Abstract
The targeting of nucleases to specific DNA sequences facilitates genome editing. Recent work demonstrated that the CRISPR-associated (Cas) nuclease Cas9 can be targeted to sequences in vitro simply by modifying a short7 CRISPR RNA (crRNA) guide. Here we use this CRISPR-Cas system to introduce marker-free mutations in Streptococcus pneumoniae and Escherichia coli. The approach involves re-programming Cas9 by using a crRNA complementary to a target chromosomal locus and introducing a template DNA harboring a desired mutation and an altered crRNA recognition site for recombination with the target locus. We exhaustively analyze Cas9 target requirements to define the range of targetable sequences and show strategies for editing sites that do not meet these requirements. Alone or together with recombineering, CRISPR assisted editing induces recombination at the targeted locus and kills non-edited cells leading to a recovery of close to a 100% of edited cells. Multiple crRNA can be used to modify several loci simultaneously. Our results show that CRISPR-mediated genome editing only requires programming of the crRNA and template sequences and thus constitutes a useful tool for genetic engineering.
The understanding of gene function depends on the possibility of altering DNA sequences within the cell in a controlled fashion. Site-specific mutagenesis in eukaryotes is achieved by the use of sequence-specific nucleases that promote homologous recombination of a template DNA containing the mutation of interest. Zinc finger nucleases (ZFN)1, transcription activator-like effector nucleases (TALENs)2 and homing meganucleases3 can be programmed to cut genomes in specific locations, but this requires difficult and expensive engineering. In prokaryotic organisms, mutagenesis either introduces a selection marker in the edited locus or requires a two-step process that includes a counter-selection system 4, 5. More recently, phage recombination proteins have been used for recombineering, a technique that promotes homologuous recombination of linear DNA or oligonucleotides. Because there is no selection of mutations, recombineering efficiency can be relatively low (0.1–10% for point mutations down to 10−5–10−6 for larger modifications) 6, in many cases requiring the screening of a large number of colonies. Therefore new technologies that are affordable, easy to use and efficient are still in need for the genetic engineering of both eukaryotic and prokaryotic organisms.
Recent work on the CRISPR (clustered, regularly interspaced, short palindromic repeats) prokaryotic immunity system has led to the identification of nucleases whose sequence specificity is programmed by small RNAs7. CRISPR loci are composed of a series of repeats separated by ‘spacer’ sequences that match the genomes of bacteriophages and other mobile genetic elements8–11. The repeat-spacer array is transcribed as a long precursor and processed within repeat sequences to generate small crRNA that specify CRISPR targets (also known as protospacers)12–16. Essential for cleavage is the presence of a sequence motif immediately downstream of the target region, known as the protospacer-adjacent motif (PAM)7, 17, 18. CRISPR-associated (cas) genes usually flank the repeat-spacer array and encode the enzymatic machinery responsible for crRNA biogenesis and targeting19. Cas9 is a dsDNA nuclease that uses a crRNA guide to specify the site of cleavage7, 18. Loading of the crRNA guide into Cas9 occurs during the processing of the crRNA precursor and requires a small RNA antisense to the precursor, the tracrRNA, and RNAse III (ref. 14). In contrast to ZFNs or TALENs, the change of Cas9 target specificity does not require protein engineering, therefore this enzyme is ideal for biotechnological applications, especially genome editing20–22.
We recently showed that the introduction of a CRISPR system in Streptococcus pneumoniae targeting a chromosomal locus leads to the killing of the transformed cells23. We observed that occasional survivors contained mutations in the target region, suggesting that Cas9 dsDNA nuclease activity against endogenous targets could be used for genome editing. Here we show that marker-less mutations can be introduced through the transformation of a template DNA that will recombine in the genome and eliminate Cas9 target recognition. Re-programming Cas9 with different crRNAs allows for the introduction of multiple mutations at the same time. We also characterize in detail the sequence requirements for Cas9 targeting and show that the approach can be combined with recombineering for genome editing in Escherichia coli.
RESULTS
Cas9 nuclease activity against endogenous targets can be exploited for genome editing
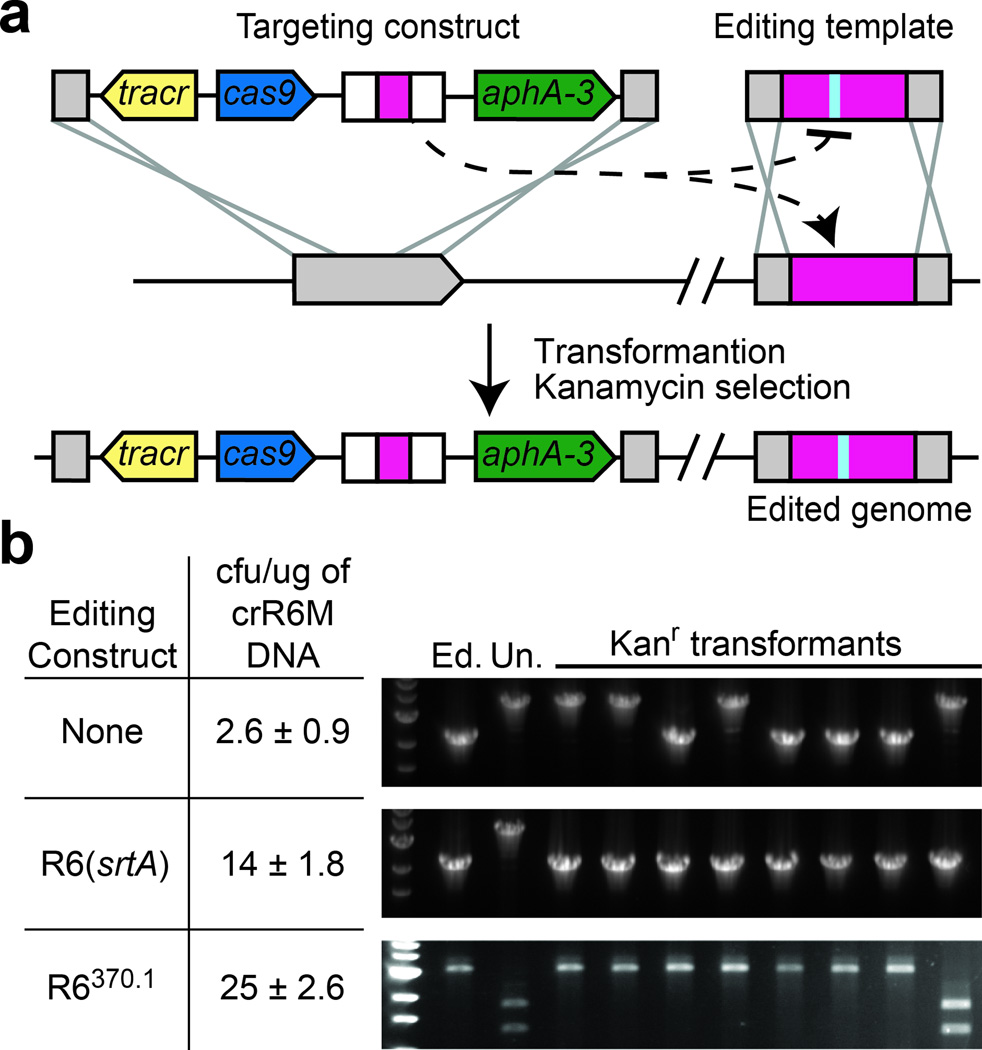
S. pneumoniae crR6 contains a Cas9-based CRISPR system that attacks a target present in the chromosome of S. pneumoniae R68232.5 cells but not the same target containing a PAM mutation in R6370.1 cells23 (Supplementary fig. 1a). We observed that transformation of the CRISPR-Cas system from genomic DNA of crR6 cells is approximately 10-fold less efficient in R68232.5 than in R6370.1 cells (Supplementary fig. 1b). Genetic analysis of eight R68232.5 transformants (Supplementary fig. 1b) revealed that the great majority are the product of a double recombination event that eliminates the toxicity of Cas9 targeting by replacing the 8232.5 target with the wild-type srtA locus (without a protospacer) present in genomic crR6 DNA. These results are proof that the concurrent introduction of a CRISPR system targeting a genomic locus (the targeting construct) together with a template for recombination into the targeted locus (the editing template) can lead to targeted genome editing (Fig. 1a).
