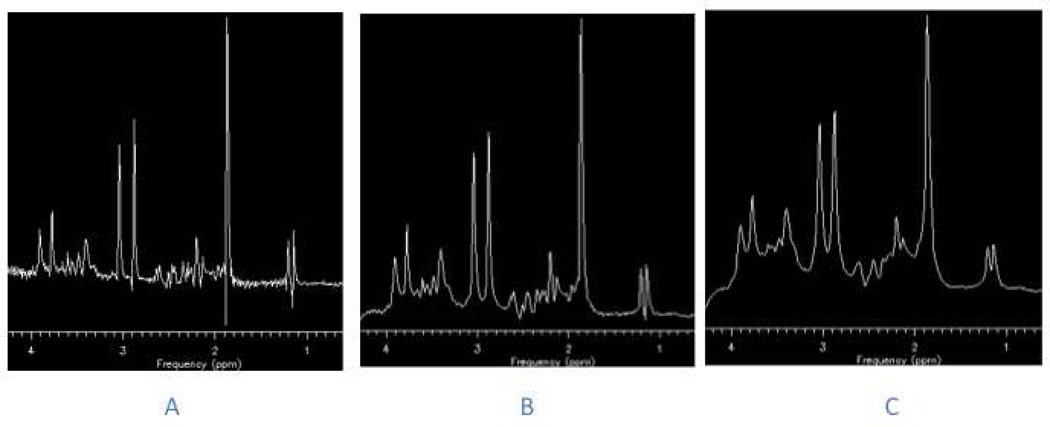
Figure 3.
The effects of line broadening (also referred to as spectral apodization) on a proton spectrum obtained using a water-based phantom with lactate, NAA, Glu, Cr, Cho, and mI included. A) No line broadening; B) minimal line broadening applied and C) excessive line broadening applied to the spectra. Note in C) the distortion of the baseline.
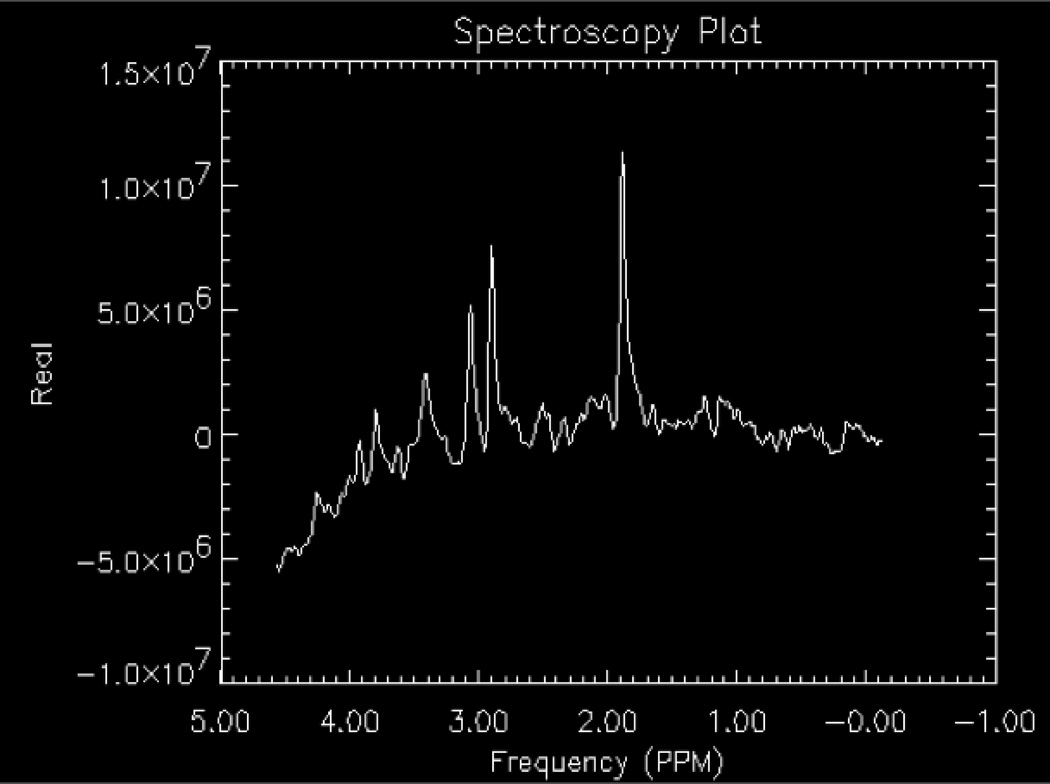
2.3-Shimming
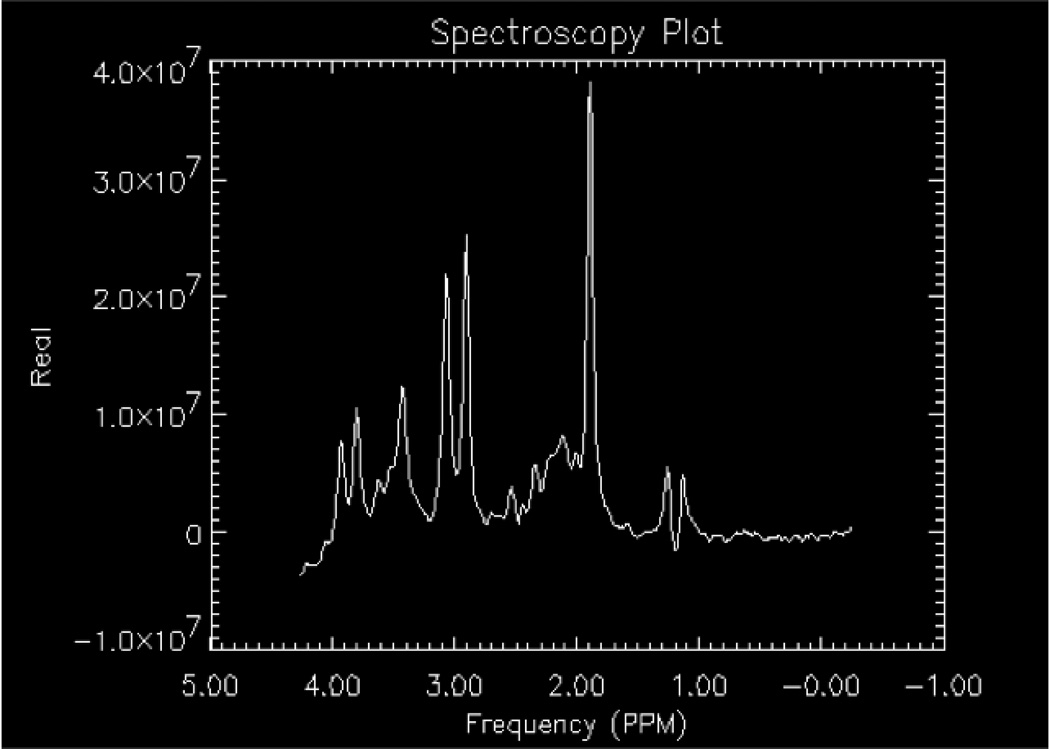
Shimming is the process of homogenizing the magnetic field by applying direct current offsets to gradient coils (usually found as first order shim coils on clinical MRI systems, higher order shims on research MRI systems) via automated software packages provided by the system manufacturers or research groups (an example is FASTMAP11). Shimming can be conducted on the raw time domain signal or the frequency (post-Fourier transformed) signal. Shimming is performed to enhance the sensitivity and the resolution of the metabolite signal by narrowing the peak widths and increasing the SNR (Figs.4 a–b). Shimming also allows for improved water suppression, as a narrower water peak is more easily nulled as the center frequency can be optimized. Single voxel spectra are less susceptible to the effects of large variations in magnetic field inhomogeneity. Thus, it is easier to shim on a relatively small, 8 cm3 volume compared to an entire region, slice or volume. Regions of interest in spectroscopic imaging or multivoxel acquisitions are more difficult to shim, as distortions in the magnetic field arise from tissue-air, tissue-bone, tissue-cerebrospinal fluid (CSF), etc. interfaces have large magnetic susceptibility differences as the tissues are magnetized differently. A compromise in homogeneity is inherent across the volume of interest for spectroscopic imaging as the shimming algorithm balances the need for higher shim settings around the sinus and frontal lobes compared to regions in the cerebrum away from ventricles, bone, etc.
Figure 4.
