Abstract
CD1 proteins evolved to present diverse lipid antigens to T cells. In comparison to MHC proteins, CD1 proteins exhibit minimal allelic diversity as a result of non-synonymous single nucleotide polymorphisms (SNPs). However, it is unknown if common SNPs in gene regulatory regions affect CD1 expression and function. We report surprising diversity in patterns of inducible CD1a expression on human dendritic cells (DCs), spanning the full range from undetectable to high density, a finding not seen with other CD1 isoforms. CD1a-deficient DCs failed to present mycobacterial lipopeptide to T cells but had no defects in endocytosis, cytokine secretion, or expression of co-stimulatory molecules after LPS treatment. We identified a SNP in the 5’ untranslated region (rs366316) that was common and strongly associated with low CD1a surface expression and mRNA levels (p=0.03 and p=0.001, respectively). Using a CD1a promoter-luciferase system in combination with mutagenesis studies, we found that the polymorphic allele reduced luciferase expression by 44% compared to the wild-type variant (p<0.001). Genetic regulation of lipid antigen presentation by varying expression on human DCs provides a mechanism for achieving population level differences in immune responses despite limited structural variation in CD1a proteins.
Introduction
Major histocompatibility complex (MHC) genes are among the most polymorphic in the human genome. For example, the human MHC Class I locus consists of three genes, HLA-A, HLA-B, and HLA-C, each of which is represented by more than one thousand variant alleles(1). This variation is important to achieve population level diversity in the adaptive immune response to pathogens, which co-evolve with their human hosts. Within an individual, the breadth of the adaptive immune response is further enhanced by the ability of T cells to rearrange and combine genes encoding antigen receptors. Therefore, MHC allelic diversity and T cell receptor (TCR) sequence diversity ensure the ability of T cells to recognize a wide array of peptide antigens.
T cells have also evolved the capacity to recognize diverse lipids in the context of CD1 proteins(2, 3). CD1 heavy chains are homologous to MHC Class I and bind non-covalently to β-2 microglobulin(4). The human CD1 locus contains five genes (CD1A, CD1B, CD1C, CD1D, and CD1E) clustered on chromosome 1(5). CD1a, CD1b, CD1c, CD1d, and CD1e proteins differ in which lipid antigens they bind, their patterns of expression on cells, and trafficking within cells(6). Notably, the mouse and rat genomes contain only orthologs of human CD1D(7-9), so mice provide a convenient experimental model for CD1d only. The other CD1 proteins are largely unexplored in the context of human immunology.
Despite their structural homology to MHC Class I heavy chain genes, CD1 genes exhibit limited allelic diversity as a result of non-synonymous polymorphisms. In early studies, Southern blots revealed largely conserved CD1 sequences among human and inbred mouse strains(5, 7, 10). Human sequence diversity in exon 2 and exon 3, which code for the alpha-1 and alpha-2 lipid-antigen binding domains, respectively, is limited with only two variant alleles in CD1a, CD1d and CD1e, and zero variants in CD1b and CD1c(11, 12). Four other rare variants of CD1e have also been reported in single individuals(13, 14). More recent data derived from The International Haplotype Mapping Project (www.hapmap.org) and 1000 Genomes Project (www.1000genomes.org) reveal that common genetic variation exists in the CD1 locus, but this variation is enriched outside of protein coding exons and thus does not alter protein structure and lipid binding. Taken together, these studies suggest that modulation in the T cell response to lipids is not achieved by the diversity of CD1 coding region alleles but might be influenced by genetic variation in non-coding regions of the gene.
The CD1a protein presents lipopeptide antigens to T cells and is expressed on Langerhans cells, thymocytes, and certain subsets of dendritic cells(6, 15). Data regarding the importance of CD1a function for human health are limited. Individual T cell clones that recognize host or pathogen derived lipids have been described and provide isolated examples of CD1a-presented lipids(16-18). Mammalian sulfatide is recognized by a CD4+ T cell clone, which produces TNF-α and shows a Th-1 phenotype upon TCR engagement(18, 19). Mycobacterial dideoxymycobactin is recognized by a CD8+ T cell clone, which produces IFN-γ and IL-2 upon stimulation and is capable of lysing cells infected with M. tuberculosis(4, 17, 20). More recently, CD1a-autoreactive T cells have been identified in the blood of human donors, in some cases with precursor frequencies as high as 0.1 – 10% of all peripheral blood T cells(21, 22). These studies have begun to reveal population-level differences in CD1a-restricted T cell responses, but the immunologic mechanisms underlying this variation remain unexplored.
Here, we report that CD1a-deficiency on in-vitro derived DCs is a common phenotype, detected in 15 percent of study subjects. A common polymorphism in the 5’ UTR of CD1a is strongly associated with both low surface expression and mRNA levels, and this polymorphism directly regulates gene expression in a promoter-luciferase assay. These studies provide a transcriptional regulatory mechanism for population level differences in T-cell responses to lipids antigens that does not depend on non-synonymous allelic diversity in the CD1 locus.
Materials and Methods
Human Subjects
The Seattle study group consisted of 122 healthy individuals who provided blood for genotyping, and 33 also provided blood for functional studies. In this cohort, 68 (55%) were female, and self-described ethnic composition was 90 (73%) Caucasian, 27 (22%) Asian, and 5 (5%) Other.
Ethics
All protocols were approved by human subject review committees at the University of Washington.
SNP Selection for Genotyping
We used data from the International Haplotype Mapping Project (www.hapmap.org, version 3, release 2) to select SNPs within ten kilobases of CD1a from Europeans of Caucasian descent (CEU). Haplotype-tagging SNPs with a minimum allele frequency of 4% and R2 cutoff of 0.80 for linkage disequilibrium were identified using Haploview v4.2 (www.broad.mit.edu/haploview).
Genomic techniques
Genomic DNA was prepared from saliva (Genotek) or peripheral blood (Qiagen). Multiplex genotyping was performed using allele-specific primer extension on the MassARRAY (Sequenom) platform(23, 24). Single SNP genotyping was performed using TaqMan SNP Genotyping Assay (Applied Biosystems). Genotypes were confirmed in a subset of individuals by DNA sequencing or by genotyping on an alternate platform.
Quantitative RT-PCR
RNA was extracted from DCs by Trizol (Invitrogen) and ethanol precipitation. Single-stranded cDNA was generated using Multiscribe reverse transcriptase (Invitrogen). Real time PCR was performed on StepOnePlus Real Time PCR System (ABI) using ABI primer probe sets CD1A-FAM (Hs00233332_m1), CD1C-FAM (Hs00233332_m1), and GAPDH-JOE (402869).
