Abstract
Huntingtin Associated Protein 1 (HAP1) is a binding partner for huntingtin, the protein responsible for Huntington’s Disease. In mammals, HAP1 is mostly found in brain where it is expressed in neurons. Although several functions have been proposed for HAP1, its role has not yet been clearly established. Here we report on the identification of a HAP1 C.elegans homolog called T27A3.1. T27A3.1 shows conservation with rat and human HAP1 as well as with Milton, a Drosophila HAP1 homolog. To determine the cellular expression of T27A3.1 (multiple isoforms; a-e), we generated several transgenic worm lines expressing a fluorescent reporter protein (GFP and DsRed2) or full length T27A3.1a/c isoforms fused to GFP under the control of the promoter for T27A3.1. We have found that T27A3.1 is expressed in many cell types including a subset of chemosensory neurons in the head and tail. These include the amphid chemosensory neurons ASKL and R, ASIL and R, ADFL and ASEL; the phasmid neurons PHBL and R; and the CAN neurons which are required for worm survival. Furthermore, we show that the subcellular localization of T27A3.1a/c resemble that of mammalian HAP1 and that T27A3.1a/c localize to stigmoid body like structures.
Keywords: Huntington, localization, amphid, phasmid, stigmoid body
Introduction
Huntingtin associated protein 1 (HAP1) was initially identified through its interactions with huntingtin (htt), the protein mutated in Huntington’s Disease (HD) (Li, 1995). HD is a hereditary neurodegenerative disorder characterized by massive neuronal loss in the striatum and cortex. The HD mutation consists of an expansion of a trinucleotide repeat (CAGn) in the IT15 gene (HDCRG, 1993), which is translated into a polyglutamine stretch near the N-terminal region of htt (Gutekunst, 1995). Expansion of the polyglutamine tract in htt causes HAP1 to exhibit a higher binding affinity to htt (Li, 1995). More recently HAP1 has been shown to be a modifier of HD with a polymorphism associated with delayed age of onset (Metzger, 2008). Despite these studies, the role of HAP1 in disease pathogenesis remains unclear.
In humans, two HAP1 isoforms have been identified and are found in the central nervous system (Li, 1998b). In mouse, there are three HAP1 transcripts resulting from alternative splicing: HAP1-A, HAP1-B and HAP1-C (Nasir, 1998; Nasir, 1999). In the developing mouse, HAP1 transcripts are found in the neuroepithelial tissue after embryonic day 8.5 (Dragatsis, 2000). Postnatally, HAP1 mRNAs have been identified in the brain, with levels highest in the olfactory bulb, hypothalamus, brain stem, striatum, cerebellum, hippocampus, and colliculi (Bertaux, 1998; Dragatsis, 2000; Li, 1995; Li, 1996; Page, 1998). In the adult rodent, expression of HAP1 proteins is restricted to the nervous system and HAP1-A and B are selectively expressed in neurons (Gutekunst, 1998; Li, 1995; Li, 1996; Martin, 1999; Page, 1998).
Electron microscopy and protein-protein interaction studies suggest a role of HAP1 in organelle trafficking (Engelender, 1997; Gutekunst, 1998; Kittler, 2004; Li, 1998a; Li, 1995; Li, 2002; Martin, 1999). HAP1 may also play a role in calcium signaling as suggested by its interaction with inositol-(1,4,5) triphosphate receptor type 1 (InsP3R1) (Tang, 2003). Based upon studies in which HAP1 was overexpressed, HAP1 also interacts with NeuroD (Marcora, 2003), and duo/P-CIP10/Kalirin-7 (Colomer, 1997; Penzes, 2001) and participates in BDNF transport (Gauthier, 2004). Overexpression of HAP1A in neuroblastoma cells result in process outgrowth suggesting a role in neuronal differentiation (Li, 2000).
Homozygous disruption of the HAP1 locus results in early postnatal lethality (Chan, 2002; Gorska-Andrzejak, 2003), which can be partially rescued by manipulation of the litter composition (Dragatsis, 2004). The early death of the pups demonstrates that HAP1 is indirectly critical for early stages of postnatal development. Interestingly, one of the HAP1 knockout models showed neurodegeneration occurring specifically in the hypothalamus (Li, 2003).
We have identified a C.elegans HAP1 homolog and demonstrated that it belongs to an even broader family of proteins. Using reporter constructs we have determine which cells express this gene. Surprisingly, although it is expressed in some neurons including chemosensory neurons, it is also expressed in several other cell types.
Material and Methods
C.elegans strains and culture
C.elegans strain N2 (wildtype) and rrf-3(pk1426) were used for these studies. evIs111=integrated Ex[F25B3.3::gfp;dpy-20(+)], generously provided by Dr. Joe Culotti, was used for confirmation of neuronal expression. Animals were raised at 20°C on solid NGM medium and fed OP50 E.coli (Brenner, 1974).
T27A3.1 promoter GFP and DsRed2 fusion constructs
To determine the expression pattern of the C.elegans HAP1 homolog, a plasmid was constructed in which 4kb of genomic sequence, produced by PCR, was fused in-frame to GFP within the promoter-less GFP vector pPD95.77 (vector kindly provided by A. Fire). This genomic sequence includes 4 kb upstream of the predicted initiator methionine codon of the largest mRNA (T27A3.1a), exon 1, intron 1 and a portion of exon 2 sequence (Figure 1A). For most C. elegans genes, 4 kb is usually a sufficient promoter sequence. Intron 1 was included in our construct because, in a number of cases, the first intron is important for proper expression of the gene. PCR reactions were performed using the Triple Master kit (Eppendorf) according to manufacturor’s instructions. PCR products were cut using HindIII and XbaI and ligated into pPD95.77 to generate pCAG50. Insertion of the promoter was confirmed by restriction digest and gel electrophoresis. Forward and reverse primers for the 4kb promoter were 5’-GCACAAGCTTCTGCCCAAAATAGGTGGTAGGTAG-3’ and 5’-GTACTCTAGAGGAGTTTCGAGTTGGAGTTGG-3’, respectively. For injection into evIs111 and confirmation of neuronal expression, GFP was substituted with DsRed2 excised from pDsRed2-C1 (Clontech). pDsRed2-C1 was cut with BclI, blunted with Klenow, cut with AgeI, and inserted into pCAG50, which had been previously cut with EcoRI, blunted, and cut with AgeI. This yielded the pCAG51 plasmid for DsRed2 expression driven by 4kb of DNA upstream of T27A3.1a.