The effects of shimming on a proton spectrum obtained using a water-based phantom with lactate, NAA, Glu, Cr, Cho, and mI included. A) Optimized shimming; B) Distorted shimming. Note the loss of baseline resolution for creatine (3.0 ppm) and choline (3.2 ppm).
2.4-Suppression
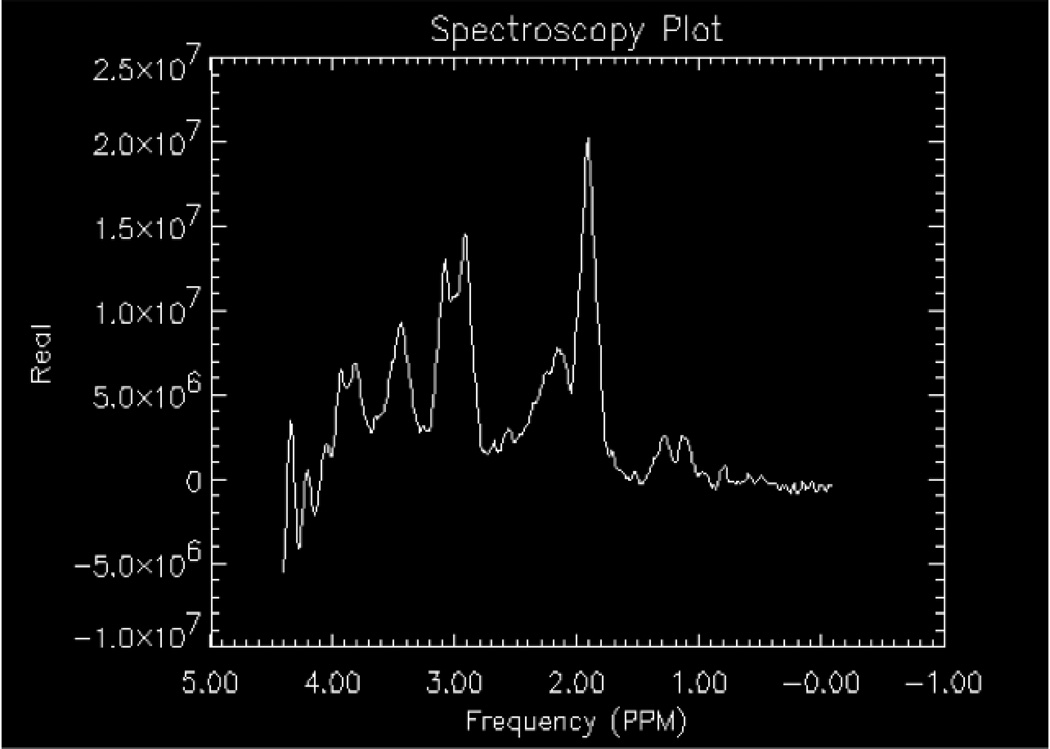
Briefly, proton MRS requires suppression of signals from water and fat. Water has a concentration level on the order of 80 Molar (M) while most metabolites of interest are at the 1–10 millimolar (mM) level. Water suppression is implemented using pulse sequences such as chemically selective saturation (CHESS12), WET13, VAPOR14, etc. The most commonly implemented sequence on clinical MR scanners, CHESS, has three narrow, frequency selective pulses applied along with a dephasing gradient to suppress the water (Fig.5). For fat, signals can be nulled with the application of a sequence with inversion pulses or simply avoided with placement of a volume selective localization pulse sequence (described in the next section) of the region of interest. To further eliminate signal from scalp and sinuses, outer volume suppression (OVS) employs the use of very selective suppression (VSS) pulses.15–17 The OVS are graphically prescribed using the imaging sequences to guide locations and angles around the scalp by the technologist. Although the VSS pulses can be used with body coil excitation, they have relatively low radiofrequency peak power. These pulses have relatively large bandwidths and sharp transition bands, which minimize chemical shift errors as the edges of the selected volume are better defined thus minimizing chemical shift misregistration. Also known as chemical shift displacement artifact, misregistration occurs when the signal is excited from metabolites which do not originate from exactly the same volume. This arises due to the variation in chemical shift within a spectrum, as the metabolites on one end of the spectrum experience different excitation profiles from the other. The localization pulse sequence selection strongly influences this artifact with a stimulated echo based approach minimizing displacement.