In-vitro generation of dendritic cells and phenotyping
Monocytes were isolated from peripheral blood mononuclear cells by positive selection using CD14 microbeads (Miltenyi Biotech) and magnetic cell separation. Monocytes were incubated in RPMI + 10% fetal calf serum (GIBCO) supplemented with L-glutamine and GM-CSF (100ng/ml) + IL-4 (20ng/ml) (Peprotech) for three days to generate DCs. For experiments in Figure 2, monocytes were also cultured with IL-4 from R&D Systems at a concentration of 100ng/ml or cultured for six days. Differentiation of monocytes into DCs was confirmed by staining with CD14-FITC (BioLegend). To examine surface expression of CD1 isoforms, cells were stained with antibodies against CD1 in PBS supplemented with 0.2% bovine serum albumin and 0.09% sodium azide. Antibodies were CD1a-PE (BioLegend), CD1b-FITC (BioLegend), CD1c-PE (BioLegend). Secondary staining for CD1a was performed by first blocking cells with PBS + 5% hAB serum and then staining with OKT6 (ATCC) followed by PE Goat Anti-Mouse Ig (BD). Median fluorescence intensity (MFI) was calculated for each CD1 isoform and corrected by subtraction of isotype control. To assess intracellular CD1a, cryopreserved DCs were thawed, washed, and first stained with the LIVE/DEAD Fixable Dead Cell Stain Kit for 633nm Excitation (Invitrogen) before being treated with FACS Perm II (BD) for 10 minutes. Cells were washed and then stained with phalloidin-FITC (Life Technologies), CD1a-PE, and CD1a-APC.
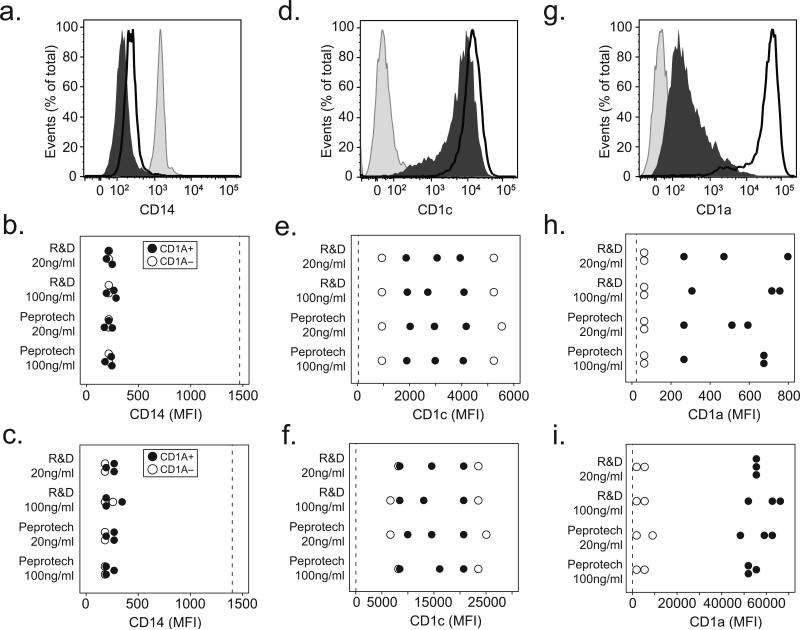
Figure 2. CD1a-deficiency does not reverse with increased IL-4 concentrations or extended time in culture.
Human monocytes were cultured with IL-4 sourced from either Peprotech or R&D Systems at a concentration of either 20ng/ml or 100ng/ml for either three or six days. Cells were harvested and stained for flow cytometry with fluorescently conjugated antibodies against CD14, CD1a, or CD1c. (a) Shown are expression profiles from unstimulated monocytes (shaded histogram) or cells cultured for six days from a CD1a-sufficient donor (dark line), and a CD1a-deficient donor (dark histogram) for CD14. Results from five subjects are combined and shown as the median fluorescence intensity (MFI) of CD14 after (b) three days of culture or (c) six days of culture. (d) Representative CD1c expression from six day cultured cells or combined data showing CD1c MFI after (e) three days or (f) six days of culture. (g) Representative CD1a expression from six day cultured cells or combined data for CD1a after (h) three days or (i) six days of culture. Expression levels on unstimulated monocytes (dotted line) are shown for comparison. All data are representative of two independent experiments.
Data was acquired on FACS Canto (BD) equipped with 488nm and 633nm lasers and analyzed using FlowJo v9.3.2 (TreeStar Inc.).
Cellular assays
To test the capacity of DCs to present lipid antigen to T cells, DCs were generated from cryopreserved monocytes and plated in triplicate in co-culture with T cell clone CD8-2 (E:T = 1:2) and the mycobacterial lipopeptide dideoxymycobactin(20). Synthetic dideoxymycobactin(25) was dried under a sterile nitrogen stream, sonicated into media and added to a final concentration of 10 nM. Cultures were incubated overnight, and supernatants were harvested for IFN-γ ELISA. To test the capacity of DCs to endocytose particulate antigen, DCs were incubated with FITC-conjugated paraformaldehyde fixed E. coli BioParticles (Molecular Probes) at a concentration of 10 mg/ml, FITC-conjugated bovine serum albumin (Molecular Probes) at a concentration of 10 mg/ml, or FITC-conjugated dextran (Sigma) at a concentration of 100 mg/ml for one hour at either 37°C or 4°C as a control. Median fluorescence intensity (MFI) was obtained and fold induction was calculated as MFI37C/MFI4C. To test the capacity of DCs to mature after stimulation, DCs were incubated in the presence of Ultra Pure LPS (List Biological Labs, Inc.) 10ng/ml or media overnight. Expression of co-stimulatory markers was assessed by flow cytometry and fold induction for each marker was calculated as MFILPS/MFIMedia. Antibodies were CD40-APC (BioLegend), CD80-PE/Cy7 (BioLegend), CD83-FITC (BioLegend), CD86-PerCp/Cy5.5 (BioLegend), HLA-DR-APC/Cy7 (BioLegend).
Statistics
Statistical analyses were performed using Stata Statistical Software: Release 11 (StataCorp LP, College Station, TX). Simple linear correlation between continuous variables was described using the Pearson ‘r’ correlation coefficient. The non-parametric Kruskal-Wallis or Mann-Whitney test was used to compare continuous variables stratified by genotypes. P-values for two-tailed hypothesis testing are reported except where specifically noted. Function ‘pwld’ was used to calculate R2 measurements of linkage disequilibrium between polymorphisms.
Cloning and Mutagenesis
Genomic DNA was isolated from whole blood using QIA-Amp (Qiagen, Inc.). We separately amplified 998 bases and 555 bases proximal to the CD1a translation start site using an Eppendorf Mastercycler Gradient 5331. We performed mutagenesis using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). Two sets of primers were used: the first for mutating rs366316 from C to T (C→T), and the second for mutating rs366316 from T to C (T→C). Primers and PCR parameters are detailed in Table 1.
Table 1.
List of primers used to perform cloning and mutagenesis
| Construct | Primer | Sequence | Tm | Cycles |
|---|---|---|---|---|
| 998 | Forward Reverse |
5′–CTGTATGGAACTAACACTCCAA–3′ 5′–CATTTGCAGATGTTATTTCC–3′ |
59 C | 35 |
| 555 | Forward Reverse |
5'–GGTGTTAGTTTGTACTGTCGC–3' 5'–CATTTGCAGATGTTATTTCC–3' |
65 C | 35 |
| C→T Mutant | Forward Reverse |
5'–CCAGAGGGAAATGAG[A]GACTGAG–3' 5'–GATGCCTACTCAGTC[T]CTCATTT–3' |
55 C | 18 |
| T→C Mutant | Forward Reverse |
5'–CCAGAGGGAAATGAG[G]GACTGAG–3' 5'–GATGCCTACTCAGTC[C]CTCATTT–3' |
55 C | 18 |
Cloning was performed using the pCR2.1 TOPO TA Cloning Kit according to manufacturer's instructions (Invitrogen). DNA was extracted from bacterial pellets using the QIAprep Spin Miniprep Kit (Qiagen). Sequencing was performed on an Applied Biosystems 3730XL DNA Analyzer.