Figure 1.
Cas9 nuclease activity against endogenous targets can be exploited for genome editing. (a) Concept of CRISPR directed genome editing. The CRISPR targeting construct directs interference against a chromosomal locus and is co-transformed with an editing template that recombines with the target to abolish the interference. Kanamycin-resistant transformants that survive CRISPR attack contain modifications introduced by the editing template. (b) Transformation of crR6M DNA in R68232.5 cells with either: no editing template, the R6 srtA or the R6370.1 editing templates. Transformation efficiency is calculated as colony forming units (cfu) per µg of crR6M DNA; the mean values with standard deviations from at least three independent experiments are shown. PCR analysis is performed on 8 clones in each transformation. “Un.” indicates the unedited srtA locus of strain R68232.5; “Ed.” shows the editing template. R68232.5 and R6370.1 targets are distinguished by restriction with EaeI.
To simplify CRISPR-mediated genome editing we deleted cas1, cas2 and csn2, which were shown to be dispensable for CRISPR interference14, 24, and generated strain crR6M (Supplementary fig. 1a). This strain retains the same properties of crR6 (Supplementary fig. 1b). To increase the efficiency of CRISPR editing and demonstrate that a template DNA of choice can be used to control the mutation introduced, we co-transformed R68232.5 cells with PCR products of either the wild-type srtA gene or the mutant R6370.1 target. This resulted in a 5- to 10-fold increase of the frequency of transformation compared with genomic DNA alone (Fig. 1b). The efficiency of editing was also substantially increased, with 8/8 transformants tested containing a wild-type srtA copy and 7/8 containing the PAM mutation present in the R6370.1 target (Fig. 1b and Supplementary fig. 2a). Taken together, the results shown in Figure 1 demonstrate the potential of CRISPR-assisted genome editing.
Analysis of Cas9 target requirements
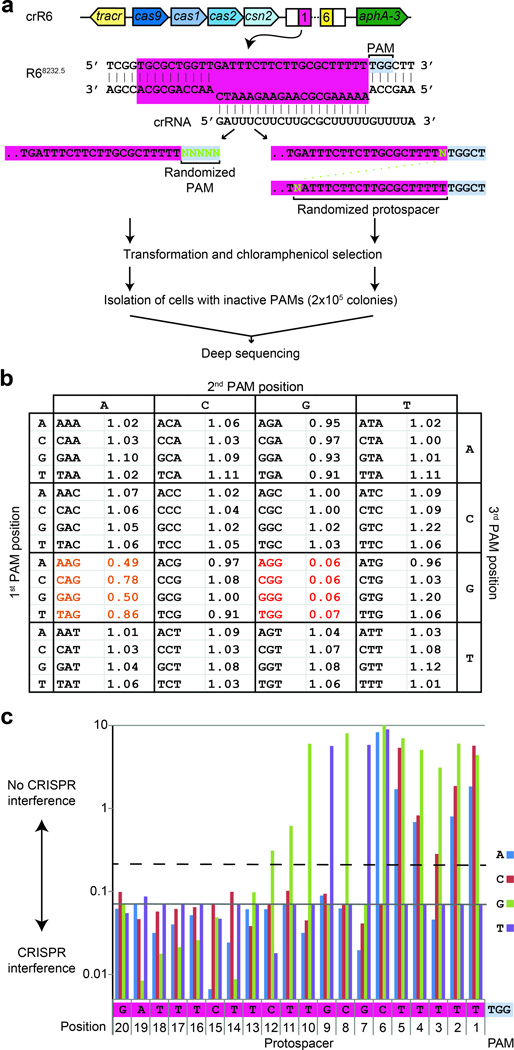
To introduce specific changes in any position of the genome the editing template must carry mutations that abolish CRISPR targeting. This is easy to achieve when the deletion of the target or its replacement by another sequence (gene insertion) is sought. When the goal is to produce gene fusions or to generate single-nucleotide mutations, the abolishment of Cas9 nuclease activity will only be possible by introducing mutations in the editing template that alter either the PAM or the protospacer sequences. To determine the constraints of CRISPR-mediated editing, we performed an exhaustive analysis of PAM and protospacer mutations that abrogate CRISPR targeting. Previous studies proposed that S. pyogenes Cas9 requires an NGG PAM immediately downstream of the protospacer7, 14, 18. However, because only a very limited number of PAM-inactivating mutations have been described so far7, 14, 18, 23, we conducted a systematic analysis to find all the 5-nucleotide sequences following the protospacer that eliminate CRISPR targeting. We used randomized oligonucleotides to generate all the possible 1,024 PAM sequences in a heterogeneous PCR product that was transformed into crR6 or R6 cells. Constructs carrying functional PAMs are expected to be recognized and destroyed by Cas9 in crR6 but not R6 cells (Fig. 2a). More than 2×105 colonies were pooled together to extract DNA and use it as template for the co-amplification of all targets. PCR products were deep sequenced and the functionality of each PAM was estimated by the relative proportion of its reads in the crR6 sample over the R6 sample. Analysis of the first three bases of the PAM, averaging over the two last bases, clearly shows that the NGG pattern is under-represented in crR6 transformants (Fig. 2b). Furthermore, the next two bases have no detectable effect on the NGG PAM (Supplementary text), demonstrating that the NGGNN sequence is sufficient to license Cas9 activity. Partial targeting was observed for NAG PAM sequences (Fig. 2b). Also the NNGGN pattern partially inactivates CRISPR targeting (Supplementary Table1), indicating that the NGG motif can still be recognized by Cas9 with reduced efficiency when shifted by 1 bp. These data shed light onto the molecular mechanism of Cas9 target recognition, but more importantly, they reveal that NGG (or CCN on the complementary strand) sequences are sufficient for Cas9 targeting and that NGG to NAG or NNGGN mutations in the editing template should be avoided. Owing to the high frequency of these tri-nucleotide sequences (once every 8 bp), this means that almost any position of the genome can be edited. Indeed, we tested ten randomly chosen targets carrying various PAMs and all were found to be functional (Supplementary Fig. 3).