Figure 5.
The effects of water suppression on a proton spectrum obtained using a water-based phantom with lactate, NAA, Glu, Cr, Cho, and mI included. A) Under-suppressed, non-optimized spectrum; B) Over-suppressed spectrum with over-rotation of the third CHESS pulse.
2.5-Approaches and localization
The easiest clinical approach for proton spectroscopic analysis is known as single volume element (voxel) proton MR spectroscopy (SVS). In its basic form on clinical MRI scanners, the SVS technique uses either a spin echo (point resolved spectroscopy-PRESS) or stimulated echo (stimulated echo acquisition mode- STEAM) based volume selective localization pulse sequence providing a cubic-based volume of interest typically 4–8 cc in volume. Crusher field gradient pulses are applied to remove signals (free induction decays from a single pulse, spin echoes generated from two pulses) arising from outside of the voxel. As the name implies, SVS is limited to a single acquisition in a given region. While the SVS technique does not interrogate multiple regions of interest simultaneously, it does afford relatively good signal to noise and homogenous peaks due to a limited region of interest for shimming within a few minutes of scan time. “Multivoxel” or “magnetic resonance spectroscopic imaging (MRSI)” or “spectroscopic imaging” generates spectra from a larger number of usually smaller sized voxels with broader anatomical coverage providing a choice of single slices, multiple slices in multiple dimensions and orientations. Volumes of interest are much larger and are also usually obtained using PRESS localization. The MRSI spectra can be transformed into color maps yielding metabolite images. Historically, long acquisition times, complicated post-processing, poor homogeneity across the regions of interest sampled, partial volume effects with signal contamination from outside a voxel, and relatively low resolution (upon comparison with MR imaging) for metabolite mapping were significant limitations for the early implementation of MRSI. Recent advances provide improved accuracy in localization, speed and resolution for spectroscopic imaging. The technical development of fast spectroscopic imaging continues to focus on implementing k-space sampling strategies (ellipsoid k-space encoding, echo planar spectroscopic imaging (EPSI), spiral spectroscopic imaging, parallel imaging reconstruction such as SENSE) for spectroscopic acquisition without artifacts from eddy currents, especially for short echo times, and spectral aliasing effects in spectral fitting due to limitations in gradient rise times.2
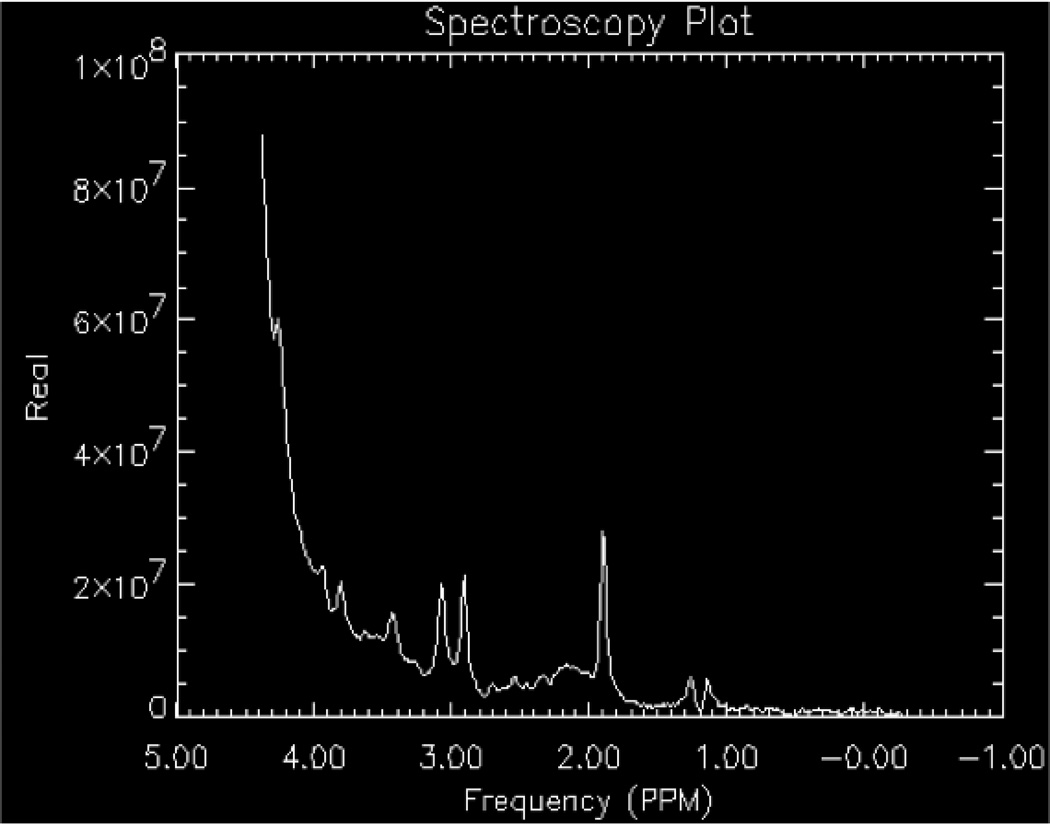
During the prescan, several steps are performed to optimize the scanner including: identifying the center of the water frequency, shimming the region of interest, setting the power for applying the pulses in the localization and suppression sequences. If power levels for the pulses are not properly set, the voxel or region of interest sampled will not match what was graphically prescribed nor will it be properly water suppressed (Fig.6).
Figure 6.