Constructing pGL4 Expression Vectors
Wild type and mutated 5’UTR sequences as well as the minimal promoter from pGL4.14[luc2/minimal/Hygro] (Promega) were isolated by digestion with XhoI and HindIII restriction endonucleases (New England BioLabs) and gel purification (QIAquick Gel Extraction Kit). Ligation was performed with T4 Ligase (New England BioLabs) using equal amounts of insert and vector. Endotoxin free plasmid DNA of these constructs and of the control vectors pGL4.51 [luc2/CMV/Neo], pGL4.14 [luc2/minimal/Hygro], and pGL4.73 [hRluc/SV40] (Promega) was generated using the NucleoBond Xtra Midi EF plasmid preparation kits (Macherey-Nagel).
Transfection and Luminometry
HEK293T cells were plated in 96-well flat bottom plates (BD Falcon) at a density of 10,000 cells/well in DMEM (GIBCO) supplemented with 10% Fetal Bovine Serum (GIBCO). The next day, cells were transfected with pGL4 expression vectors using X-tremeGENE HP DNA Transfection Reagent (reagent to plasmid DNA ratio 3:1 and 2:1) (Roche Applied Science). After 24 hours, cells were lysed using 50ul Passive Lysis Buffer (Promega), and luminescence from firefly (luc2) and Renilla (hRluc) luciferase was assessed using the Dual Luciferase Reporter Assay System (Promega). Luminometry was performed on a Veritas Microplate Luminometer (Turner Biosystems) and values are reported as the ratio of firefly to Renilla luciferase (RLU).
Phenotyping of peripheral blood dendritic cells
Whole blood collected in ACD tubes from healthy donors was stained to determine the level of CD1a and CD1c expression on peripheral blood dendritic cells using a previously described method for staining(26). The cells were gated to distinguish dendritic cell populations by first gating lymphocytes and monocytes using CD45 and side scatter. Then, monocytes were excluded by gating CD14 negative cells. Dendritic cells were distinguished by gating HLA-DR positive and CD3/CD19/CD20/CD56 negative cells and subsequently excluding any possible contaminating inflammatory monocytes or NK cells by excluding any CD16 positive cells. Plasmacytoid and myeloid dendritic cells were then distinguished by their CD11c and CD123 expression, and CD1a and CD1c expression were examined for each subset as shown.
Results
We measured inducible CD1 surface expression using flow cytometry on monocyte-derived dendritic cells (DCs) generated after activation with GM-CSF and IL-4. Initial screening of blood bank donors for induction of CD1a, CD1b, and CD1c revealed two donors that induced CD1b and CD1c but not CD1a, a reproducible finding that was not attributable to media or culture conditions such as the lot of fetal calf serum or density of cells in culture (data not shown). Therefore, we undertook a more formal analysis of CD1a induction on monocytes in 19 healthy adults. We found more than 100-fold variability in the absolute expression intensity of CD1a among donors, a pattern that was not linked to variable expression of the other two inducible CD1 isoforms, CD1b and CD1c (Fig. 1a, 1b and Supplementary Fig. 1).
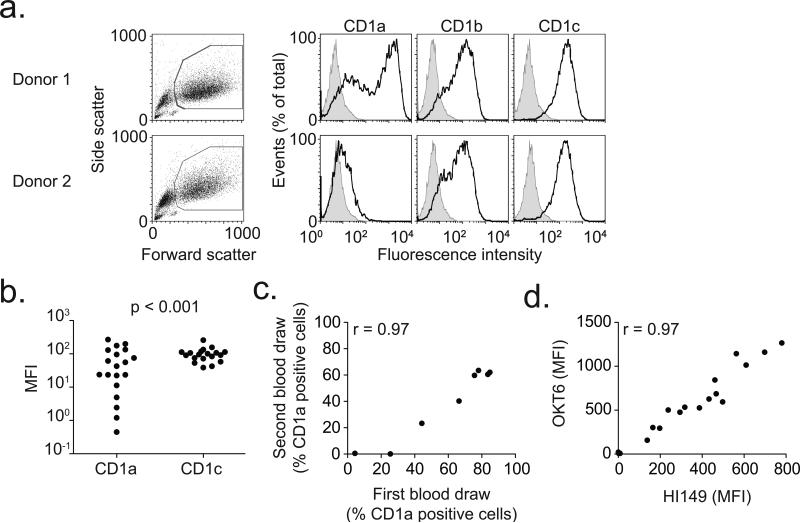
Figure 1. Deficiency of CD1a on human dendritic cells.
(a) Viable monocyte-derived dendritic cells, identified by high forward and side scatter profiles, were stained with fluorescently-conjugated antibodies against CD1a, CD1b, and CD1c (dark lines) as well as isotype control antibody (shaded histogram). Shown are representative plots from two donors. (b) Median fluorescence intensity (MFI) of CD1a and CD1c is shown for 19 healthy blood donors. In four donors, CD1a MFI is less than 10. P-value reflects Bartlett's test for non-homogeneity of variances. (c) Simple linear correlation between CD1a staining results of sequential blood draws for eight subjects. Data are represented as % positive cells rather than MFI to adjust for temporal variation in flow cytometry calibration. (d) Simple linear correlation of staining between two antibodies (OKT6 and HI149) that bind CD1a.
Several lines of evidence indicated that donor-specific factors contributed to varied CD1a surface density rather than the conditions of culture or measurement(27, 28). First, CD1b and CD1c, two other inducible forms of CD1, showed high density among all donors (Fig. 1b and Supplementary Fig. 1). Second, the high or low levels of CD1a were reproduced upon repeated blood collection (r=0.97, Fig. 1c). Third, low expression of CD1a was consistent over at least three experiments and seen with two monoclonal antibodies, OKT6 and HI149 (r=0.97, Fig. 1d), suggesting that low staining was not a result of differences in epitope binding. These data identify a CD1a-specific effect in which we observed high variance of CD1a surface density on DCs among all donors and identified three donors (15%) without detectable CD1a surface protein. We herein refer to the phenotype of extremely low or absent CD1a surface staining as ‘CD1a-deficiency.’
Previous studies suggest that the induction of CD1a is particularly sensitive to the presence of IL-4 and time in culture (29, 30). Therefore, we examined the effect of two sources and two concentrations of IL-4 as well as three days and six days of culture. Based on suppression of CD14 expression, we confirmed that monocytes differentiated into DCs in all conditions tested (Figure 2a, 2b and 2c). Similarly, we observed induction of CD1c in all donors and in all conditions tested (Figure 2d, 2e, and 2f). Among CD1a-sufficient donors, we confirmed increased CD1a expression after six days in culture compared to three days, but there was no effect of the source or concentration of IL-4. However, among CD1a-deficient donors, neither the source of IL-4, concentration of IL-4, nor extended time in culture could reverse the low expression of CD1a (Figure 2g, 2h and 2i). Together, these data further confirm that CD1a-deficiency is a donor-specific phenomenon that is independent of culture conditions.