Figure 2.
Analysis of PAM and seed sequences that eliminate CRISPR interference. (a) PCR products with randomized PAM sequences or randomized seed sequences (in green) were transformed in crR6 cells. These cells express Cas9 loaded with a crRNA that targets a chromosomal region of R68232.5 cells (highlighted in pink) that is absent from the R6 genome. More than 2×105 chloramphenicol-resistant transformants, carrying inactive PAM or seed sequences, were combined for amplification and deep sequencing of the target region. (b) Relative proportion of number of reads after transformation of the random PAM constructs in crR6 cells (compared to number of reads in R6 transformants). The relative abundance for each 3-nucleotide PAM sequence is shown. Severely underrepresented sequences (NGG) are shown in red; partially underrepresented one in orange (NAG) (c) Relative proportion of number of reads after transformation of the random seed sequence constructs in crR6 cells (compared to number of reads in R6 transformants). The relative abundance of each nucleotide for each position of the first 20 nucleotides of the protospacer sequence is shown. High abundance indicates lack of CRISPR targeting, i.e. a CRISPR inactivating mutation. The grey line shows the level of the WT sequence. The dotted line represents the level above which a mutation significantly disrupts interference (Supplementary text).
Another way to disrupt Cas9-mediated cleavage is to introduce mutations in the protospacer region of the editing template. It is known that point mutations within the ‘seed sequence’ (the 8 to 10 protospacer nucleotides immediately adjacent to the PAM) can abolish CRISPR targeting7, 25, 26. However the exact length of this region is not known and it is unclear whether mutations to any nucleotide in the seed can disrupt Cas9 target recognition. We followed the same deep sequencing approach described above to randomize the entire protospacer sequence involved in base pair contacts with the crRNA and to determine all sequences that disrupt targeting. Each position of the 20 matching nucleotides14 in the spc1 target present in R68232.5 cells (Fig. 1a) was randomized and transformed into crR6 and R6 cells (Fig. 2a). Consistent with the presence of a seed sequence, only mutations in the 12 nucleotides immediately upstream of the PAM abrogated CRISPR targeting (Fig. 2c). However, different mutations displayed markedly different effects. The distal (from the PAM) positions of the seed (12 to 7) tolerated most mutations and only one particular base substitution abrogated targeting. In contrast, mutations to any nucleotide in the proximal positions (6 to 1, except 3) eliminated Cas9 activity, although at different levels for each particular substitution. At position 3, only two substitutions affected CRISPR activity and with different strength. We conclude that, although seed sequence mutations can prevent CRISPR targeting, there are restrictions regarding the nucleotide changes that can be made in each position of the seed. Moreover, these restrictions can most likely vary for different spacer sequences. Therefore we believe that mutations in the PAM sequence, if possible, should be the preferred editing strategy. Alternatively, multiple mutations in the seed sequence could be introduced to prevent Cas9 nuclease activity.
Cas9-mediated genome editing in S. pneumoniae
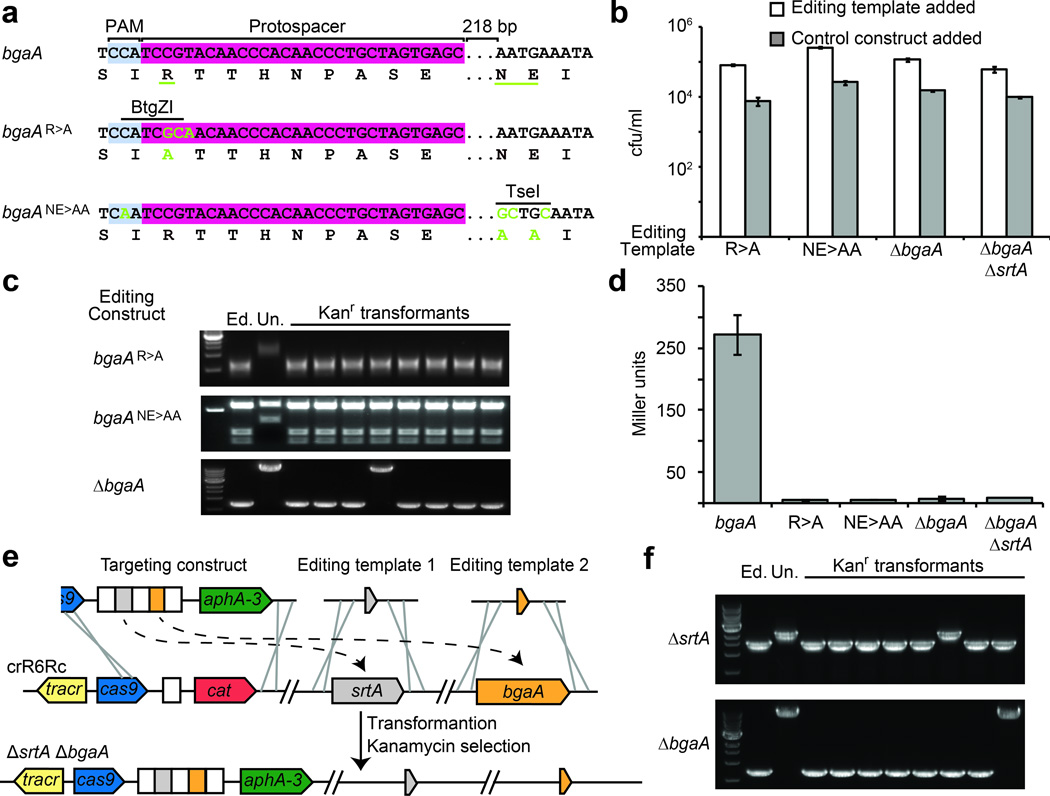
To develop a rapid and efficient method for targeted genome editing, we engineered strain crR6Rk, a strain in which spacers can easily be introduced by PCR (Supplementary fig. 4). We decided to edit the β-galactosidase (bgaA) gene of S. pneumoniae, whose activity can be easily measured27. We introduced alanine substitutions of amino acids belonging to the active site of this enzyme: R481A (R>A mutation) and N563A,E564A (NN>AA mutation). To illustrate different editing strategies, we designed mutations of both the PAM sequence and the protospacer seed. In both cases we used the same targeting construct, a region encoding part of the active site of β-galactosidase that is adjacent to a TGG PAM sequence (CCA in the complementary strand, Fig. 3a). The R>A editing template created a three-nucleotide mismatch on the protospacer seed sequence (CGT to GCA, also introducing a BtgZI restriction site). In the NE>AA editing template we simultaneously introduced a synonymous mutation that creates an inactive PAM (TGG/CCA to TTG/CAA) along with mutations that are 218 nt downstream of the protospacer region (AAT GAA to GCT GCA, also generating a TseI restriction site). This last editing strategy demonstrates the possibility of using a remote PAM to make mutations in places where a proper target might be hard to choose. For example, although the S. pneumoniae R6 genome, which has a 39.7% GC content, contains on average one PAM motif every 12 bp, some PAM motifs are separated by up to 194 bp (Supplementary fig. 5). In addition we designed a ΔbgaA in-frame deletion of 6664 bp. In all three cases, co-transformation of the targeting and editing templates produced 10-times more kanamycin-resistant cells than co-transformation with a control editing template containing wild-type bgaA sequences (Fig. 3b). We genotyped 24 transformants (8 for each editing experiment) and found that all but one incorporated the desired change (Fig. 3c). DNA sequencing also confirmed not only the presence of the introduced mutations but also the absence of secondary mutations in the target region (Supplementary Figs. 1b,c). Finally we measured β-galactosidase activity27 to confirm that all edited cells displayed the expected phenotype (Fig. 3d).