The effects of power scaling on a proton spectrum obtained using a water-based phantom with lactate, NAA, Glu, Cr, Cho, and mI included. The optimization of pulse power during prescan optimizes localization and water suppression. When values are not optimized, the voxel location can be distorted. Two window scalings are displayed in this figure.
Another challenge is devising the optimal methods for combining metabolite signals from multiple channels afforded from parallel techniques while preserving sensitive phase information. Spectra from each coil can be combined with weighting factors such as deriving from the first point of the unsuppressed water free induction decay signal from each coil or other schemes such as SENSE sensitivity encoding for fast MRI18 and also nonparametric, semiparametric and parametric processing tools with available prior knowledge.19
3-Post processing
In clinical settings, metabolite ratios are often employed both for SVS or MRSI rather than obtaining absolute metabolite concentrations due to variations in magnetic field homogeneity, radiofrequency (RF) coil homogeneity and patient loading, susceptibility of tissues, composition of tissues and changes in the receiver gain which are not easily accounted for given the time constraints of imaging a patient in a typical clinical MRI scanner. Often the internal creatine signal is employed as the reference metabolite, as both creatine and phosphocreatine are sampled on proton MRS. While it is recognized that many pathologies (neoplasms, infarcts and metabolic diseases) may have alterations in the creatine and phosphocreatine concentration, it is among the primary three metabolites (NAA, Cr and Cho) the most consistent for the majority of conditions.
Each MRI manufacturer supplies software on the scanner or workstation which employs various algorithms to treat the raw data with zero-filling, line broadening, eddy current corrections, Fourier transformation, phasing, baseline correction, peak fitting, intensity and area approximations. These software packages are either fully automated with no selection of parameters or semi-automated requiring a technologist to select a post-processing protocol where parameters can be modified. These software packages are acceptable post-processing for clinical MRS within an institution or for comparison with similar scanners. However, if more rigorous determinations of concentration are necessary, there are other widely recognized software packages that work with data from the major clinical MRI vendors. Linear combination of modeled spectra20 (LCModel) is a commercially available software package that compares a library of model spectra with the user’s proton MRS data in frequency domain (meaning data are analyzed after Fourier transformation). The model spectra in the LCModel library are obtained from either in vitro solutions of individual metabolites using the same field strength, pulse sequence and other general parameters or via computer simulations.21 This software allows for the entire spectrum to be modeled simultaneously. Vendor software usually performs fitting on individual peaks independent of the relationship with other peaks from the same molecule within the spectrum. With the known concentrations from the model metabolite solutions, one can think of the LCModel software as mixing and matching various combinations of the pure metabolite spectra to fit the user’s spectral data. This software allows concentration estimates to be generated almost instantaneously, but does require extra off-line processing with extracting the raw spectral data and exporting to a separate workstation, as the software is not usually installed on the scanner.
While LCModel can fit short and long echo spectra, a freely downloadable software package known as jMRUI can provide spectral processing of long echo proton MRS data in the time domain (meaning data are decay signals measured in seconds prior to Fourier transformation).22, 23 With jMRUI, user’s data are fitted using prior knowledge of frequencies, line widths, coupling patterns, etc. The software allows editing of unwanted signals, such as artifacts or water. LCModel and jMRUI not only provide signal intensity estimates, but also a measure of the quality of the fitting with the Cramer-Rao lower bound as an error estimate. While these software packages are improvements over vendor-supplied software, they do not fully obtain “absolute” concentrations. Users often report the levels of a metabolite in “institutional units”. This could mean the output from LCModel untouched or additional corrections such as CSF correction, relaxation rate corrections, tissue composition, etc. The literature provides details about performing additional corrections for proton MRS quantification.24–26 The author would like to remind the reader not to be discouraged if only vendor supplied software is available. Qualitative and semi-quantitative interpretation of MRS is usually sufficient to support a diagnosis. On the other hand, monitoring certain therapeutic responses requires a more rigorous, research style of quantitation.
4-Interpretation
The interpretation of an MR spectrum primarily arises from variations in chemical ratios and/or concentrations, which alter peak area for the endogenous metabolites, the appearance of pathologic metabolites (i.e., lactate, alanine, etc.) and exogenous compounds (i.e., propanediol, mannitol) if administered.
The three main metabolite entities, NAA, Cr and Cho, are commonly observed independent of TE and standard localization technique. The key metabolites observed with proton MRS are listed according to their spectral locations (relative to water located at 4.7 ppm) with their known roles briefly described further below.
THE NAA RESONANCE
N-acetyl aspartate and N-acetyl-aspartyl-glutamate at 2.0 ppm. N-acetyl aspartate comprises a majority of the NAA resonance neurospectroscopy. Regional variations of NAA and N-acetyl-aspartyl-glutamate (NAAG), the other primary component of the resonance have been reported.27 NAA is second only to glutamate as the most abundant free amino acid in adult brain.28 Although discovered by Tallan in 1956, the function of this amino acid is not fully known.29, 30 Studies in the developing rat brain show that NAA is synthesized in the mitochondria from aspartate and acetyl Co-A.31 After transport across the mitochondrial membrane, it is cleaved in the cytosol by aspartoacylase into aspartate and acetate.32 This suggests NAA may function as a transporter of acetyl groups across the mitochondrial membrane for lipogenesis during development.33 However, NAA is almost exclusively localized to neurons. It is found in cell cultures of oligodendroglia progenitors but absent from mature glial cells, CSF and blood.34, 35
Studies36 indicate other possible roles for NAA:
NAA may operate as neuronal osmolyte serving as a co-transport substrate for a molecular water pump removing excess water from neurons.
NAA plays a role in neuronal energy metabolism of mitochondria and specific brain fatty acids, respectively.
NAA provides a reservoir for glutamate.37
NAA serves as a substrate for NAAG biosynthesis.
NAAG, a dipeptide derivative of NAA and glutamate, is the most abundant brain peptide. With a concentration of approximately 1 mM, NAAG is found in neurons, oligodendrocytes and microglia. NAAG signals astrocytes about the state of neurostimulation of the neurons changing requirements for vascular energy supplies and metabolic waste removal.