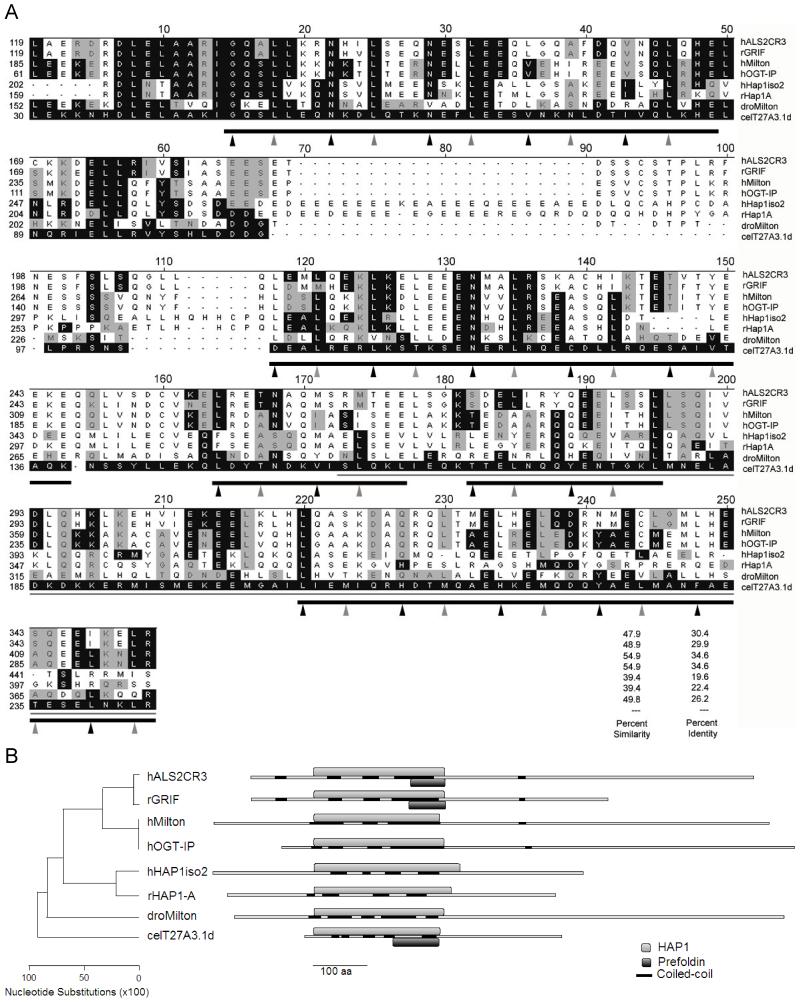
Figure 1. Comparison of an N-terminal fragment of T27A3.1d to the most closely related sequences in the databases.
(A) Clustal W alignment of human ALS2CR3 (hALS2CR3), rat GRIF (rGRIF), human Milton (hMilton), human OGT (hOGT-IP), human HAP1 isoform 2 (hHAP1iso2), rat HAP1-A (rHAP1A), Drosophila Milton (dMilton), and C.elegans T27A3.1d (celT27A3.1d) are shown. The black shaded boxes show identical amino acids and gray shaded boxes show conserved amino acids as they compare to T27A3.1d. The thin gray line represents a putative region of rHAP1-A responsible for binding to htt in yeast. The thick gray lines represent the putative 5 coiled coil regions in T27A3.1d identified by Lupa’s method (Coil at http://www.ch.embnet.org/software/COILS_form.html, window=21). Amino acids in a and d positions in the coiled coil regions are indicated by arrows. T27A3.1d maintains a higher proportion of preferred residues in the a and d positions than rHAP1-A. T27A3.1d more closely resembles human and Drosophila Milton proteins in these a and d positions. Accession numbers are O60296 for hALS2CR3, Q8R2H7 for rGRIF, BAA82994 for hMilton, NP_055780 for hOGT-IP, NP_817084 for hHAP1iso2, AAC52327 for rHAP1A, AY03001 for dMilton, and AAO21402 for T27A3.1d. The percent similarity and identity between T27A3.1d and the other proteins in the alignment are listed.
(B) Schematic of the phylogenetic tree illustrating the relative distance between each of the proteins based on their above shown alignment in DNAStar. Also shown are the relative positions of the aligned N-terminal region shown in A designated as HAP1, the coiled coil domains identified using the Coil online software, and the Prefoldin domain suggested by Pfam.
Full length T27A3.1 GFP fusion constructs
To determine the subcellular localization if T27A3.1 we generated a plasmid in which 4 kb of the genomic
Generation of transgenic lines
Young adult, hermaphrodite N2 were injected with pCAG50 at 40ng/μl along with pRF4 at 80ng/μl. pRF4 encodes a dominant version of rol-6, which confers a rolling phenotype and is used as a transformation marker. These experiments yielded 3 GFP expressing lines carrying sfEx27[4kbT27A3.1pro::gfp; rol-6]. Each of the lines gave 50-90% GFP expressing worms in subsequent generations.
DiD
For each T27A3.1 pro::GFP line, worms from one entire plate were collected and rinsed twice in M9 (6g Na2HPO4, 3g KH2PO4, 5g NaCl, 0.25g MgSO4.7H20 per Liter) buffer (Brenner, 1974). Worms were then resuspended in M9 containing 3,3 ′-dioctadecyloxacarbocyanine (DiD at 0.01 mg/ml; Molecular Probes). The next day, worms were rinsed twice in M9, resuspended in M9 and fed overnight on OP50 seeded plates. The next day, worms were collected, fixed with 4% paraformaldehyde in 0.1M phosphate buffer and visualized by fluorescence microscopy for GFP (488 nm) and DiD (594 nm).
Neuronal expression confirmation
Young adult, hermaphrodite N2 and evIs111 were injected with pCAG51 at 40ng/ul along with pRF4 at 80ng/ul as described above, resulting in one N2 and two evIs111 DsRed2 expressing lines carrying sfEx28[4kbT27A3.1pro::DsRed2; rol-6]. Each of the lines obtained gave 50-90% DsRed2 worms in subsequent generations.