Figure 3.
Introduction of single and multiple mutations using CRISPR-mediated genome editing. (a) Nucleotide and amino acid sequences of the wild-type and edited (green nucleotides; underlined amino acid residues) bgaA. The protospacer, PAM and restriction sites are shown. (b) Transformation efficiency of cells transformed with targeting constructs in the presence of an editing template or control. (c) PCR analysis for 8 transformants of each editing experiment followed by digestion with BtgZI (R>A) and TseI (NE>AA). Deletion of bgaA is revealed as a smaller PCR product. (d) Miller assay to measure the β-galactosidase activity of WT and edited strains. (e) For a single-step double deletion the targeting construct contains two spacers (in this case matching srtA and bgaA) and is co-transformed with two different editing templates (f) PCR analysis for 8 transformants to detect deletions in srtA and bgaA loci. 6/8 transformants contained deletions of both genes.
CRISPR-mediated editing can also be used to generate multiple mutations for the study of biological pathways. To illustrate this possibility, we introduced mutations in the sortase-dependent pathway that anchors surface proteins to the envelope of Gram-positive bacteria28. We introduced a sortase deletion by co-transformation of a chloramphenicol-resistant targeting construct and a ΔsrtA editing template (Supplementary Figs. 6a and b), followed by a ΔbgaA deletion using a kanamycin-resistant targeting construct that replaces the previous one. In S. pneumoniae, β-galactosidase is covalently linked to the cell wall by sortase27. Therefore deletion of srtA results in the release of the surface protein into the supernatant while the double deletion has no detectable β-galactosidase activity (Supplementary fig. 7c). This sequential selection can be iterated as many times as required to generate multiple mutations. More importantly, CRISPR-based genome engineering presents the possibility of introducing both mutations at the same time. To do this we simply designed a targeting construct containing two spacers, each matching one gene, and co-transformed it with both editing templates at the same time (Fig. 3e). Genetic analysis of transformants showed that editing occurred in 6/8 cases (Fig. 3f). Notably, the remaining two clones each contained a ΔsrtA or a ΔbgaA deletion, suggesting the possibility of performing combinatorial mutagenesis using CRISPR editing. Finally, to eliminate the CRISPR sequences, we introduced a plasmid containing a target and a spectinomycin resistance gene along with genomic DNA from the wild-type strain R6. Spectinomycin-resistant transformants that retain the plasmid eliminated the CRISPR sequences (Supplementary fig. 7a,d).
Mechanism and efficiency of editing
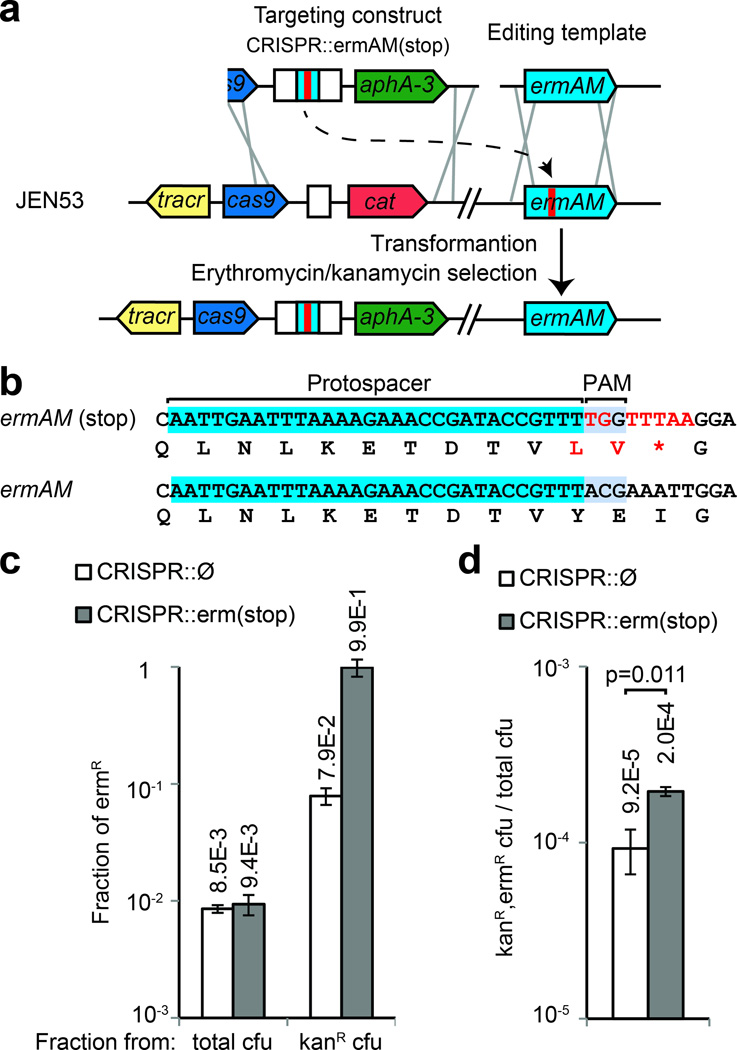
To understand the mechanisms underlying CRISPR-assisted genome editing, we designed an experiment where the editing efficiency can be measured independently of Cas9 targeting. We integrated the ermAM erythromycin resistance gene in the srtA locus, and introduced a premature stop codon using CRISPR-mediated editing (Supplementary fig. 6). The resulting strain (JEN53) contains an ermAM(stop) allele and is sensitive to erythromycin. This strain can be used to assess the efficiency at which the ermAM gene is repaired by measuring the fraction of cells that restore antibiotic resistance with or without the use of CRISPR targeting. JEN53 was transformed with an editing template that restores the wild-type allele, together with either a kanamycin-resistant CRISPR construct targeting the ermAM(stop) allele [CRISPR::ermAM(stop)] or a control construct without a spacer (CRISPR::Ø) (Fig. 4a,b). In the absence of kanamycin selection, the fraction of edited colonies (resistant to erythromycin) is on the order of 10−2 (Fig. 4c). However, if kanamycin selection is applied, the fraction of edited colonies increases to about 10−1 when the control construct is co-transformed, and to almost 1 in the presence of CRISPR targeting (Fig. 4c). This result shows that selection for the recombination of the CRISPR locus co-selected for recombination in the ermAM locus independently of CRISPR targeting, suggesting that a subpopulation of cells is more prone to transformation and/or recombination. In addition to this co-selection, CRISPR targeting enhanced the fraction of erythromycin-resistant, edited cells to 99 % (Fig. 4c). To determine if CRISPR selection is caused by the killing of non-edited cells, we compared the kanamycin-resistant colony forming units (cfu) obtained after co-transformation of JEN53 