The NAA resonance is widely regarded as a marker for neuronal injury and death.38 A majority of pathological conditions and diseases demonstrate reduced NAA. A leukodystrophy in which NAA is elevated is Canavan´s disease. NAA accumulation in the mitochondria arises when a genetic defect affects aspartoacylase, which is responsible for transporting NAA into the cytosol. It may also be elevated in Pelizaeus Merzbacher disease, Salla disease and hypermetabolic conditions. There is one reported incidence of a patient without brain NAA.39
THE Cr RESONANCE
Creatine and phosphocreatine appear at 3.0 and 3.9 ppm. For the interested reader, Wyss and Kaddurah-Daouk40 published a comprehensive review of creatine and creatinine metabolism. Creatine and phosphocreatine are compounds involved in the regulation of cellular energy metabolism. Phosphocreatine serves as a reserve for high-energy phosphates in the cytosol of muscle and neurons and buffers cellular ATP/ADP reservoirs.41 The enzyme, creatine kinase, converts creatine to phosphocreatine using ATP. Although regarded as an internal standard, it has been demonstrated in certain pathologies such as stroke, tumor and head injury to be reduced as well as absent in creatine deficiency syndromes.
THE Cho RESONANCE
Choline compounds: choline, acetylcholine, phosphocholine, cytidine diphosphate choline and glycerophosphocholine composite appear at 3.2 ppm. MRS detects the trimethylammonium residues of choline and mobile choline-containing compounds. Changes in the Cho level are inherently linked to membrane biochemistry. Choline is regarded as a product of myelin breakdown. Cell proliferation associated with tumor growth is responsible for the elevation in choline observed by proton MRS. Mobile choline-containing compounds contribute to the choline signal. These primarily include the intracellular pools of the membrane precursor phosphocholine (PC), membrane breakdown product glycerol (GPC) and a small portion (5%) of free choline for an approximate total observable choline concentration approximating 1–2 mM. While phosphatyidylcholine is a major membrane constituent produced in all cells, it does not contribute significantly to the observable Cho signal. For white matter diseases, elevations of the resonance reflect the precursors of myelin synthesis as well as the degradation products upon myelin degradation and/or destruction. Increased choline levels observed in tumors arise from increased cellular density and proliferation of membrane phospholipids.42
On short echo MRS (TE 35 msec or less), elevations of lipids, macromolecules, neurotransmitters, myo-inositol and lactate can be appreciated in vivo as these metabolites have relatively short T2 values at commonly encountered field strengths (1.5, 3T, 4T).
THE mI RESONANCE
Myo-inositol, myo-inositol-monophosphate and glycine appear at 3.5 ppm. Myo-inositol is important because it may relate to intracellular sodium content and glial activation. Changes in mI levels have been correlated with osmolarity conditions in the brain.43 The myo-inositol (mI) resonance at 3.5 ppm is largely comprised of myo-inositol (5 mM concentration in the brain) along with myo-inositol-monophosphate and the α-protons of glycine.44 In the brain, myo-inositol is also believed to be primarily located in glial cells and absent from neurons.34 Elevations of mI may be attributed to gliosis and reactive astrocytosis in certain pathologies.
THE amino acid (AA) PEAKS
Glutamine (Gln), glutamate (Glu), gamma aminobutyric acid (GABA), and aspartate form a composite set of peaks between 2.2 and 2.6 ppm with additional components at 3.6 ppm. In addition, glucose (Glc) has peaks at 3.43 and 3.80 ppm. Glu is a neurotransmitter and is the most abundant amino acid in the human brain. Gln is found primarily in cerebral astrocytes. Glutamine is the primary derivative for glutamate. Increases in the composite of glutamate and glutamine (referred to as GLX) are indications of destructive processes. Glc dominates the fuel supply for brain in accordance with blood flow. Increases in glucose concentration have been reported in cases of diabetes mellitus and hypoxic encephalopathy. GABA is an important inhibitory neurotransmitter derived from Glu via decarboxylation with its ultimate source considered Gln.
THE LIPX PEAKS
Lipids have broad multiple peaks at 0.8 ppm and 1.3 ppm which include acetate and macromolecular proteins. Macromolecules have multiple peaks centered at 0.9, 2.05 and 3.0 ppm. Lipids are shown to be elevated as well as lactate and alanine in a variety of tumors. The significance of the macromolecule peaks is less certain, but changes in the levels are associated with stroke, MS and tumors.45 Membrane changes may be reflected by alterations in lipid levels. In neonatal rat macromolecule peaks were present before myelination occurred. In the pediatric brain, regions sampled prior to myelination demonstrate elevated lipid signals.
THE LACTATE RESONANCE is a doublet at 1.3 ppm. The lactate resonance, demonstrated in proton MR spectroscopy, represents the endpoint of anaerobic glycolysis. In pathologies such as stroke, high-grade tumors and abscess, this finding is consistent with our understanding of each of the biochemical processes taking place in the body. However, the presence of lactate in low-grade tumors without regions of necrosis and other metabolic disorders is explained as alterations in metabolism to provide alternative energetic pathways. It is sometimes necessary to differentiate lactate from lipid/macromolecule peaks in short echo MRS studies. The interaction of the lactate methyl and methine proton peaks (occurs at 7 Hertz), referred to as J-coupling, provides for a distinct double-peak (doublet) pattern and location indicative of lactate in long echo proton MRS studies. Using long echo times such as 144 and 288 milliseconds (msec) (multiples of 1/7 Hz), lactate can be distinguished from lipid signals. Sampling with an echo time 144 msec inverts the resonance below the baseline due to spin or J-coupling. While this distinguishes the lactate doublet peak from macromolecules and lipids, the signal intensity is diminished which is especially problematic in low concentrations (on the order of 5 mM or below) and within some technical circumstances (i.e. errors in pulse generation & reception).46 The doublet peak emerges above the baseline at an echo time of 288 msec. The peak should now represent lactate. Technical issues with signal transmission and reception may cause the doublet to be asymmetric. Shimming problems may broaden the signal so as to smooth the doublet. At a TE 288 msec, if the signal arises from lipids, it should correspond anatomically with high signal from fat on imaging, otherwise it could be lactate. The appearance of lactate is pathologic as it classically represents anaerobic glycolysis. Lactate signal may also possibly reflect mitochondrial impairment, as an inflammatory response and macrophage infiltration depending upon the condition.