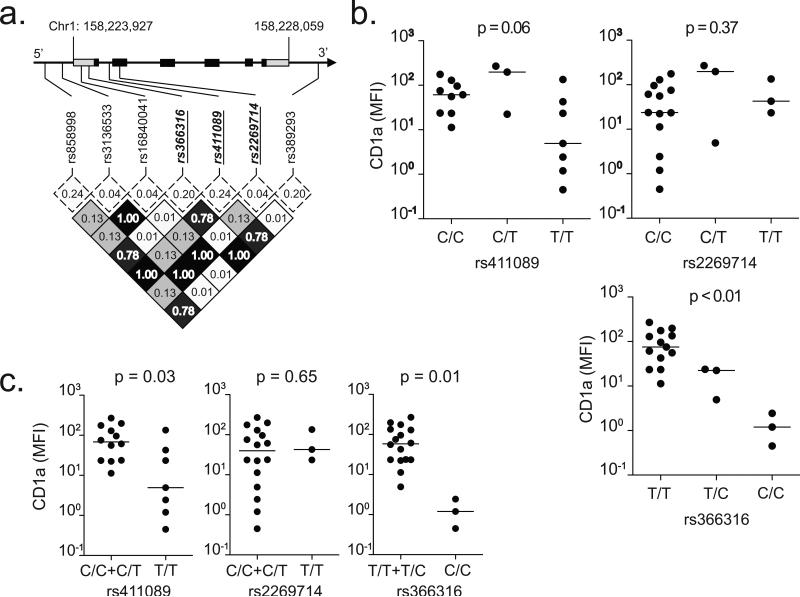
The host-specific nature of the effect led us to hypothesize that this phenotype was genetic. Because the expression of CD1b and CD1c did not vary substantially among donors, we considered defects in β-2 microglobulin unlikely and instead focused on potential defects near or within the CD1a heavy chain. There are seven common SNPs located near the CD1a heavy chain. Only one polymorphism codes for an amino acid substitution, and the rest are located in regulatory or non-coding regions (Table 2). Because of the high degree of linkage disequilibrium among these SNPs, all of the major haplotypes of an individual are defined by only three SNPs (Fig. 3a). When we stratified expression levels by genotype, rs411089 and rs366316 but not rs2269714 were associated with CD1a-deficiency (Fig. 3b). Of note, rs2269714 codes for one of the two most common allelic variants of CD1a. Our data confirm published studies showing rs2269714 is not associated with defects in surface expression on transfected cells and extends this finding to include DCs(31). The association appeared strongest with the minor homozygous genotypes, so we performed a recessive model analysis and found that the minor homozygous genotypes of rs411089 and rs366316 were associated with low CD1a expression (p=0.03 and p=0.01, respectively) (Fig. 3c). By contrast, there was no association between rs2269714 and CD1a expression (p=0.65, Fig. 3c), and there was no difference in CD1b or CD1c expression when stratified by any SNP (Supplemental Fig. 2a and 2b). Thus, rs411089 and rs366316 are genetic markers for CD1a-deficiency.
Table 2.
List of single nucleotide polymorphisms (SNPs) in CD1A. SNPs were tabulated from dbSNP (www.ncbi.nlm.nih.gov/SNP) based on human genome assembly GRCh37.p5, build 37.3. SNPs with a minor allele frequency greater than 4% in at least one HapMap population were studied.
| SNP Name | Position on Chromosome 1 | Alleles | Location in CD1A | Effect |
|---|---|---|---|---|
| rs858998 | 158220575 | C or T | Promoter | Unknown |
| rs3136533 | 158223060 | G or A | Promoter | Unknown |
| rs16840041 | 158224059 | G or A | 5' UTR | Unknown |
| rs366316 | 158224282 | A or G | 5' UTR | Unknown |
| rs411089 | 158224825 | C or T | Intron 1 | Unknown |
| rs2269714 | 158224904 | C or T | Exon 2 | Thr -> Ile |
| rs389293 | 158228392 | G or A | 3' | Unknown |
Figure 3. Association between CD1a SNPs and expression.
(a) Linkage disequilibrium plot of SNPs in CD1a coding region ± 10kB flanking regions among CEU population from HapMap. CD1a spans 4132 bases on chromosome 1 and consists of six exons (black squares) and two untranslated regions (gray squares). Minor allele frequencies (dotted box) and linkage disequilibrium as measured by R2 values (shaded box) are indicated. Three haplotype-tagging SNPs selected for genotyping are emphasized in bold italic underline. (b) CD1a median fluorescence intensity (MFI) stratified by SNP genotypes. The bar indicates the median value. The nonparametric Kruskal-Wallis test was used to determine statistical significance for a genotypic model. (c) Recessive model analysis combines SNP genotypes ‘AA’ and ‘Aa.’
Having identified donors with CD1a-deficiency on in-vitro derived DCs, we next examined the expression of CD1a on peripheral blood DCs ex-vivo. It had been previously reported that CD1a is normally expressed on a subset of CD11c+ peripheral blood myeloid DCs(32), though later work revealed that the antibody clone actually bound to CD1b/c(33). Therefore, we first used K562 cells that had been stably transfected to express CD1a to validate the specificity of the CD1a staining antibody (Supplementary Fig. 3a). We stained fresh whole blood and examined expression of CD1a and CD1c on plasmacytoid and myeloid DCs (Supplementary Fig 3b). As expected, we were able to detect CD1c on the surface of myeloid DCs but not on plasmacytoid DCs. However, we were unable to detect CD1a on either DC subset indicating CD1a is not endogenously expressed on unstimulated circulating DCs. Further experiments were therefore conducted on monocyte-derived DCs.