RNA interference by injection
A 1.7 kb fragment of cDNA, contained in all T27A3.1 isoforms, was generated by PCR and cloned into the L4440 vector (Timmons and Fire, 1998) by using the BamH1 and Xho1 cloning sites. Forward and reverse primers for cloning were 5’-GTCAGGATCCCACTTGATCCGTCGTGTGCTCTC-3’ and 5’-GTCACTCGAGCCAAGCTCATTGGACCACCTGCATC-3’, respectively. Sense and antisense RNA were produced using the two T7 promoters of L4440 and components of the RiboMAX RNA synthesis kit (Promega). The RNAs were annealed and injected into the gut of wild-type hermaphrodites. F1 laid between the 19th and 43rd hour after injection were examined for defects in development, morphology, behavior and fertility (brood size).
RNA interference by feeding
On day 1, HGMP ID I-2J10 bacterial clone (MRC GeneService) was cultured in 20 ml LB broth supplemented with tetracycline (15μg/ml) and ampicillin (50 μg/ml). RNAi for pat-3, using the feeding vector pDM275, kindly provided by Don Moerman, which produces embryonic lethality at the 2-fold stage, was used as a positive control. Negative controls were fed with control bacteria (OP50). On day 2, 10 cm plates were prepared with NGM, supplemented with IPTG (1mM) and carbenecillin (25 μg/ml) then seeded with bacterial cultures prepared on day 1. On day 3, twenty, L4 N2 or rrf-3 hermaphodites were transferred to plates seeded with the various RNA producing bacteria. On day 4, the adults were transferred to new plates, also seeded on day 2. F1 laid between the 19th and 43rd hour were examined for phenotypes.
Microscopy and analysis
Images were acquired using either a Leica inverted scope with Simple PCI software or a Zeiss AxioSkop 2 upright microscope with Axioncam HR (black/white) or MR (color) cameras and Axiovision software (Carl Zeiss USA, Thornwood, NY). For more precise localization, images were captured with a two photon Zeiss LSM 510 laser scanning confocal microscope coupled to a Zeiss 100M Axiovert (Carl Zeiss USA, Thornwood, NY). After scanning-in the images, they were processed with Adobe Photoshop. The identity of the GFP positive cells was determined by comparing our images with online picture sources through the C. elegans Wormbase server (www.wormbase.org) and descriptions in other publications.
Results
Sequence conservation between T27A3.1d and other proteins
To evaluate the similarity between T27A3.1 and HAP1 we compared the amino acid sequences of T27A3.1d (cDNA sequence confirmed) with that of human Hap1 isoform 2 and rat HAP1-A. Pairwise Clustal W alignment was performed using the MegAlign component of the DNA Star software (DNASTAR, inc., Madison, WI). T27A3.1d was chosen for this analysis because it received the highest score and E value (5.4e-11) in a BLASTP search of the C.elegans genome using human HAP1 isoform 2 and rat HAP1-A as queries. Although full-length T27A3.1d protein shows only 16% identity and is 48% conserved relative to rHAP1-A, there is stronger similarity within the N-terminal region (Figure 1A). In a pairwise comparison, the region between residues 35 and 243 of T27A3.1d shows 23.1% identity and 47.1% conservation with human HAP1 isoform 2. This same region shows higher similarity with rat HAP1 with 22.6% identity and 49.5% conservation. Notably, all T27A3.1 isoforms (discussed later in text) contain this region. Within this region of T27A3.1d, the segment corresponding to residues 36 to 95 shows the most similarity to rHAP1-A with 45% identity and 70% conservation.
As expected, T27A3.1d also shows close resemblance to Milton, a previously proposed homolog of HAP1 in Drosophila (Gorska-Andrzejak, 2003; Stowers, 2002). Pairwise comparison of full-length T27A3.1d with Drosophila Milton (dMilton: aa 25-1114) shows 25% identity and 62% conservation. As for the HAP1 proteins, the similarity is stronger in the N-terminal region with the two proteins (T27A3.1d aa 30-243; dMilton aa 152-373) showing 26.3% identity and 59.6% conservation in a segment of 139 amino acids.
When a BLASTP search of T27A3.1d is performed against the Human Genome Database (http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html), the first two proteins showing the highest matching scores are “amyotrophic lateral sclerosis 2 chromosomal region candidate gene protein 3” (hALS2CR3) and “O-GlcNAc transferase-interacting protein of 106 kDa” (hOGT-IP). Human HAP1 isoform 2 gives the third highest match. Interestingly, the same scenario applies when the search is performed using dMilton. A Clustal W alignment including these proteins as well as T27A3.1d, HAP1, Milton and the rat ortholog of hALS2CR3 (GRIF; “GABA receptor interacting factor” (Beck, 2002)) reveals a region of high similarity in their N-terminal region (Figure 1A). Within this N-terminal region, the alignment suggests that T27A3.1d is more closely related to human Milton and OGT-IP followed by Drosophila Milton, rat GRIF, human ALS2CR3, and rat and human HAP1 proteins as shown by percent conservation and a phylogenetic tree (Figure 1A,B).
Because rat OGT-IP has been shown to interact with OGT substrates and has itself also been shown to be an OGT substrate (Iyer, 2003), we scanned the T27A3.1d sequence for glycosylation sites. A PROSITE scan of T27A3.1 revealed 3 potential N-glycosylation sites at residues 139-142, 288-291, and 397-400. This suggests that like OGT-IP, T27A3.1d could be glycosylated by OGT. No such residues were identified by PROSITE in HAP1 proteins.
Rat GRIF, which is an ortholog of hALS2CR3 protein, associates with GABA(A) receptor α1 and β2 subunits in adult rat brain (Beck, 2002). In fact, the region of rGRIF critical for GABA(A) receptor β2 binding (residues 124-283) is contained within the N-terminal region shown to have the most similarity with T27A3.1 in our alignment (Figure 1A). Recently, rHAP1A has also been shown to associate with GABA(A) receptor β1- β3 subunits via residues between 220-520 (Kittler, 2004), which also overlap with our conserved region (Figure 1A). The region of GABA receptor binding common to rGRIF and rHAP1A corresponds to the region of T27A3.1d between residues 105 and 175 (Figure A1). Based on our family alignment, this portion of T27A3.1 shows 40% and 41% conservation with rGRIF and rHAP1A respectively, suggesting that T27A3.1 proteins might also interact with GABA receptor subunits.