Contrast-enhanced sequences are frequently a vital component of MRI studies, especially in neoplastic and infectious diseases. If proton MRS can be performed prior to contrast administration, the likelihood of diagnostic spectra is improved over MRS obtained after contrast administration. Gadolinium based contrast agents that quickly accumulate in a lesion can broaden line widths to the degree where the spectra are not diagnostic. However, in a study of adult brain tumors in which proton MRS was acquired both before and after contrast administration, a blinded review failed to show a significant difference in the MR spectroscopic diagnosis related to the presence of contrast. Individual ratios changed pre versus post; however, no systemic change was appreciated in the study. Thus, MRS should not be abandoned if the only option is to obtain spectra after contrast administration.
5-Normal development
During the first 2–3 years of brain development, metabolite levels vary primarily due to the myelination process.47 Defining normal metabolite concentration levels is heavily dependent on the MRS technique and operating parameters. Most sites either develop their own normative databases or rely on the literature and the work of others as automated MRS acquisition protocols now allow MRS acquisitions to be more closely replicated between sites. In both the adult and pediatric setting, differences in metabolite levels are also found between gray and white matter as well as between the different lobes of the brain (frontal versus occipital).48
6-Normal aging
During the last decades of life (>60 years), there is an observable and region specific reduction in brain tissue volume with age. With this volume loss, metabolite changes are expected with the typical aging process. Haga et al. reviewed the literature for studies evaluating healthy aging with proton MRS.49 Beginning with 231 potentially relevant studies, a meta-analysis identified from four studies with extractable metabolite data a decrease in frontal NAA levels approaching significance, and significant increases in parietal choline and creatine levels. The authors identified the need for large scale studies with rigorous approaches for defining healthy participants, performing tissue segmentation, reporting absolute metabolite concentrations in multiple brain regions.
7-Protocols
7.1-Diffuse disorders, disorders without focal lesions
Location, Echo Times Diffuse disorders, especially those without focal lesions, can be assessed using either SVS or MRSI approaches. The choice should be based on the available software, skill and experience of the technologists at the institution for the given approach.
A standard MRS location sampled for gray matter assessment in infants and children includes the left basal ganglia composed with the caudate, internal capsule, globus pallidus and putamen with minimal inclusion of the thalamus. As children become adolescents, iron deposition within the basal ganglia widens the spectral peaks, which may affect quantitation. At this point, the standard MRS location is often across the hemispheres within the parasagittal cortex of the occipital or parietal lobes.
A standard location chosen to assess myelination and white matter disorders is the frontal lobe for pediatric populations. Sampling within this region will inevitably have some gray matter involvement. For practical purposes, all of the metabolite levels, other than choline, will remain unaffected with this mixture of composition. In adults, sampling within the centrum semiovale is appropriate. A key pitfall of these locations is the inclusion of the lateral ventricles, which results in a selective decrease in the creatine signal. This change will artificially elevate metabolite ratios.
If a radiologist were asked to perform only one MR imaging sequence to evaluate a patient, a variety of opinions would be expressed. For spectroscopist, the same analogy is found when asking what one echo time should be acquired within an MRS examination. Short echo MRS affords many more metabolites which ultimately are useful in diagnosis. However, the baseline is not as flat as the MRS acquired with long echo sequences. Thus, the spectral fitting and interpretation is more complex. The flatter baseline is also a key reason why MRSI usually employs long echo pulse sequences. The long echo is primarily heralded for its ability to demonstrate lactate peaks. However, to answer the question of which echo to employ if only one is allowed, the short echo MRS is the preferred approach by the author of this article, especially for SVS studies, as often one finds useful changes in the peaks with short T2 values for diagnosis that would not be present on long echo studies.
7.2-Focal lesions
Sampling within focal lesions is relatively straightforward; however, there are some pitfalls to be aware of in the process. Again, the SVS or MRSI approach and choice of echo time is site dependent. For most scenarios, the ideal protocol would employ a short echo SVS sequence followed by a long echo, either a 144 or 288 millisecond echo time MRSI with inclusion of the focal lesion, adjacent parenchyma and when possible, the contralateral hemisphere. This approach provides for a full compliment of metabolite sampling, determining lesion extent and providing an internal comparison. Focal lesions which are very small or would likely need reproducible follow-up over time (such as a demyelinating lesion) should be studied with an MRSI approach as this method is not as technically demanding with respect to precise voxel positioning as it can allow for voxel shifting in post-processing procedures and produce very small voxel sizes. If MRSI is not in use at a site, SVS can be usually performed with voxel dimensions of approximately 1.5 cm per side. (One should maintain a total volume of 4–8 cc, so one voxel dimension could potentially be as small as 1 cm while others 2 cm and/or larger). While partial voluming with “unaffected”, parenchyma adjacent to the focal lesion may occur, one can usually appreciate significant changes in choline or other metabolite levels, especially if a separate reference SVS is acquired in the contralateral hemisphere at an equivalent, symmetrical location.
In some pediatric patients with cerebral abscess, sampling within the core of the lesion can be accomplished to visualize succinate, acetate, valine and other amino acids. However, in adults, the central core of an abscess is usually necrotic. Cerebral abscess and regions with large necrotic portions such as high grade gliomas can be technically challenging due to large lipid peaks altering shimming, and quantification of other metabolites. For qualitative assessment, the y-axis scale of the spectrum is usually automatically matched to visualize the height of the largest peak. (Some manufacturers provide a scale for qualitative comparison, but others do not). In a necrotic pathology, the lipid peak often becomes the largest peak in the spectrum.
8-Summary
The goal of this article was to explain the principals behind proton MRS in a practical fashion. Understanding the principals behind proton MRS can provide exceptional benefit to the neuroradiologist when MRS is applied in the clinical setting.