cells with the CRISPR::ermAM(stop) or CRISPR::Ø constructs. We counted 5.3 times less kanamycin-resistant colonies after transformation of the targeting construct (2.5×104/4.7×103, Supplementary fig. 8a), a result that suggests that indeed CRISPR targeting leads to the killing of non-edited cells. Finally, because the introduction of dsDNA breaks in the bacterial chromosome is known to trigger repair mechanisms that increase the rate of recombination of the damaged DNA29, we investigated whether Cas9 cleavage induces recombination of the editing template. We counted 2.2 times more colonies after co-transformation with the CRISPR::erm(stop) than with the CRISPR::Ø construct (Fig. 4d), indicating that there is a modest induction of recombination. Taken together, these results show that co-selection of transformable cells, induction of recombination by Cas9 cleavage and CRISPR selection against non-edited cells, each contribute to the high efficiency of genome editing in S. pneumoniae.
Figure 4.
Mechanisms underlying CRISPR directed editing. (a) A stop codon was introduced in the erythromycin resistance gene ermAM to generate strain JEN53. The wild-type sequence can be restored through CRISPR-directed editing by targeting the stop codon with the CRISPR::ermAM(stop) construct, and using the ermAM wild-type sequence as an editing template. (b) Mutant and wild-type ermAM sequences. (c) Fraction of erythromicyn-resistant (ermR) cfu calculated from total or kanamycin-resistant (kanR) cfu, in the presence or absence of CRISPR targeting. (d) Fraction of total cells that acquire both the CRISPR construct and the editing template, in the presence or absence of CRISPR targeting. Co-transformation of the CRISPR targeting construct produced more transformants: t-test, t=0.011. In all cases the values show the mean±s.d. for three independent experiments.
As cleavage of the genome by Cas9 should kill non-edited cells, one would not expect to recover any cells that received the kanamycin resistance–containing Cas9 cassette but not the editing template. However, in the absence of the editing template we recovered many kanamycin-resistant colonies after transformation of the CRISPR::ermAM(stop) construct (Supplementary fig. 8a). Cells that ‘escape’ CRISPR-induced death produce a background that determines a limit of the method. This background frequency can be calculated as the ratio of CRISPR::ermAM(stop)/CRISPR::Ø cfu, 2.6×10−3 (7.1×101/2.7×104) in this experiment, meaning that if the recombination frequency of the editing template is less than this value, CRISPR selection will not efficiently recover the desired mutants above the background. To understand their origin, we genotyped 8 of these background colonies and found that 7 contained deletions of the targeting spacer (Supplementary fig. 8b) and one harbored a presumably inactivating mutation in Cas9 (Supplementary fig. 8c).
Cas9-assisted genome editing in E. coli
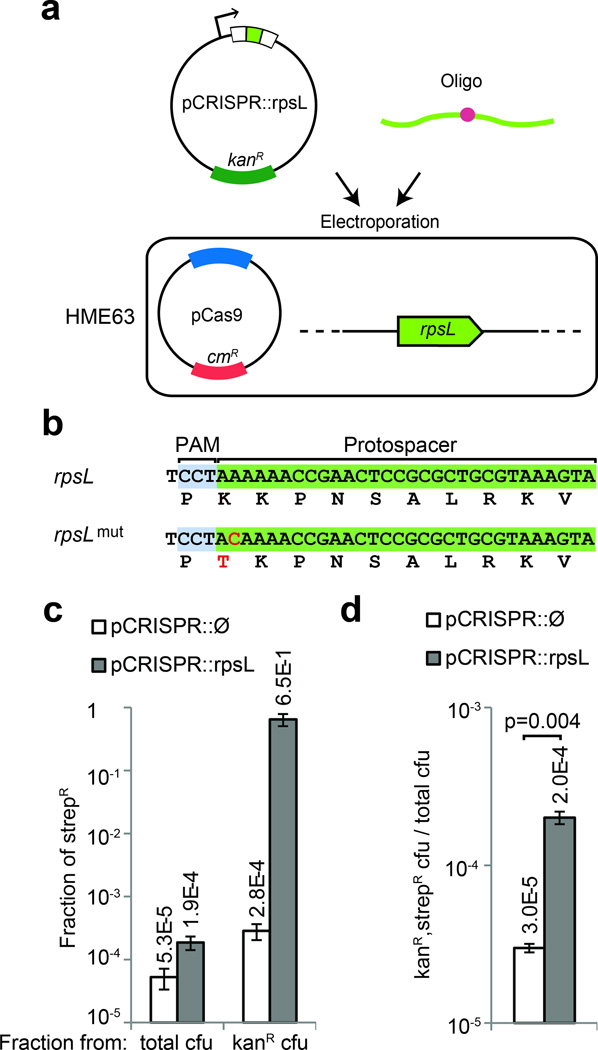
The activation of CRISPR interference through the chromosomal integration of a CRISPR-Cas system is only possible in organisms that are highly recombinogenic. To develop a more general method that is applicable to other microbes, we decided to perform CRISPR-assisted genome editing in E. coli using a plasmid-based CRISPR-Cas system. Two plasmids were constructed: a pCas9 plasmid carrying the tracrRNA, Cas9 and a chloramphenicol resistance cassette (Supplementary fig. 9), and a pCRISPR kanamycin-resistant plasmid carrying the array of CRISPR spacers. To measure the efficiency of editing independently of CRISPR selection, we sought to introduce an A to C transversion in the rpsL gene that confers streptomycin resistance30. We constructed a pCRISPR::rpsL plasmid harboring a spacer that would guide Cas9 cleavage of the wild-type, but not the mutant rpsL allele (Fig. 5b). The pCas9 plasmid was first introduced into E. coli MG1655 and the resulting strain was co-transformed with the pCRISPR::rpsL plasmid and W542, an editing oligonucleotide containing the A to C mutation. We were only able to recover streptomycin-resistant colonies after transformation of the pCRISPR::rpsL plasmid, suggesting that Cas9 cleavage induces recombination of the oligonucleotide (Supplementary fig. 10). However, the number of streptomycin-resistant colonies was two orders of magnitude lower than the number of kanamycin-resistant colonies, presumably CRISPR ‘escapers’. Therefore, in these conditions, CRISPR targeting facilitates the introduction of the mutation, but not enough to select the mutant cells above the background of ‘escapers’.