Conservation of domain organization in T27A3.1d and other proteins
We used Pfam (www.sanger.ac.uk/Software/Pfam/), InterProScan (www.ebi.ac.uk/InterProScan/) and ProDom (protein.toulouse.inra.fr/prodom/current/html/home.php) to identify the presence of characterized protein domains in T27A3.1, and to further analyze its potential homology to HAP1 based on domain similarities. Pfam recognized a domain within T27A3.1d between amino acids 1 and 246 that is also present in human, rat, and mouse HAP1 homologs (score: 437, E value: 1.4e−128). This segment overlaps with the region of high identity and conservation found using our previous alignment and is designated the “HAP1_N domain” by Pfam. Despite the fact that the HAP1_N represents an N-terminal conserved region found in several HAP1homologs, little is known about its function. In addition to the HAP1_N domain, InterProScan identified a region towards the C-terminal segment of the HAP1_N domain as a Prefoldin domain (residues 162-243). Prefoldin is part of a molecular chaperone system that promotes the correct folding of nascent polypeptide chains (Hansen, 1999; Hartl, 2002).
Because HAP1 is known to contain several coiled coil domains which have been shown to participate in protein-protein interactions, we also looked for the presence of these domains in T27A3.1. According to Coil (http://www.ch.embnet.org/software/COILS_form.html, window=21), T27A3.1d contains several coiled coil regions (Figure 1A-B). The first coiled coil region is in the segment of T27A3.1d most similar to rHAP1-A. Interestingly, within this region T27A3.1d maintains a higher proportion of preferred non-polar residues in the a and d positions of the heptad repeat than rHAP1-A (Figure 1A). In these a and d positions, T27A3.1d also resembles human and Drosophila Milton proteins.
The presence of HAP1_N and coiled coil domains in both T27A3.1 and HAP1 suggest that these proteins not only show conservation at the amino acid level, they also have similar domains. This might suggest that the two proteins have similar functions such as specific protein-protein interactions, and/or have similar binding partners. Additionally, the presence of these domains in T27A3.1 and HAP1 mammalian proteins provides another level of confidence that these proteins are homologs and play similar roles in different species.
Molecular cloning of the T27A3.1 promoter
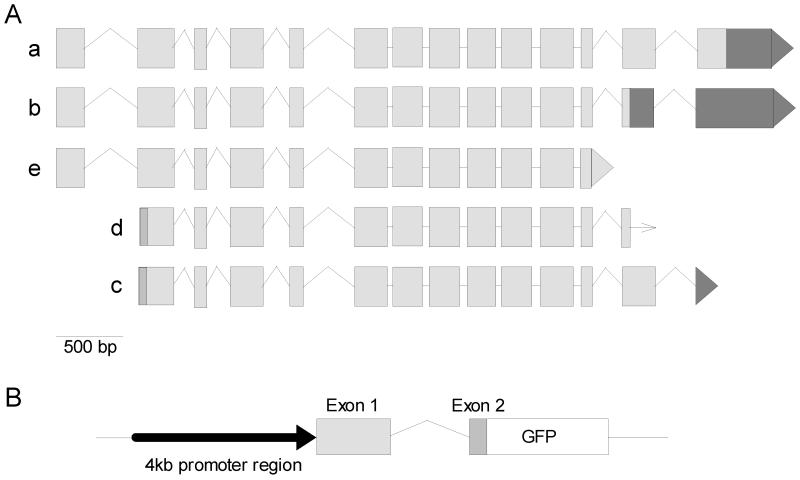
The T27A3.1 gene from C.elegans is located on chromosome 1 and has multiple, predicted isoforms. According to Wormbase, there are 5 potential isoforms of T27A3.1 (Figure 2). Two of these isoforms have been confirmed by cDNAs including the smallest isoform T27A3.1d. The largest confirmed isoform (T27A3.1c) is 3.4kb in length (unspliced) and includes 14 exons. Larger isoforms (a and b, partially confirmed by cDNAs) may exist, totaling nearly 4.3 kb (unspliced) and including an additional, 5’ exon. Isoform T27A3.1e, which contains fewer 3’ exons than its larger counterparts, is currently only a predicted isoform. To determine which cells express T27A3.1, we fused a 5.1 kb genomic fragment to GFP that includes 4 kb upstream of the ATG of isoform a, together with exon 1, intron 1 and a part of exon 2. The way the fragment was designed, it should also serve as a promoter region for the two isoforms d and c that do not share exon 1 with isoforms a, b and e (Figure 2). Although less coding sequence from these isoforms will be represented in the final construct, the fragment will incorporate greater upstream sequence and utilize their respective start codons. The 4kb region upstream from the start plus exon 1 through a segment of exon 2 of T27A3.1a (these exons, of the partially confirmed isoform, have been confirmed) was PCR amplified from C. elegans genomic DNA and inserted into the promoter-less green fluorescent protein (GFP) reporter vector, pPD95.77.
Figure 2. T27A3.1 isoforms.
(A) Splicing patterns of the five hypothetical isoforms of T27A3.1 (a – e, grey boxes indicate exons). According to Wormbase, isoforms c and d have been fully confirmed by cDNA, whereas isoforms a, b, and e were only partially confirmed by cDNAs. These isoforms differ in their amino and carboxyl termini.
(B) Schematic T27A3.1pro::gfp construct including 4kb upstream of exon 1, exon 1, intron 1 and a portion of exon 2 of T27A3.1 (a,b,e), or a portion of exon 1 of T27A3.1(c and d), GFP, and the pPD95.77 vector.