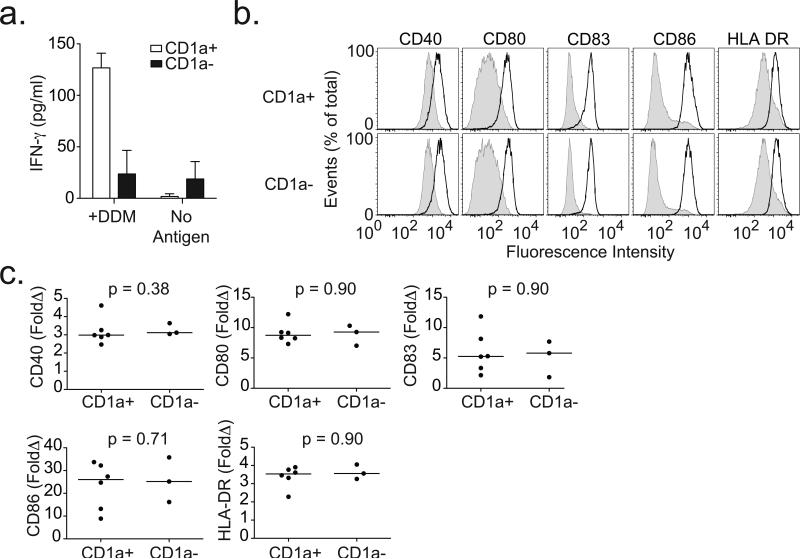
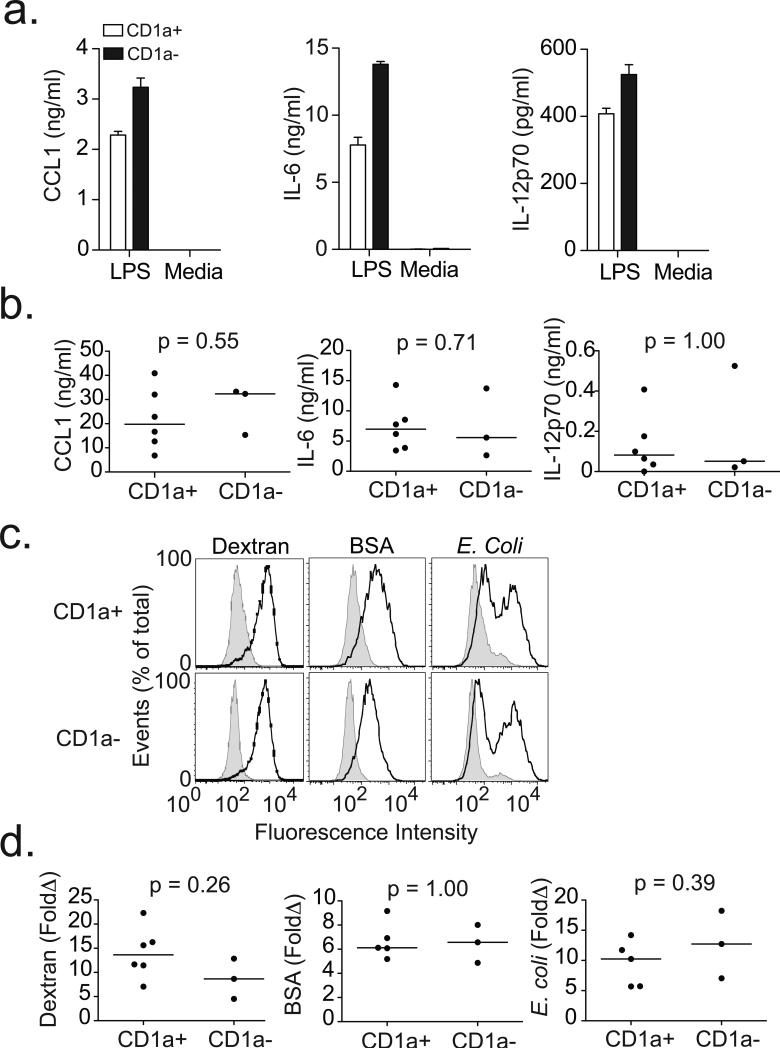
The identification of three donors with CD1a-deficiency allowed mechanistic investigation of the extreme phenotype. First, we evaluated CD1a-sufficient and CD1a-deficient DC presentation of lipid antigen to T cells by co-incubating DCs with dideoxymycobactin and antigen-specific T cells. Only CD1a-expressing DCs stimulated the release of IFN-γ from T cells (Fig. 4a). Next, we stimulated DCs with lipopolysaccharide (LPS) and examined expression of co-stimulatory molecules as well as secretion of cytokines. Compared to media, LPS stimulation resulted in the expected increase in expression of CD40, CD80, CD83, CD86, and HLA-DR; however, we did not find any difference based on CD1a expression (Fig. 4b and 4c). Similarly, LPS induced the production of IL-12p70, IL-6, and CCL1, though again there was no difference in the analysis stratified by CD1a expression (Fig. 5a and 5b). Finally, we exposed cells to fluorescently-conjugated particles to assess endocytic and phagocytic capacity. We observed no difference in the uptake of dextran, bovine serum albumin, or E. coli based on CD1a expression (Fig. 5c and 5d). Our data show that CD1a-deficient DCs are selectively impaired in their ability to present lipid antigen to T cells, but appear to maintain other important aspects of DC function.
Figure 4. CD1a-deficient dendritic cells fail to present antigen to T cells but are able to induce expression of co-stimulatory molecules.
(a) CD1a-sufficient (CD1a+) or CD1a-deficient (CD1a-) dendritic cells were co-incubated with T cell clone CD8-2 in the presence or absence of mycobacterial lipopeptide antigen dideoxymycobactin (DDM). IFN-γ levels were measured in overnight supernatants by ELISA for one donor per group. Shown are mean and standard deviation for triplicate measurements. (b) Shown are expression profiles for cells derived from two healthy blood donors and treated with media (shaded) or LPS (dark line) overnight and then stained with antibodies against co-stimulatory molecules and HLA-DR. (c) Results from nine subjects are combined and shown as fold change (LPS/Media) in median fluorescence intensity (MFI) for CD40, CD80, CD83, CD86, and HLA-DR. All data are representative of two or three independent experiments.
Figure 5. CD1a-deficient dendritic cells do not exhibit defects in cytokine production or endocytic capacity.
(a) Supernatants from DCs exposed to media or LPS overnight were assessed for cytokines by ELISA. Histograms show mean and standard deviation of triplicate measurements from two healthy blood donors. (b) Results from nine subjects are combined and shown as mean values of triplicate measurements with media values subtracted. (c) DCs were exposed to FITC-conjugated dextran, bovine serum albumin (BSA), or E. coli for one hour at either 4°C (shaded histogram) or 37°C (dark line) to assess endocytic capacity. Shown are histograms from two healthy blood donors. (d) Results from nine subjects are combined and shown are fold change (37°C/4°C) in MFI for each particle. All data are representative of two or three independent experiments.
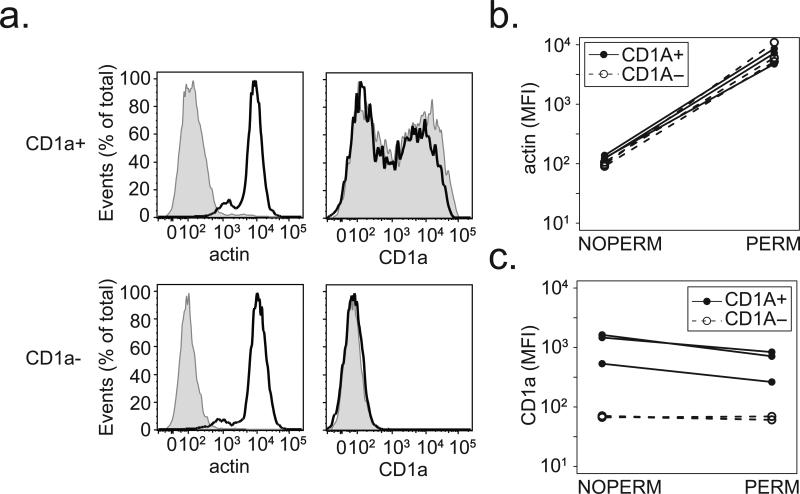
We then sought to determine the mechanism by which a SNP might affect CD1a expression at the cell surface. Previous studies have revealed that most CD1a is localized to the cell surface, and the primary mechanism of CD1a expression at the cell surface is via transcription of new protein rather than altered trafficking (34-36). However, the lack of cell surface CD1a staining led us to consider the possibility that a trafficking defect could result in the intracellular accumulation of mature CD1a. We stained permeabilized DCs from CD1a-sufficient and CD1a-deficient donors and noted that intracellular actin was detected in all donors only after permeabilization (Figure 6a and 6b). Among CD1a-sufficient donors, CD1a staining in permeabilized cells was qualitatively similar to that of unpermeabilized cells. However, among CD1a-deficient donors there was no additional CD1a detected upon permeabilization (Figure 6c). These data reveal that the CD1a-deficiency phenotype is not the result of a defect in protein trafficking to the cell surface.