Figure 5.
CRISPR-directed editing in E. coli. (a) A pCRISPR plasmid targeting the gene to edit can be transformed in the HME63 recombineering strain containing pCas9, together with an oligonucleotide specifying the mutation. (b) A K42T mutation conferring streptomycin resistance was introduced in the rpsL gene (c) Fraction of streptomicyn-resistant (strepR) cfu calculated from total or kanamycin-resistant (kanR) cfu, in the presence or absence of CRISPR targeting. (d) Fraction of total cells that acquire both the pCRISPR plasmid and the editing oligonucleotide, in the presence or absence of CRISPR targeting. Co-transformation of the pCRISPR targeting plasmid produced more transformants: t-test, t=0.004. In all cases the values show the mean±s.d. for three independent experiments.
To improve the efficiency of CRISPR-assisted editing in E. coli, we combined it with recombineering, using CRISPR targeting to select for the desired mutations. The pCas9 plasmid was introduced into the recombineering strain HME63 (ref. 31), which contains the Gam, Exo and Beta functions of the λ-red phage. The resulting strain was co-transformed with the pCRISPR::rpsL plasmid (or a pCRISPR::Ø control) and the W542 oligonucleotide (Fig. 5a). The recombineering efficiency was 5.3×10−5, calculated as the fraction of total cells that become streptomycin-resistant in the absence of CRISPR targeting (Fig. 5c). In contrast, in the presence of CRISPR targeting the percentage of mutant cells increases to 65 ± 14 % (Fig. 5c and Supplementary fig. 2f). Selection occurs through CRISPR killing of non-edited cells; we indeed measured a reduction of about three orders of magnitude in cfu after transformation of the pCRISPR::rpsL plasmid when compared with the control plasmid (4.8×105/5.3×102, Supplementary fig. 11a). To measure the rate of CRISPR inactivation, an important parameter of our method, we transformed both plasmids in the absence of the editing oligonucleotide (Supplementary fig. 11a). This background of CRISPR ‘escapers’, measured as the ratio of pCRISPR::rpsL/pCRISPR::Ø cfu in the absence of W542, was 2.5×10−4 (1.2×102/4.8×105). Genotyping eight of these escapers revealed that in all cases there was a deletion of the targeting spacer (Supplementary fig. 11b). This background is higher than the recombineering efficiency of the rpsL mutation, 5.3×10−5, which suggested that in order to obtain 65 % CRISPR-assisted editing, Cas9 cleavage should induce oligonucleotide recombination. To confirm this, we compared the number of kanamycin- and streptomycin-resistant cfu after transformation of the pCRISPR::rpsL and pCRISPR::Ø (Fig. 5d). As in the case for S. pneumoniae, we observed a modest induction of recombination, about 6.7 fold (2.0×10−4/3.0×10−5). Taken together, these results indicate that CRISPR provides a method for selecting mutations introduced by recombineering.
DISCUSSION
Here we show that CRISPR-Cas systems can be used for targeted genome editing in bacteria by the co-introduction of a targeting construct that kills wild-type cells and an editing template that at the same time eliminates CRISPR interference and introduces the desired mutations. Different types of mutations (insertions, deletions or scar-less single-nucleotide substitutions) can be generated. Multiple mutations can be introduced at the same time. The specificity and versatility of CRISPR-assisted editing rely on several unique properties of the Cas9 nuclease: (i) its target specificity can be programmed with a small RNA, without the need of enzyme engineering, (ii) target specificity is very high, determined by a 20 bp RNA-DNA interaction with low probability of non-target recognition, (iii) almost any sequence can be targeted, the only requirement being the presence of an adjacent NGG sequence, (iv) almost any mutation in the NGG sequence, as well as mutations in the seed sequence of the protospacer, eliminates targeting.
We show that CRISPR-assisted genome engineering works not only in highly recombinogenic bacteria such as S. pneumoniae, but also in other organisms, the requirement being the possibility to introduce plasmids. We tested such a plasmid-based system in E. coli and combined it with the recombineering of mutagenic oligonucleotides. To use this methodology in other microbes where recombineering is not a possibility, the editing template could also be introduced in a plasmid. In addition, because accumulated evidence indicates that the activation of CRISPR ‘auto-immunity’ leads to cell death in many bacteria 23, 32, 33 and archaea34, 35, it is possible to envision the use of endogenous CRISPR-Cas systems for editing purposes.
In both S. pneumoniae and E. coli, we observed that although editing is facilitated by a co-selection of transformable cells and a small induction of recombination in the target site by Cas9 cleavage, the mechanism that contributes the most to editing is the selection against non-edited cells by CRISPR targeting. Therefore the major limitation of the method is the presence of a background of cells that escape CRISPR auto-immunity and lack the desired mutation. We showed that these ‘escapers’ arise primarily through the deletion of the targeting spacer, presumably after the recombination of the repeat sequences that flank the targeting spacer. Future improvements will focus on the engineering of flanking sequences that can still support the biogenesis of functional crRNAs but that are sufficiently different from one another to eliminate recombination. Alternatively, the direct transformation of chimeric crRNAs7 could be explored. In the particular case of E. coli, the construction of the CRISPR-Cas system is not possible if this organism is also used as a cloning host. We solved this issue by placing Cas9 and the tracrRNA on a different plasmid than the CRISPR array. In addition, the engineering of an inducible system could circumvent this limitation.
Although new DNA synthesis technologies provide the ability to cost-effectively create any sequence with a high throughput, it remains a challenge to integrate synthetic DNA in living cells to create functional genomes. Recently, the co-selection MAGE strategy was shown to improve the mutation efficiency of recombineering by selecting a subpopulation of cells that has an increased probability to achieve recombination at or around a given locus36. In this method, the introduction of selectable mutations is used to increase the chances of generating nearby non-selectable mutations. As opposed to the indirect selection provided by this strategy, the use of CRISPR targeting makes it possible to directly select for the desired mutation and to recover it with a high efficiency. These technologies add to the toolbox of genetic engineers, and together with DNA synthesis, they are likely to substantially advance both our ability to decipher gene function and to manipulate organism for biotechnological purposes. While our work was in review, two studies reported the CRISPR-assisted engineering of mammalian genomes37, 38. It is expected that these crRNA-directed genome editing technologies will have broad implications on basic and medical sciences.