T27A3.1 expression
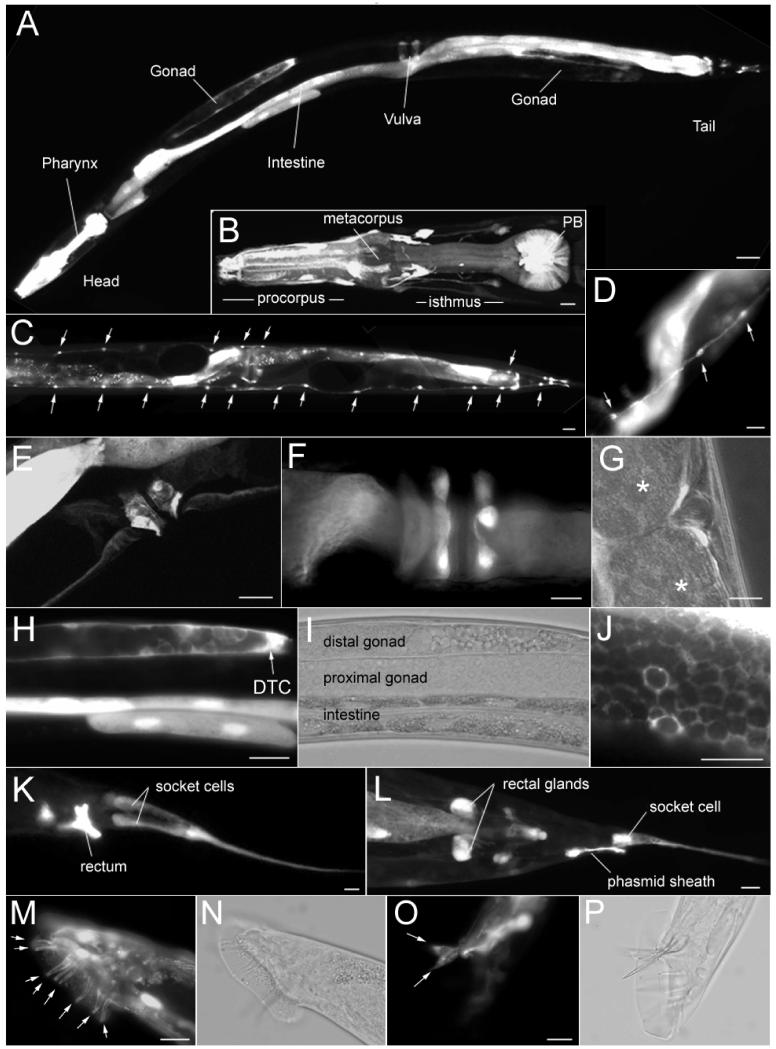
We studied the tissue distribution of T27A3.1 gene expression by analyzing the expression of the GFP reporter gene under control of 4kb of the T27A3.1 putative promoter (Figure 2B). The construct, pCAG50, was injected into the gonads of young N2 hermaphrodite worms. GFP expression was seen in the F1 generation, which indicated successful injection and incorporation into the dividing cells. Several independently derived lines of GFP expressing worms were established from their F1 generation and characterized for their pattern of cellular expression. Each transgenic line displayed similar expression patterns. GFP expression was visible late in embryogenesis but before morphogenesis and continued through the larval stages into adulthood. In adults, expression was found in a variety of cell types: GFP was found in the cells of the pharynx (Figure 3A-B), in the epithelial cells of the intestine (Figure 3A,H), in the seam cells that line the sides of the worm (Figure 3C-D), in cells of the vulval region (Figure 3E-G), in the somatic gonads (Figure 3H-J), and in cells of the tail region (Figure 3A, K-P).
Figure 3. T27A3.1 is expressed in many cell types.
(A) Micrograph showing expression of T27A3.1pro::gfp in an hermaphrodite sfEx27 adult worm. GFP is visible in cells in various organs including the pharynx, the intestine, the gonads, as well as cells in the vulval and tail regions.
(B) Micrographs of T27A3.1pro::gfp expression in the pharynx of an hermaphrodite sfEx27 adult worm. GFP positive cells are located within the posterior bulb, in what appears to be muscle cells. GFP is also visible in cells around the pharyngeal bulb as well as in processes along the bulb (arrows) and in the nerve ring (arrowheads).
(C) Micrographs showing GFP expression in the seam cells (arrows) along the body of an hermaphrodite sfEx27 adult worm.
(D) Higher magnification of GFP expression in the seam cells (arrows).
(E) Lateral view of the vulval region of an hermaphrodite sfEx27 adult worm showing GFP in cells surrounding the vulva (arrowheads).
(F) Ventral view of the vulva region showing GFP in cells surrounding the vulva (arrowheads).
(G) Lateral view of the vulval region showing the position of the GFP positive cells relative to embryos (*).
(H) Micrographs of T27A3.1pro::gfp expression in the gonads of an hermaphrodite sfEx27 adult worm. GFP is visible in the DTC cell as well as in the intestine.
(I) Same as H but shown in phase contrast mode.
(J) High magnification of fish-net like GFP expression in the distal sheath cell pair 1 of the gonads.
(K) Micrographs of the tail region of a T2A3.1pro::gfp adult sfEx27 hermaphrodite. GFP is visible in the rectum and in socket cells.
(L) In the tail region, GFP is also found in the rectal glands and phasmid sheath cells.
(M) Micrograph of the tail region of a T2A3.1pro::gfp adult male sfEx27 showing GFP expression in the rays (arrows).
(N) Same as H in phase contrast mode.
(O) In the male tail, GFP is also visible in the spicules (arrows).
(P) Same as H in phase contrast mode. Scale bars: A: 100 μm; B-D: 20 μm; E-J: 10 μm; K: 40 μm; L: 20 μm; M-P: 10 μm.
In the pharyngeal bulbs, the morphology and striated appearance of GFP positive cells is consistent with muscle cell characteristics. In the vulval region, the GFP positive cells did not appear to be neurons or muscle cells and their identity remains unclear. In the gonads, GFP expression was visible in the distal tip cell (DTC) as well as in the distal sheath cell pair 1 which can be identified by it’s fish-net like appearance (Figure 3H-J). In the tail, GFP positive cells most likely include the rectal gland cells, the rectum epithelial cells, and phasmid sheath cells (Phsh) and socket cells (Phso1 and 2) that are associated with sensory receptors and create a protective environment for the cilium (Figure 3K-L).
In males, GFP expression was found in the bilateral sensory rays and in the spicules (Figure 3M-P). These cells have been shown to have specific roles in guiding the execution of male mating behavior (Liu, 1995).
Cells with neuronal-like processes were visible immediately following the embryonic stage and remained through the life of the worm. GFP positive cells were visible in the head anterior and posterior ganglia, which contain most of the C.elegans neurons as well as other associated cells. GFP positive neuronal-like processes were also found in the nerve ring encircling the isthmus of the pharynx (Figure 3B). Whether the GFP positive cells were indeed neurons could not be determined solely on their localization. However, the finding of GFP positive processes in the nerve ring suggested that at least some neurons were expressing T27A3.1. When a shorter promoter region, only 2 kb of genomic DNA upstream from the start codon of T27A3.1a, was used to drive the expression of GFP, a similar expression pattern was seen, however, fewer GFP expressing neurons were visible (data not shown). This data suggest that the larger 4 kb promoter region contains regulatory elements necessary for specific neuronal expression that are not contained within the smaller 2 kb promoter segment.