Figure 6. CD1a-deficiency is not the result of a defect in protein trafficking.
DCs were permeabilized and stained for intracellular actin and CD1a. (a) Shown are expression profiles comparing permeabilized (dark line) and unpermeabilized (shaded histogram) cells from a CD1a-sufficient (CD1a+) and CD1a-deficient (CD1a-) donor. Results from three CD1a-sufficient and three CD1a-deficient donors are combined and shown as the difference in median fluorescence intensity (MFI) between permeabilized (PERM) and unpermeabilized (NOPERM) cells for (b) intracellular actin and (c) CD1a. All data are representative of two independent experiments.
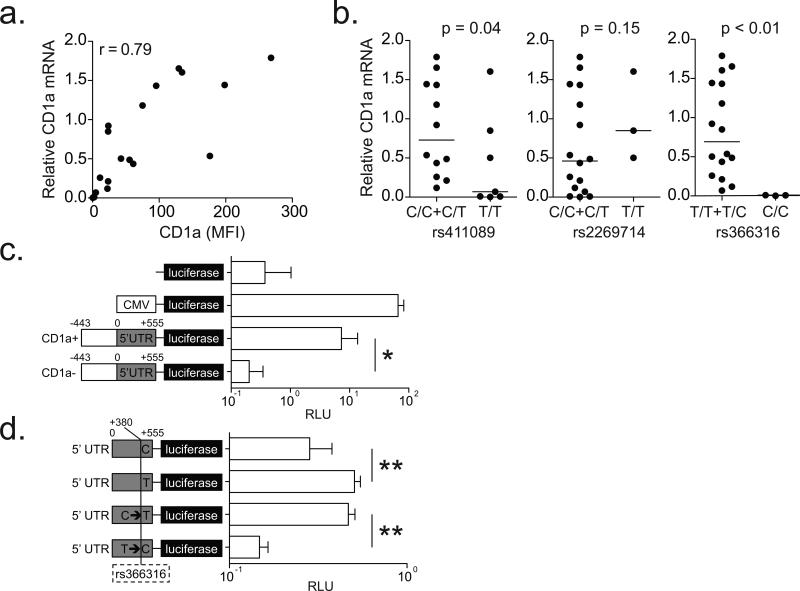
In myeloid cells, CD1a expression is an inducible phenomenon that is controlled by transcription, but the regulatory factors involved are poorly understood(36, 37). We found a strong positive linear correlation between CD1a mRNA and surface staining (r=0.79, Fig. 7a). When we stratified CD1a mRNA by genotype, we found that the minor homozygous genotypes of rs411089 and rs366316 were associated with lower transcript level (Fig. 7b), a pattern similar to what we had observed for surface staining (Fig. 3c). Again, there was no association with rs2269714 (Fig. 7b), and the level of CD1c mRNA was not associated with any SNP (Supplementary Fig. 2c). Since rs366316 is located in the 5’ UTR of CD1a, we hypothesized that one or more promoter variants might directly regulate CD1a transcription. To study the function of CD1a promoter variants, we developed a promoter-luciferase assay to compare the activity of promoters cloned from a one CD1a-sufficient and one CD1a-deficient subject. The cloned DNA contains 998 bases composed of 555 bases of the 5’ UTR as well as an additional 443 bases upstream of the transcription start site. We found that the CD1a-sufficient promoter construct showed more than thirty fold higher luciferase expression than the CD1a-deficient promoter construct (p=0.003) (Fig 7c). This result was consistent with reduced transcription of CD1a (Fig 7b) but did not show a causal relationship between rs366316 and gene expression. We analyzed the 998 base pair promoter sequence from a total of three CD1a-deficient and six CD1a-sufficient individuals and found eleven variants, including eight SNPs, two deletions, and one insertion (data not shown). However, none of these variants correlated as strongly with CD1a-deficiency as rs366316, suggesting this SNP might account for the difference in luciferase expression. We repeated the cloning with a focus on the 5’ UTR and performed site-directed mutagenesis of rs366316. These constructs allowed us to compare normalized luciferase expression among naturally occurring and mutated 5’ UTR sequences. We found that the T allele showed on average 56% higher expression than the C allele, whether it was naturally occurring (p<0.001) or the result of mutagenesis (p<0.001). Further, luciferase expression by the C→T mutant approximated that of the natural T allele, and expression by the T→C mutant substantially reduced promoter activity (Fig 7d). Our data reveal that the C variant of rs366316 is causally related to reduced luciferase expression in our assay. Because rs366316 is strongly associated with CD1a-deficiency, these data suggest rs366316 directly regulates CD1a gene expression.
Figure 7. CD1a-deficiency is the result of transcriptional regulation by a SNP in the 5’ UTR.
(a) CD1a mRNA levels were normalized to that of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Simple linear correlation between relative CD1a mRNA levels and surface protein expression in DCs is shown. (b) Relative CD1a mRNA levels stratified by SNP genotypes are shown with results of one-sided hypothesis testing. (c) HEK293T cells were transfected with firefly luciferase-expressing plasmids (pGL4) under the control of various promoter sequences: none, cytomegalovirus (CMV), 998 bp from CD1a-sufficient (CD1a+), or CD1a-deficient (CD1a-). Cells were co-transfected with Renilla luciferase under a SV40 promoter, and data are reported as relative luciferase units (RLU) which is calculated by dividing the firefly luciferase signal by the Renilla luciferase signal. (d) HEK293T cells were transfected with firefly luciferase-expressing plasmids (pGL4) under the control of 555 bp CD1A 5’ UTR with variants of rs366316: natural C allele, natural T allele, C mutated to T (C→T), or T mutated to C (T→C). All data are representative of two or three independent experiments. * p=0.003 and ** p<0.001 by Welch two sample t-test.
Discussion
Here we report the discovery of three aspects of CD1a function in humans. First, we demonstrate the surprisingly diverse and donor-specific capacity for CD1a induction on dendritic cells. Second, we identify individuals whose dendritic cells lack detectable CD1a expression and are unable to present mycobacterial lipid antigen to T cells. Third, we associate two common SNPs with functional CD1a-deficiency and demonstrate that rs366316, which is located in the 5’ UTR of CD1a, is causally associated with gene expression. In the MHC antigen presenting system, high rates of amino acid sequence variation among individuals control patterns of peptide antigen presentation and T cell activation. In contrast, our data reveal that activation of CD1a-restricted T cells is influenced by genetically determined differences in surface expression despite amino acid sequence conservation.