Online Methods
Strains and culture conditions
S. pneumoniae strain R639 was provided by Dr. Alexander Tomasz. Strain crR6 was generated in a previous study23. Liquid cultures of S. pneumoniae were grown in THYE medium (30g/l Todd-Hewitt agar, 5 g/l yeast extract). Cells were plated on tryptic soy agar (TSA) supplemented with 5 % defibrinated sheep blood. When appropriate, antibiotics were added as followings: kanamycin (400 µg/ml), chloramphenicol (5 µg/ml), erythromycin (1 µg/ml) streptomycin (100 µg/ml) or spectinomycin (100 µg/ml). Measurements of β-galactosidase activity were made using the Miller assay as previously described27.
E. coli strains MG1655 and HME63 (derived from MG1655, Δ(argF-lac) U169 λ cI857 Δcro-bioA galK tyr 145 UAG mutS<>amp)31 were provided by Jeff Roberts and Donald Court, respectively. Liquid cultures of E. coli were grown in LB medium (Difco). When appropriate, antibiotics were added as followings: chloramphenicol (25 µg/ml), kanamycin (25 µg/ml) and streptomycin (50 µg/ml).
S. pneumoniae transformation
Competent cells were prepared as described previously23. For all genome editing transformations, cells were gently thawed on ice and resuspended in 10 volumes of M2 medium supplemented with 100 ng/ml of competence-stimulating peptide CSP1 40, and followed by addition of editing constructs (editing constructs were added to cells at a final concentration between 0.7 ng/µl to 2.5 µg/ul). Cells were incubated 20 min at 37 °C before the addition of 2 µl of targeting constructs and then incubated 40 min at 37 °C. Serial dilutions of cells were plated on the appropriate medium to determine the colony forming units (cfu) count.
E. coli Lambda-red recombineering
Strain HME63 was used for all recombineering experiments. Recombineering cells were prepared and handled according to a previously published protocol6. Briefly, a 2 ml overnight culture (LB medium) inoculated from a single colony obtained from a plate was grown at 30 °C. The overnight culture was diluted 100-fold and grown at 30 °C with shaking (200rpm) until the OD600 is from 0.4–0.5 (approximately 3 hrs). For Lambda-red induction, the culture was transferred to a 42 °C water bath to shake at 200rpm for 15 min. Immediately after induction, the culture was swirled in an ice-water slurry and chilled on ice for 5–10 min. Cells were then washed and aliquoted according to the protocol. For electro-transformation, 50 µl of cells were mixed with 1mM of salt-free oligos (IDT) or 100–150 ng of plasmid DNA (prepared by QIAprep Spin Miniprep Kit, Qiagen). Cells were electroporated using 1mm Gene Pulser cuvette (Bio-rad) at 1.8kV and were immediately resuspended in 1 ml of room temperature LB medium. Cells were recovered at 30 °C for 1–2 hrs before being plated on LB agar with appropriate antibiotic resistance and incubated at 32 °C overnight.
Preparation of S. pneumoniae genomic DNA
For transformation purposes, S. pneumoniae genomic DNA was extracted using the Wizard Genomic DNA Purification Kit, following instructions provided by the manufacturer (Promega). For genotyping purposes, 700ul of overnight S. pneumoniae cultures were pelleted, resuspended in 60ul of lysozyme solution (2mg/ml) and incubated 30min at 37°C. The genomic DNA was extracted using QIAprep Spin Miniprep Kit (Qiagen).
Strain construction
All primers used in this study are provided in table S2. To generate S. pneumoniae crR6M, an intermediate strain, LAM226, was made. In this strain the aphA-3 gene (providing kanamycin resistance) adjacent to the CRISPR array of S. pneumoniae crR6 strain was replaced by a cat gene (providing chloramphenicol resistance). Briefly, crR6 genomic DNA was amplified using primers L448/L444 and L447/L481, respectively. The cat gene was amplified from plasmid pC194 (ref. 41) using primers L445/L446. Each PCR product was gel-purified and all three were fused by SOEing PCR42 with primers L448/L481. The resulting PCR product was transformed into competent S. pneumoniae crR6 cells and chloramphenicol-resistant transformants were selected. To generate S. pneumoniae crR6M, S. pneumoniae crR6 genomic DNA was amplified by PCR using primers L409/L488 and L448/L481, respectively. Each PCR product was gel-purified and they were fused by SOEing PCR with primers L409/L481. The resulting PCR product was transformed into competent S. pneumoniae LAM226 cells and kanamycin-resistant transformants were selected.
To generate S. pneumoniae crR6Rc, S. pneumoniae crR6M genomic DNA was amplified by PCR using primers L430/W286, and S. pneumoniae LAM226 genomic DNA was amplified by PCR using primers W288/L481. Each PCR product was gel-purified and they were fused by SOEing PCR with primers L430/L481. The resulting PCR product was transformed into competent S. pneumoniae crR6M cells and chloramphenicol-resistant transformants were selected.
To generate S. pneumoniae crR6Rk, S. pneumoniae crR6M genomic DNA was amplified by PCR using primers L430/W286 and W287/L481, respectively. Each PCR product was gel-purified and they were fused by SOEing PCR with primers L430/L481. The resulting PCR product was transformed into competent S. pneumoniae crR6Rc cells and kanamycin-resistant transformants were selected.
To generate JEN37, S. pneumoniae crR6Rk genomic DNA was amplified by PCR using primers L430/W356 and W357/L481, respectively. Each PCR product was gel-purified and they were fused by SOEing PCR with primers L430/L481. The resulting PCR product was transformed into competent S. pneumoniae crR6Rc cells and kanamycin-resistant transformants were selected.
To generate JEN38, R6 genomic DNA was amplified using primers L422/L461 and L459/L426, respectively. The ermAM gene (specifying erythromycin resistance) was amplified from plasmid pFW15 43 using primers L457/L458. Each PCR product was gel-purified and all three were fused by SOEing PCR with primers L422/L426. The resulting PCR product was transformed into competent S. pneumoniae crR6Rc cells and erythromycin-resistant transformants were selected.
S. pneumoniae JEN53 was generated in two steps. First JEN43 was constructed as illustrated in Figure S5. JEN53 was generated by transforming genomic DNA of JEN25 into competent JEN43 cells and selecting on both chloramphenicol and erythromycin.