T27A3.1 expression in neurons
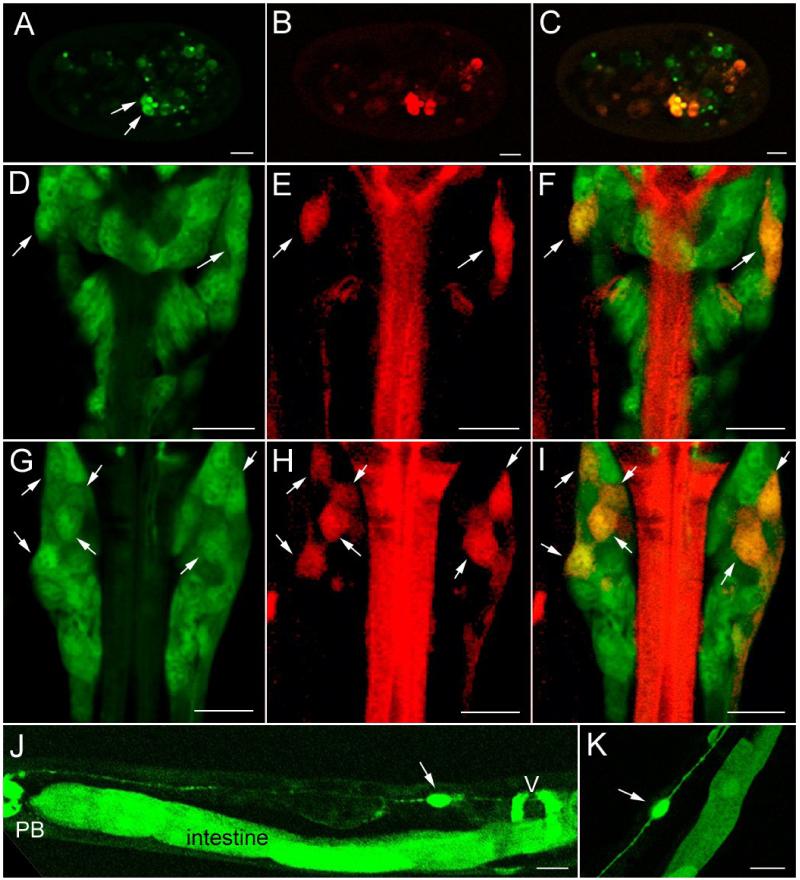
To confirm the neuronal expression pattern of T27A3.1 we developed a new construct where the original, GFP reporter was replaced by the red fluorescent protein DsRed2. This new construct, pCAG51, was injected into evIs111, an integrated strain expressing GFP exclusively in all neurons (pan-neural::gfp; J. Culotti, personal communication) and has been used by other investigators as a pan-neuronal marker (Altun-Gultekin, 2001). Several T27A3.1pro::DsRed2 positive cells in the head region of the worms were also pan-neural::gfp positive, demonstrating that T27A3.1 is indeed expressed in a subset of neurons (Figure 4). T27A3.1pro::DsRed2 positive neurons were first visible late in embryogenesis (Figure 4A-C). Although there were some variations between worms within and between lines, up to 7 neurons on each side of the head of the worm expressed T27A3.1 (Figure 4D-I). We were able to identify another pair of T27A3.1 expressing neurons along the body of the worm as CAN L and R (Figure 4J-K). These neurons are easily identified since they are the only bipolar neurons, with their cell bodies located anterior to the vulval region, sending one process to the head and one to the tail along the excretory canal. The CAN neurons are involved in the maintenance of homeostatic balance and are necessary for the survival of worms (Forrester, 1997; Strange, 2003).
Figure 4. T27A3.1 is expressed in some neurons.
(A-C) Confocal micrographs showing localization of T27A3.1pro::DsRed2 expression in sfEx28 embryos. DsRed2 expression (red) overlaps with some of the GFP positive neurons (green).
(D-I) Confocal micrographs showing T27A3.1pro::DsRed2 expression in the head region of an adult sfEx28. Panels E and H show T27A3.1pro::DsRed2 expression alone (red), panels D and G show pan neuronal GFP expression alone (green) and panel F and I show overlapping expression of T27A3.1pro::DsRed2 and pan neuronal marker GFP (yellow). Arrows indicate T27A3.1pro::DsRed2 expressing neurons.
(J) Micrograph showing T27A3.1pro::gfp expression in an adult sfEx27. GFP is visible in a canal neuron anterior to the vulva (v).
(K) Another view of T27A3.1pro::gfp expression in a bipolar canal neuron in an adult sfEx27. Scale bars: 10 μm.
T27A3.1 expression in chemosensory neurons
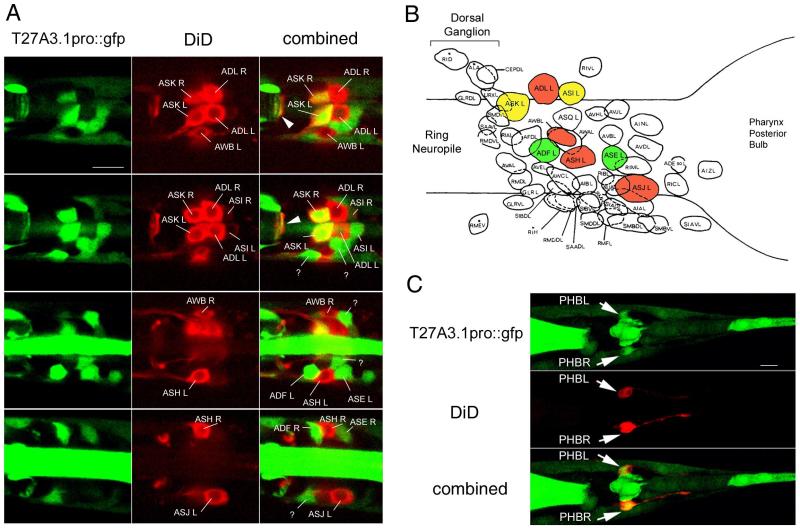
In the head and the tail region some chemosensory neurons have nerve endings (cilia) in direct contact with the exterior environment. When worms are exposed to a solution containing lipophilic dyes, the dyes penetrate the nerve endings and are transported back into cell bodies where they accumulate. This ability to back-fill is specific to 6 pairs of amphid chemosensory neurons located in the head and to 2 pairs of phasmid sensory neurons located in the tail. To determine whether the T27A3.1pro::gfp neurons were chemosensory amphid and/or phasmid neurons, we stained young adult worms using the lipophilic dye, carbocyanine DiD. We found co-localization of DiD staining with GFP in the head and the tail (Figure 5). Based on their positions, the chemosensory neurons expressing T27A3.1 are ASK L, ASK R, ASI L and ASI R (Figure 5A). In addition, at least two other T27A3.1 expressing neurons located anterior and posterior to the DiD filled, ASH L neuron were identified as ADF L and ASE L chemosensory neurons (Figure 5A). Other T27A3.1 expressing neurons and their processes were found alongside DiD stained chemosensory neurons and will require further analysis to be identified. In the tail region, T27A3.1pro::gfp was found in the more anterior DiD filled neurons corresponding to PHB L and PHB R phasmid neurons (Figure 5C).