We found that CD1a-deficient DCs were selectively impaired in their ability to present lipid antigen to T cells, but showed no defects in endocytosis, cytokine secretion, or expression of co-stimulatory molecules after LPS treatment. On the other hand, studies of DCs sorted by CD1a expression from a single individual revealed higher phagocytic capacity and decreased production of IL-12p70 and CCL1 from DCs with low CD1a expression(27, 38, 39). The authors associate the relative lack of CD1a expression with developmental arrest in the transition from monocyte to immature dendritic cell. In these studies, culture conditions, such as the presence of serum lipoproteins, was shown to influence the induction of CD1a as well as DC function more generally. Our data are compatible with this observation because we would not expect SNPs in cis with CD1a to affect aspects of DC development and function that might be more susceptible to modulation by external factors.
We determined that rs366316 and rs411089 were associated with CD1a-deficiency in monocyte-derived dendritic cells within a primarily Caucasian population. We attempted to extend these studies to peripheral blood myeloid DCs but were unable to detect CD1a on the surface of these cells. Whether CD1a-deficiency exists within dermal DCs, Langerhans cells, or thymocytes is the focus of future studies. The frequency of these SNPs varies by ethnicity (data not shown), so the prevalence of CD1a-deficiency in other populations may differ. Also, multiple transcription factors have been shown to modulate gene expression within the CD1a promoter(37), so it is possible that CD1a-deficiency may correlate with a different SNP in a different population.
Because rs366316 is located in the 5’ UTR of CD1a, it may influence CD1a surface expression by reducing translation. Our data do not exclude this possibility, though we chose to focus on the effect of rs366316 on transcription. We found rs366316 was strongly associated with reduced CD1a mRNA levels as well as reduced luciferase expression in transfection studies. Polymorphisms in the 5’ UTR of a gene can affect gene expression by altering transcription factor binding sites or DNA methylation sites, or by reducing the stability of the transcript and increasing mRNA degradation(40, 41). Notably, GATA transcription factor binding sites located within the 5’ UTR of CD1a but downstream of rs366316 have been shown to affect luciferase expression in a system similar to ours(37). Finally, our data leave open the possibility that multiple SNPs may be involved in regulating CD1a transcription. We show rs366316 is in high linkage disequilibrium with rs411089, which is located in the first intron and also associated with decreased mRNA levels. It is therefore possible that a ‘deficiency haplotype’ of multiple SNPs within CD1a are collectively responsible for the molecular mechanism of CD1a deficiency.
Our data suggest that donor-specific variation in CD1a expression can modulate CD1a-restricted T cell activation. Recently, high frequencies of CD1a-autoreactive T cells were reported from most but not all healthy blood donors(21, 22). Thus, it is possible that genetically determined variation in CD1a expression could account for difference in frequencies of CD1a-autoreactive T cells. We also found that CD1a-deficient cells had a selective functional deficiency in lipid antigen presentation to T cells. By linking a genetic polymorphism to this phenotype, our data lay the foundation for genetic association studies that seek to elucidate the role of CD1a in human disease susceptibility. Previous studies have attempted to do this with CD1a coding region polymorphisms that are not associated with any functional deficiency. These studies have been underpowered and failed to replicate(42). Instead, we propose that future studies should focus on rs411089 and rs366316. For example, it has been postulated that IL-22 is important for the pathogenesis of psoriasis(43), and it was recently demonstrated that autoreactive CD1a-restricted T cells in the skin produce IL-22(22). In principle, one could compare the SNP frequency in patients afflicted with psoriasis and compare this to the frequency observed in healthy controls. More detailed studies examining the skin lesions from patients with different genotypes would also be possible. Since CD1a also presents mycobacterial lipopeptides to T cells, similar studies could be performed in cohorts of patients with tuberculosis.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Marta Janer and Sarah Li at the Institute for Systems Biology (Seattle, WA) for assistance with genotyping.
Footnotes
Author Contributions
CS, TH, EAN, TYC, MJM, DBM, and TRH designed the experiments. CS, MS and TYC conducted all the experiments except peripheral blood DC phenotyping, which was performed by TH and EAN. All authors analyzed the data. CS, DBM, and TRH wrote the manuscript with contributions from all authors.
Competing Financial Interests
All authors state that they do not have any competing financial interests with the work presented here.
This work was supported by the NIH (1K24AI089794 to TRH, K08-AI89938 to CS, R01 AI049313 to DBM), the Burroughs Wellcome Foundation (TRH and DBM), The Irvington Institute Fellowship Program of the Cancer Research Institute (CS).
References
- 1.Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol. 2007;314:3–26. doi: 10.1007/978-3-540-69511-0_1. [DOI] [PubMed] [Google Scholar]
- 3.Kasmar A, Van Rhijn I, Moody DB. The evolved functions of CD1 during infection. Current opinion in immunology. 2009;21:397–403. doi: 10.1016/j.coi.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Rhijn I, Zajonc DM, Wilson IA, Moody DB. T-cell activation by lipopeptide antigens. Current opinion in immunology. 2005;17:222–229. doi: 10.1016/j.coi.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Martin LH, Calabi F, Milstein C. Isolation of CD1 genes: a family of major histocompatibility complex-related differentiation antigens. Proc Natl Acad Sci U S A. 1986;83:9154–9158. doi: 10.1073/pnas.83.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988;7:3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balk SP, Bleicher PA, Terhorst C. Isolation and expression of cDNA encoding the murine homologues of CD1. J Immunol. 1991;146:768–774. [PubMed] [Google Scholar]
- 9.Ichimiya S, Kikuchi K, Matsuura A. Structural analysis of the rat homologue of CD1. Evidence for evolutionary conservation of the CD1D class and widespread transcription by rat cells. J Immunol. 1994;153:1112–1123. [PubMed] [Google Scholar]
- 10.Martin LH, Calabi F, Lefebvre FA, Bilsland CA, Milstein C. Structure and expression of the human thymocyte antigens CD1a, CD1b, and CD1c. Proc Natl Acad Sci U S A. 1987;84:9189–9193. doi: 10.1073/pnas.84.24.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. 1999;54:122–127. doi: 10.1034/j.1399-0039.1999.540202.x. [DOI] [PubMed] [Google Scholar]