To generate S. pneumoniae JEN62, S. pneumoniae crR6Rk genomic DNA was amplified by PCR using primers W256/W365 and W366/L403, respectively. Each PCR product was purified and ligated by Gibson assembly. The assembly product was transformed into competent S. pneumoniae crR6Rc cells and kanamycin-resistant transformants were selected.
Plasmid construction
pDB97 was constructed through phosphorylation and annealing of oligonucleotides B296/B297, followed by ligation in pLZ12spec (ref. 44) digested by EcoRI/BamHI. We fully sequenced pLZ12spec and deposited its sequence in genebank (accession: KC112384).
pDB98 was obtained after cloning the CRISPR leader sequence was cloned together with a repeat-spacer-repeat unit into pLZ12spec. This was achieved through amplification of crR6Rc DNA with primers B298/B320 and B299/B321, followed by SOEing PCR of both products and cloning in pLZ12spec with restriction sites BamHI/EcoRI. In this way the spacer sequence in pDB98 was engineered to contain two BsaI restriction sites in opposite directions that allow for the scar-less cloning of new spacers.
pDB99 to pDB108 were constructed by annealing of oligonucleotides B300/B301 (pDB99), B302/B303 (pDB100), B304/B305 (pDB101), B306/B307 (pDB102), B308/B309 (pDB103), B310/B311 (pDB104), B312/B313 (pDB105), B314/B315 (pDB106), B315/B317 (pDB107), B318/B319 (pDB108), followed by ligation in pDB98 cut by BsaI.
The pCas9 plasmid was constructed as follow. Essential CRISPR elements were amplified from Streptococcos pyogenes SF370 genomic DNA with flanking homology arms for Gibson Assembly. The tracrRNA and Cas9 were amplified with oligos HC008 and HC010. The leader and CRISPR sequences were amplified HC011/HC014 and HC015/HC009, so that two BsaI type IIS sites were introduced in between two direct repeats to facilitate easy insertion of spacers.
pCRISPR was constructed by subcloning the pCas9 CRISPR array in pZE21-MCS1 through amplification with oligos B298+B299 and restriction with EcoRI and BamHI. The rpsL targeting spacer was cloned by annealing of oligos B352+B353 and cloning in the BsaI cut pCRISPR giving pCRISPR::rpsL.
Generation of targeting and editing constructs
Targeting constructs used for genome editing were made by Gibson assembly45 of Left PCRs and Right PCRs (table S3). Editing constructs were made by SOEing PCR 42 fusing PCR products A (PCR A), PCR products B (PCR B) and PCR products C (PCR C) when applicable (table S3). The CRISPR::Ø and CRISPR::ermAM(stop) targeting constructs were generated by PCR amplification of JEN62 and crR6 genomic DNA respectively, with oligos L409 and L481.
Generation of targets with randomized PAM or protospacer sequences
The 5 nucleotides following the spacer 1 target were randomized through amplification of R68232.5 genomic DNA with primers W377/ L426. This PCR product was then assembled with the cat gene and the srtA upstream region that were amplified from the same template with primers L422/W376. 80 ng of the assembled DNA was used to transform strains R6 and crR6.
Samples for the randomized targets were prepared using the following primers: B280-B290/L426 to randomize bases 1–10 of the target and B269-B278/L426 to randomize bases 10–20. Primers L422/B268 and L422/B279 were used to amplify the cat gene and srtA upstream region to be assembled with the first and last 10 PCR products respectively. The assembled constructs were pooled together and 30 ng was transformed in R6 and crR6. After transformation, cells were plated on chloramphenicol selection. For each sample more than 2×105 cells were pooled together in 1 ml of THYE and genomic DNA was extracted with the Promega Wizard kit. Primers B250/B251 were used to amplify the target region. PCR products were tagged and run on one Illumina MiSeq paired-end lane using 300 cycles.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vincent Fischetti and Chad Euler for plasmid pLZ12spec, Don Court for the HME63 strain and Jeff Roberts for the MG1655 strain. D.B. is supported by a Harvey L. Karp Discovery Award and the Bettencourt Schuller Foundation. D.C. is supported by the Medical Scientist Training Program. F.Z. is supported by a NIH Director’s Pioneer Award (DP1MH100706), Transformative R01, the Keck, McKnight, Gates, Damon Runyon, Searle Scholars, Klingenstein, and Simons Foundations, Bob Metcalfe, Mike Boylan, and Jane Pauley. L.A.M is supported by the Searle Scholars Program, the Rita Allen Scholars Program, a Irma T. Hirschl Award and a NIH Director’s New Innovator Award (1DP2AI104556-01).
Footnotes
The authors have no conflicting financial interests.
AUTHOR CONTRIBUTIONS
WJ, DB and LAM designed the experiments; WJ, DB and DC performed experiments; WJ, DB, FZ and LAM wrote the paper.
Supplementary Materials:
Supplementary text
Figures S1-S11
Tables S1-S3
REFERENCES
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 3.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 4.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 2001;67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 9.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 10.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends. Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl. Acad. Sci. U.S.A. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012 doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrangou R. RNA-mediated programmable DNA cleavage. Nat. Biotechnol. 2012;30:836–838. doi: 10.1038/nbt.2357. [DOI] [PubMed] [Google Scholar]
- 21.Brouns SJ. Molecular biology. A Swiss army knife of immunity. Science. 2012;337:808–809. doi: 10.1126/science.1227253. [DOI] [PubMed] [Google Scholar]
- 22.Carroll D. A CRISPR Approach to Gene Targeting. Mol. Ther. 2012;20:1658–1660. doi: 10.1038/mt.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011 doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiedenheft B, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U.S.A. 2011 doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zahner D, Hakenbeck R. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 2000;182:5919–5921. doi: 10.1128/jb.182.20.5919-5921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motamedi MR, Szigety SK, Rosenberg SM. Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosaka T, et al. The novel mutation K87E in ribosomal protein S12 enhances protein synthesis activity during the late growth phase in Escherichia coli. Mol. Genet. Genomics. 2004;271:317–324. doi: 10.1007/s00438-004-0982-z. [DOI] [PubMed] [Google Scholar]
- 31.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R, Qimron U. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J. Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer S, et al. An archaeal immune system can detect multiple Protospacer Adjacent Motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012;287:33351–33363. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudbergsdottir S, et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang HH, et al. Genome-scale promoter engineering by coselection MAGE. Nat Methods. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013 In press. [Google Scholar]
- 38.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013 doi: 10.1126/science.1232033. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoskins J, et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton RM. In Vitro Recombination and Mutagenesis of DNA : SOEing Together Tailor-Made Genes. Methods Mol. Biol. 1993;15:251–261. doi: 10.1385/0-89603-244-2:251. [DOI] [PubMed] [Google Scholar]
- 43.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 44.Husmann LK, Scott JR, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infection and immunity. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.