Figure 5. T27A3.1 is expressed in some head and tail chemosensory neurons.
(A) Confocal micrographs of the head region of an adult sfEx27 stained with DiD. Each row of images represents a different dorsal to ventral view to allow us to visualize all of the DiD labeled neurons. DiD staining is shown in red and GFP in green. In the head, DiD is known to back-fill 6 pairs of amphid neurons including ASK, ASI, ADL, AWB, ASH and ASJ neurons which can be identified based on their respective positions. T27A3.1pro::gfp is expressed in the ASK and ASI pairs of chemosensory neurons. In addition to being present in some amphid neurons, GFP was also present in ADF and ASE chemosensory neurons. Arrows indicate overlapping GFP and DiD in neuronal processes in the nerve ring.
(B) Schematic showing the position of the various neurons in the posterior ganglion modified from Wormatlas. T27A3.1pro::gfp expressing neurons are identified in green, DiD back-filled chemosensory neurons are in red, overlapping is shown in yellow.
(C) Confocal micrographs of the tail region of an adult sfEx27 hermaphrodite stained with DiD. In the tail region, DiD back-fills 2 pairs of phasmid neurons. The most anterior pair of neurons (arrows) corresponding to PHBL and PHBR shows overlapping GFP and DiD. Scale bars: A and C: 10 μm.
No lethal effect of T27A3.1 RNA interference
Since HAP1 homozygous animals die shortly following birth, we examined the effect of T27A3.1 RNA interference on worm viability. More specifically, we compared the number of laid oocytes, number of dead embryos, growth rate, and morphology between treated and non-treated worms. We found no effect of T27A3.1 RNAi by feeding the usual wildtype strain N2, which is consistent with two previously published high throughput studies (Fraser, 2000; Maeda, 2001). Because of the localization of T27A3.1 to neurons, and the known resistance of most neurons to systemic RNAi, we also performed RNAi in rrf-3(pk1426) young adults. Despite the fact that this line, in some cases, has been shown to respond better to RNAi than wild type N2 for some neuronally expressed genes (Simmer, 2003; Simmer, 2002; Timmons, 2004), we found no evidence of gross behavioral, morphological or growth defects following T27A3.1 RNAi. The bacterial clone used for silencing T27A3.1 produces an RNA spanning from exon 6 to exon 13 and therefore should silence all isoforms. We have also analyzed the effect of T27A3.1 RNAi by injection of double stranded RNA (dsRNA). For these experiments the fragment of RNA extended from within exon 2 to exon 8, a region also present in all isoforms. RNAi by injection has proven more effective for some genes and has led to the identification of phenotypes that were not evident by bacterial feeding RNAi [personal communication]. However, here again we found no obvious phenotype resulting from T27A3.1 RNA interference.
Discussion
If two proteins share a common ancestor they are called homologous. Such homologous proteins often share functional characteristics and/or a measurable similarity in amino acid sequence. Typically, a stretch of 100 amino acids bearing at least 25% identity suggests a common evolutionary origin for the two proteins. Our study shows that the T27A3.1d protein has at least 27% identity (aa 30-168) with HAP1 and Milton, suggesting a common evolutionary pathway for these proteins. Mammals have separate genes for HAP1 and Milton, but it would appear that invertebrates such as C.elegans and Drosophila have single genes (T27A3.1 and the gene encoding Milton, respectively). In C.elegans the T27A3.1 protein, seems to be equally similar to both HAP1 and Milton. We would speculate that during the course of evolution, a duplication event was followed by divergence so that distinctive functions were assumed by HAP1 and Milton. In fact, further homology studies with T27A3.1 have led us to conclude that HAP1 and Milton are part of a larger family of proteins, which include hALS2CR3 (or its ortholog, rGRIF) and OGT-IP. A T27A3.1 fragment between amino acid 30 and 243 shows 34.6% identity with human OGT-IP and 29.9% identity with hALS2CR3.
The region with the highest similarity between T27A3.1 and all of the above mentioned family members suggests that this region has a conserved biochemical function. In fact, this region overlaps with the region of HAP1 known to bind to htt. This suggests that T27A3.1 might be able to bind to a C.elegans homolog of htt. Although all the reported studies of htt in C. elegans have involved transgenic expression of a portion of htt from another organism (Faber, 1999; Parker, 2001), Wormbase identifies F21G4.6 as a potential htt homolog with 39.7% similarity to human htt (E-value: 4e−07).
If HAP1, Milton, GRIF, OGT-IP and T27A3.1 have similar functions, one might expect them to have similar or overlapping expression patterns. HAP1 expression has been studied in humans, rats, and mice where it is found only in neurons in both the central and peripheral nervous system (Dragatsis, 2000; Gutekunst, 1998; Li, 2003; Li, 1996; Martin, 1999). At this point, there are no studies describing the tissue distribution of Drosophila Milton. So far, we only know that fly heads contain Milton protein by Western blot (Gorska-Andrzejak, 2003; Stowers, 2002). The Milton silencing studies were done only in fly photoreceptor cells (Gorska-Andrzejak, 2003; Stowers, 2002). In rats, GRIF mRNA is found in the brain but also in other excitable tissues including heart, and skeletal muscle (Beck, 2002). Antibodies reactive to rat GRIF and hOGT-IP detected these proteins in all rat tissues as well as many human cell lines (Iyer, 2003). Our studies show that similar to HAP1, T27A3.1 is expressed in neurons. However, similar to GRIF and OGT-IP, expression of T27A3.1 is more widespread than that of HAP1. Thus, the widespread expression pattern of T27A3.1 is consistent with one protein serving the functions of multiple proteins in other organisms.