Key Points.
Magnetic resonance spectroscopy (MRS) provides information on neuronal and axonal viability, energetics of the cellular structures, and status of the cellular membranes.
The interpretation of an MR spectrum primarily arises from variations in chemical ratios and/or concentrations, which alter peak area for the endogenous metabolites.
The single voxel spectroscopy (SVS) technique provides a cubic-based volume of interest typically 4–8 cc in volume.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Drost DJ, Riddle WR, Clarke GD. Proton magnetic resonance spectroscopy in the brain: report of AAPM MR Task Group #9. Medical physics. 2002 Sep;29(9):2177–2197. doi: 10.1118/1.1501822. [DOI] [PubMed] [Google Scholar]
- 2.Zhu H, Barker PB. MR spectroscopy and spectroscopic imaging of the brain. Methods Mol Biol. 2011;711:203–226. doi: 10.1007/978-1-61737-992-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Costanzo A, Trojsi F, Tosetti M, et al. High-field proton MRS of human brain. Eur J Radiol. 2003 Nov;48(2):146–153. doi: 10.1016/j.ejrad.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Tran T, Ross B, Lin A. Magnetic resonance spectroscopy in neurological diagnosis. Neurol Clin. 2009 Feb;27(1):21–60. doi: 10.1016/j.ncl.2008.09.007. xiii. [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan R, Vigneron D, Sailasuta N, Hurd R, Nelson S. A comparative study of myo-inositol quantification using LCmodel at 1.5 T and 3.0 T with 3 D 1H proton spectroscopic imaging of the human brain. Magn Reson Imaging. 2004 May;22(4):523–528. doi: 10.1016/j.mri.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Kreis R. Issues of spectral quality in clinical 1H-magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004 Oct;17(6):361–381. doi: 10.1002/nbm.891. [DOI] [PubMed] [Google Scholar]
- 7.Posse S, Cuenod CA, Risinger R, Le Bihan D, Balaban RS. Anomalous transverse relaxation in 1H spectroscopy in human brain at 4 Tesla. Magn Reson Med. 1995 Feb;33(2):246–252. doi: 10.1002/mrm.1910330215. [DOI] [PubMed] [Google Scholar]
- 8.Hetherington HP, Mason GF, Pan JW, et al. Evaluation of cerebral gray and white matter metabolite differences by spectroscopic imaging at 4.1T. Magn Reson Med. 1994 Nov;32(5):565–571. doi: 10.1002/mrm.1910320504. [DOI] [PubMed] [Google Scholar]
- 9.Barker PB, Hearshen DO, Boska MD. Single-voxel proton MRS of the human brain at 1.5T and 3.0T. Magn Reson Med. 2001 May;45(5):765–769. doi: 10.1002/mrm.1104. [DOI] [PubMed] [Google Scholar]
- 10.Bartha R, Drost DJ, Menon RS, Williamson PC. Comparison of the quantification precision of human short echo time (1)H spectroscopy at 1.5 and 4.0 Tesla. Magn Reson Med. 2000 Aug;44(2):185–192. doi: 10.1002/1522-2594(200008)44:2<185::aid-mrm4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 11.Gruetter R. Automatic, localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. 1993 Jun;29(6):804–811. doi: 10.1002/mrm.1910290613. [DOI] [PubMed] [Google Scholar]
- 12.Haase A, Frahm J, Hanicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985 Apr;30(4):341–344. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 13.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B. 1994 May;104(1):1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 14.Tkac I, Starcuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999 Apr;41(4):649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 15.Duyn JH, Gillen J, Sobering G, van Zijl PC, Moonen CT. Multisection proton MR spectroscopic imaging of the brain. Radiology. 1993 Jul;188(1):277–282. doi: 10.1148/radiology.188.1.8511313. [DOI] [PubMed] [Google Scholar]
- 16.Tran TK, Vigneron DB, Sailasuta N, et al. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000 Jan;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 17.Osorio JA, Xu D, Cunningham CH, et al. Design of cosine modulated very selective suppression pulses for MR spectroscopic imaging at 3T. Magn Reson Med. 2009 Mar;61(3):533–540. doi: 10.1002/mrm.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999 Nov;42(5):952–962. [PubMed] [Google Scholar]
- 19.Sandgren N, Stoica P, Frigo FJ, Selen Y. Spectral analysis of multichannel MRS data. J Magn Reson. 2005 Jul;175(1):79–91. doi: 10.1016/j.jmr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001 Jun;14(4):260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 21.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993 Dec;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 22.Naressi A, Couturier C, Devos JM, et al. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001 May;12(2–3):141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 23.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001 Jul;31(4):269–286. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 24.Barantin L, Le Pape A, Akoka S. A new method for absolute quantitation of MRS metabolites. Magn Reson Med. 1997 Aug;38(2):179–182. doi: 10.1002/mrm.1910380203. [DOI] [PubMed] [Google Scholar]
- 25.Henriksen O. In vivo quantitation of metabolite concentrations in the brain by means of proton MRS. NMR Biomed. 1995 Jun;8(4):139–148. doi: 10.1002/nbm.1940080402. [DOI] [PubMed] [Google Scholar]
- 26.Soher BJ, van Zijl PC, Duyn JH, Barker PB. Quantitative proton MR spectroscopic imaging of the human brain. Magn Reson Med. 1996 Mar;35(3):356–363. doi: 10.1002/mrm.1910350313. [DOI] [PubMed] [Google Scholar]