- 12.Oteo M, Parra JF, Mirones I, Gimenez LI, Setien F, Martinez-Naves E. Single strand conformational polymorphism analysis of human CD1 genes in different ethnic groups. Tissue Antigens. 1999;53:545–550. doi: 10.1034/j.1399-0039.1999.530604.x. [DOI] [PubMed] [Google Scholar]
- 13.Mirones I, Oteo M, Parra-Cuadrado JF, Martinez-Naves E. Identification of two novel human CD1E alleles. Tissue Antigens. 2000;56:159–161. doi: 10.1034/j.1399-0039.2000.560208.x. [DOI] [PubMed] [Google Scholar]
- 14.Tamouza R, Sghiri R, Ramasawmy R, Neonato MG, Mombo LE, Poirier JC, Schaeffer V, Fortier C, Labie D, Girot R, Toubert A, Krishnamoorthy R, Charron D. Two novel CD1 E alleles identified in black African individuals. Tissue Antigens. 2002;59:417–420. doi: 10.1034/j.1399-0039.2002.590509.x. [DOI] [PubMed] [Google Scholar]
- 15.Nestle FO, Zheng XG, Thompson CB, Turka LA, Nickoloff BJ. Characterization of dermal dendritic cells obtained from normal human skin reveals phenotypic and functionally distinctive subsets. J Immunol. 1993;151:6535–6545. [PubMed] [Google Scholar]
- 16.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 17.Rosat JP, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, Modlin RL, Porcelli SA, Furlong ST, Brenner MB. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J Immunol. 1999;162:366–371. [PubMed] [Google Scholar]
- 18.Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nature immunology. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 20.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O'Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 21.de Lalla C, Lepore M, Piccolo FM, Rinaldi A, Scelfo A, Garavaglia C, Mori L, De Libero G, Dellabona P, Casorati G. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. European journal of immunology. 2011;41:602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- 22.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nature immunology. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanssens P, Zabeau M, Meersseman G, Remes G, Gansemans Y, Storm N, Hartmer R, Honisch C, Rodi CP, Bocker S, van den Boom D. High-throughput MALDI-TOF discovery of genomic sequence polymorphisms. Genome Res. 2004;14:126–133. doi: 10.1101/gr.1692304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 25.Young DC, Kasmar A, Moraski G, Cheng TY, Waltz AJ, Hu J, Xu X, Endres GW, Uzieblo A, Zajonc D, Costello CE, Miller MJ, Moody DB. Synthesis of dideoxymycobactin antigens presented by CD1A reveals T cell fine specificity for natural lipopeptide structures. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.M109.000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hensley TR, Easter AB, Gerdts SE, De Rosa SC, Heit A, McElrath MJ, Andersen-Nissen E. Enumeration of major peripheral blood leukocyte populations for multicenter clinical trials using a whole blood phenotyping assay. J Vis Exp. 2012:e4302. doi: 10.3791/4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gogolak P, Rethi B, Szatmari I, Lanyi A, Dezso B, Nagy L, Rajnavolgyi E. Differentiation of CD1a- and CD1a+ monocyte-derived dendritic cells is biased by lipid environment and PPARgamma. Blood. 2007;109:643–652. doi: 10.1182/blood-2006-04-016840. [DOI] [PubMed] [Google Scholar]
- 28.Smed-Sorensen A, Moll M, Cheng TY, Lore K, Norlin AC, Perbeck L, Moody DB, Spetz AL, Sandberg JK. IgG regulates the CD1 expression profile and lipid antigen-presenting function in human dendritic cells via FcgammaRIIa. Blood. 2008;111:5037–5046. doi: 10.1182/blood-2007-07-099549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 30.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony- stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oteo M, Arribas P, Setien F, Parra JF, Mirones I, Gomez del Moral M, Martinez-Naves E. Structural characterization of two CD1A allelic variants. Hum Immunol. 2001;62:1137–1141. doi: 10.1016/s0198-8859(01)00314-7. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, Inaba M, Inaba K, Toki J, Sogo S, Iguchi T, Adachi Y, Yamaguchi K, Amakawa R, Valladeau J, Saeland S, Fukuhara S, Ikehara S. A CD1a+/CD11c+ subset of human blood dendritic cells is a direct precursor of Langerhans cells. J Immunol. 1999;163:1409–1419. [PubMed] [Google Scholar]
- 33.MacDonald K, Munster D, Clark G, Vuckovic S, Hart D. Peripheral blood dendritic cell subset analysis. In: Mason D, editor. Leucocyte Typing VII. Oxford University Press; Oxford, England: 2002. pp. 315–319. [Google Scholar]
- 34.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 35.Cernadas M, Cavallari M, Watts G, Mori L, De Libero G, Brenner MB. Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J Immunol. 2010;184:1235–1241. doi: 10.4049/jimmunol.0804140. [DOI] [PubMed] [Google Scholar]
- 36.Roura-Mir C, Wang L, Cheng TY, Matsunaga I, Dascher CC, Peng SL, Fenton MJ, Kirschning C, Moody DB. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J Immunol. 2005;175:1758–1766. doi: 10.4049/jimmunol.175.3.1758. [DOI] [PubMed] [Google Scholar]
- 37.Colmone A, Li S, Wang CR. Activating transcription factor/cAMP response element binding protein family member regulated transcription of CD1A. J Immunol. 2006;177:7024–7032. doi: 10.4049/jimmunol.177.10.7024. [DOI] [PubMed] [Google Scholar]
- 38.Cernadas M, Lu J, Watts G, Brenner MB. CD1a expression defines an interleukin-12 producing population of human dendritic cells. Clin Exp Immunol. 2009;155:523–533. doi: 10.1111/j.1365-2249.2008.03853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang CC, Wright A, Punnonen J. Monocyte-derived CD1a+ and CD1a dendritic cell subsets differ in their cytokine production profiles, susceptibilities to transfection, and capacities to direct Th cell differentiation. J Immunol. 2000;165:3584–3591. doi: 10.4049/jimmunol.165.7.3584. [DOI] [PubMed] [Google Scholar]
- 40.Reynard LN, Bui C, Canty-Laird EG, Young DA, Loughlin J. Expression of the osteoarthritis-associated gene GDF5 is modulated epigenetically by DNA methylation. Hum Mol Genet. 2011;20:3450–3460. doi: 10.1093/hmg/ddr253. [DOI] [PubMed] [Google Scholar]
- 41.Landa I, Ruiz-Llorente S, Montero-Conde C, Inglada-Perez L, Schiavi F, Leskela S, Pita G, Milne R, Maravall J, Ramos I, Andia V, Rodriguez-Poyo P, Jara-Albarran A, Meoro A, del Peso C, Arribas L, Iglesias P, Caballero J, Serrano J, Pico A, Pomares F, Gimenez G, Lopez-Mondejar P, Castello R, Merante-Boschin I, Pelizzo MR, Mauricio D, Opocher G, Rodriguez-Antona C, Gonzalez-Neira A, Matias-Guiu X, Santisteban P, Robledo M. The variant rs1867277 in FOXE1 gene confers thyroid cancer susceptibility through the recruitment of USF1/USF2 transcription factors. PLoS Genet. 2009;5:e1000637. doi: 10.1371/journal.pgen.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uncini A, Notturno F, Pace M, Caporale CM. Polymorphism of CD1 and SH2D2A genes in inflammatory neuropathies. J Peripher Nerv Syst. 2011;16(Suppl 1):48–51. doi: 10.1111/j.1529-8027.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- 43.Boniface K, Guignouard E, Pedretti N, Garcia M, Delwail A, Bernard FX, Nau F, Guillet G, Dagregorio G, Yssel H, Lecron JC, Morel F. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.