- 27.Pouwels PJ, Frahm J. Differential Distribution of NAA and NAAG in Human Brain as Determined by Quantitative Localized Proton MRS. NMR in Biomedicine. 1997;10:73–78. doi: 10.1002/(sici)1099-1492(199704)10:2<73::aid-nbm448>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Matalon R, Michals K, Sebesta D, Deanching M, Gashkoff P, Casanova J. Aspartoacylase deficiency and N-acetylaspartic aciduria in patients with Canavan disease. Am J Med Genet. 1988 Feb;29(2):463–471. doi: 10.1002/ajmg.1320290234. [DOI] [PubMed] [Google Scholar]
- 29.Tallan HH, Moore S, Stein WH. N-acetyl-L-aspartic acid in brain. Journal of Biological Chemistry. 1956;219:257–264. [PubMed] [Google Scholar]
- 30.Tallan HH. Studies on the distribution of N-acetyl-L-aspartic acid in brain. Journal of Biological Chemistry. 1956;224:41–45. [PubMed] [Google Scholar]
- 31.Benuck M, D'Adamo JAF. Acetyl transport mechanisms. Metabolism of N-acetyl-L-aspartic acid in the non-nervous tissues of the rat. Biochem. Biophys. Acta. 1968;152(3):611–618. doi: 10.1016/0005-2760(68)90101-x. [DOI] [PubMed] [Google Scholar]
- 32.D'Adamo AF, Jr, Smith JC, Woiler C. The occurrence of N-acetylaspartate amidohydrolase (Aminoacylase II) in the developing rat. Journal of Neurochemistry. 1973;20:1275–1278. doi: 10.1111/j.1471-4159.1973.tb00097.x. [DOI] [PubMed] [Google Scholar]
- 33.Austin SJ, Connelly A, Gadian DG, Benton JS, Brett EM. Localized 1H NMR spectroscopy in Canavan's Disease: A Report of Two Cases. Magnetic Resonance in Medicine. 1991;19:439–445. doi: 10.1002/mrm.1910190235. [DOI] [PubMed] [Google Scholar]
- 34.Brand A, Richter-Landsberg C, Leibfritz D. Multinuclear NMR studies on the energy metabolism of glial and neuronal cells. Dev Neurosci. 1993;15(3–5):289–298. doi: 10.1159/000111347. [DOI] [PubMed] [Google Scholar]
- 35.Urenjak J, Williams SR, Gadian DG, Noble M. Specific expression of N-acetylaspartate in neurons, oligodendrocyte- type-2 astrocyte progenitors, and immature oligodendrocytes in vitro. J Neurochem. 1992;59(1):55–61. doi: 10.1111/j.1471-4159.1992.tb08875.x. [DOI] [PubMed] [Google Scholar]
- 36.Moffett JR, Tieman SB, Weinberger DR, Coyle JT, Namboodiri AMA. N-Acetylaspartate A Unique Neuronal Molecule in the Central Nervous System; 2006. Bethesda, Maryland, USA: 2004. [Google Scholar]
- 37.Clark JF, Doepke A, Filosa JA, et al. N-acetylaspartate as a reservoir for glutamate. Med Hypotheses. 2006;67(3):506–512. doi: 10.1016/j.mehy.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 38.Birken DL, Oldendorf WH. N-acetyl-L-aspartic acid: a literature review of a compound prominent in 1H-NMR spectroscopic studies of brain. Neurosci Biobehav Rev. 1989;13(1):23–31. doi: 10.1016/s0149-7634(89)80048-x. [DOI] [PubMed] [Google Scholar]
- 39.Martin E, Capone A, Schneider J, Hennig J, Thiel T. Absence of N-acetylaspartate in the human brain: impact on neurospectroscopy? Ann Neurol. 2001 Apr;49(4):518–521. [PubMed] [Google Scholar]
- 40.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000 Jul;80(3):1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 41.Miller B. A Review of Chemical Issues in 1H NMR Spectroscopy: N-Acetyl-L-aspartate, Creatine and Choline. NMR in Biomedicine. 1991;4:47–52. doi: 10.1002/nbm.1940040203. [DOI] [PubMed] [Google Scholar]
- 42.Stork C, Renshaw PF. Mitochondrial dysfunction in bipolar disorder: evidence from magnetic resonance spectroscopy research. Mol Psychiatry. 2005 Oct;10(10):900–919. doi: 10.1038/sj.mp.4001711. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, Arcinue E, Ross B. Organic osmolytes in the brain of an infant with hypernatremia. New England Journal of Medicine. 1994;331:439–442. doi: 10.1056/NEJM199408183310704. [DOI] [PubMed] [Google Scholar]
- 44.Ross BD. Biochemical considerations in 1H spectroscopy. Glutamate and glutamine; Myo-inositol and related metabolites. NMR in Biomedicine. 1991;4:59–63. doi: 10.1002/nbm.1940040205. [DOI] [PubMed] [Google Scholar]
- 45.Petroff OAC, Pleban LA, Spencer DD. Symbiosis Between In Vivo and In Vitro NMR Spectroscopy: The Creatine, N-Acetylaspartate, Glutamate, and GABA Content of the Epileptic Human Brain. Magnetic Resonance Imaging. 1995;13(8):1197–1211. doi: 10.1016/0730-725x(95)02033-p. [DOI] [PubMed] [Google Scholar]
- 46.Lange T, Dydak U, Roberts TP, Rowley HA, Bjeljac M, Boesiger P. Pitfalls in lactate measurements at 3T. AJNR Am J Neuroradiol. 2006 Apr;27(4):895–901. [PMC free article] [PubMed] [Google Scholar]
- 47.Kreis R, Ernst T, Ross BD. Development of the human brain: in vivo quantification of metabolite and water content with proton magnetic resonance spectroscopy. Magn Reson Med. 1993;30(4):424–437. doi: 10.1002/mrm.1910300405. [DOI] [PubMed] [Google Scholar]
- 48.Pouwels PJW, Frahm J. Regional Metabolite Concentrations in Human Brain as Determined by Quantitative Localized Proton MRS. Magnetic Resonance in Medicine. 1998;39:53–60. doi: 10.1002/mrm.1910390110. [DOI] [PubMed] [Google Scholar]
- 49.Haga KK, Khor YP, Farrall A, Wardlaw JM. A systematic review of brain metabolite changes, measured with 1H magnetic resonance spectroscopy, in healthy aging. Neurobiol Aging. 2009 Mar;30(3):353–363. doi: 10.1016/j.neurobiolaging.2007.07.005. [DOI] [PubMed] [Google Scholar]