T27A3.1 expressing neurons were mostly found in the head and tail of the worm located in positions suitable to act as environmental sensors. Indeed, we have shown that some of the T27A3.1 expressing neurons are chemosensory. In C.elegans, chemosensory neurons respond to environmental chemical and thermal stimuli, and participate in multiple behavioral responses, as well as developmental decisions (Albert, 1981; Riddle, 1981). All chemosensory neurons share a set of common traits. These neurons respond to environmental stimuli by extending their dendrites towards the outside. Amphid neurons also extend axons into the nerve ring, where the environmental signal is integrated. The amphid neurons expressing T27A3.1 are involved in chemotaxis to sodium (ADF, ASEL, ASI), chloride (ADF, ASER), cyclic AMP (ADF, ASE, ASI), biotin (ADF, ASE), lysine (ASE, ASK) and potassium (ASE) based on studies in which animals were tested for their responsiveness following laser ablation of specific neurons (Bargmann, 1991). Significantly, T27A3.1 is expressed in ASEL but not ASER, and in ADFL but not ADFR. Similar asymmetric expression has been described for various guanylyl cyclase isoforms, with gcy-5 expressed exclusively in ASER and gcy-6 and gcy-7 exclusively in ASEL (Chang, 2004; Yu, 1997). Likewise, the homeobox gene lim-6 is expressed in ASEL but not in ASER (Pierce-Shimomura, 2001). Asymmetry in gene expression in the chemosensory neurons might enable the worms to better detect chemical gradients (Yu, 1997), and to better discriminate between odors in complex environments (Wes, 2001). The pattern of GFP expression we observed indicates that the promoter region of all T27A3.1 isoforms, including up to 4kb of nucleotides upstream from the ATG site of exon 1, does not drive expression only in neurons of the nematode. However, it is possible that the various isoforms are expressed in different cell types and that only some of the isoforms are specifically expressed in neurons. We are now generating antibodies to determine the tissue expression patterns of the various T27A3.1 isoforms and to study their subcellular localization.
In rat neurons, HAP1 is normally localized in the cytoplasm. Immunogold localization shows that HAP1 associates with microtubules and the cytoplasmic surface of many membrane-bound organelles such as vesicles, tubular vesicles, and mitochondria (Gutekunst, 1998). HAP1 has also been shown to interact with motor proteins, and it is hypothesized that it might participate in the trafficking of proteins and organelles within processes (BlockGalara, 1997; Gutekunst, 1998; Martin, 1999). HAP1 was shown to be homologous to Milton, a protein involved in mitochondria transport in fruit flies. Thus, although there is evidence that both HAP1 and Milton are associated with mitochondria, whether T27A3.1 also associates with mitochondria or is even involved in transport is unknown. However it is interesting that a large scale yeast-two-hybrid study (Li, 2004) has identified the C. elegans gene ZC97.1, an ortholog of Metaxin-2 (Armstrong, 1997; Armstrong, 1999), a protein localized at the surface of mitochondria, as a potential binding partner of T27A3.1.
Our alignment data showing T27A3.1 as part of a family of proteins including HAP1, Milton, rGRIF and OGT is interesting for two reasons. First, there is evidence that some of these family members associate with GABA receptor subunits. rGRIF associates with GABA(A) receptor α1 and β2 subunits in adult rat brain (Beck, 2002). In fact, the region of rGRIF critical for GABA(A) receptor β2 binding (residues 124-283) is located in the N-terminal region shown to have the most similarity with T27A3.1 in our alignment (29.9 % identity and 48.9 % similarity). Second, HAP1 has also been recently shown to associate with GABA(A) receptor β1- β3 subunits via residues between 220-520 (Kittler, 2004), which overlap with the same region. Identification of GABA receptor binding regions in rGRIF and HAP1 suggest that the other family members present in our alignment, including T27A3.1 and Milton, might also interact with some of the GABA receptor subunits. Another possibility is that these proteins are part of a larger protein complex containing GABA receptors. Interestingly out of the various T27A3.1 expression neurons we have identified only the CAN neurons are suggested to express GABAA/glycine receptor-like protein GGR-2 (Fujiwara, 1996).
Our RNAi data on T27A3.1 are consistent with those of other studies, but are inconclusive. We observed no obvious effect on morphology, growth or behavior in both wildtype and RNAi hypersensitive backgrounds. If T27A3.1 has a non-redundant function in the non-neuronal cells in which it is expressed, we would have expected a phenotype. One possibility is that the phenotype may have been too subtle to detect, which mimics the rescue of the mice in a non-competitive environment. Another possibility is that if the neurons were resistant to systemic RNAi, a phenomenon commonly known in the worm, no obvious behavioral effect would have been seen. Of course it is possible that RNAi for T27A3.1 will have subtle behavioral defects in chemotaxis or thermotaxis, which will be investigated further.
Acknowledgements
We thank A. Fire (Stanford University School of Medicine) for expression vectors. We thank J. Culotti (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada) for the pan neuronal GFP marker strain.
This work was supported by a grant from NIH AR051466 (GMB), and a grant from the Emory University Research Committee.
Literature Cited
- Albert PS, Brown SJ, Riddle DL. Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol. 1981;198:435–451. doi: 10.1002/cne.901980305. [DOI] [PubMed] [Google Scholar]
- Altun-Gultekin , Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Armstrong LC, Komiya T, Bergman BE, Mihara K, Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J Biol Chem. 1997;272:6510–6518. doi: 10.1074/jbc.272.10.6510. [DOI] [PubMed] [Google Scholar]
- Armstrong LC, Saenz AJ, Bornstein P. Metaxin 1 interacts with metaxin 2, a novel related protein associated with the mammalian mitochondrial outer membrane. J Cell Biochem. 1999;74:11–22. [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Beck M, Brickley K, Wilkinson HL, et al. Identification, molecular cloning, and characterization of a novel GABAA receptor-associated protein, GRIF-1. J Biol Chem. 2002;277:30079–30090. doi: 10.1074/jbc.M200438200. [DOI] [PubMed] [Google Scholar]
- Bertaux F, Sharp AH, Ross CA, Lehrach H, Bates GP, Wanker E. HAP1-huntingtin interactions do not contribute to the molecular pathology in Huntington’s disease transgenic mice. FEBS Lett. 1998;426:229–232. doi: 10.1016/s0014-5793(98)00352-4. [DOI] [PubMed] [Google Scholar]
- BlockGalara J, Chase K, Sapp E, et al. Fast transport and retrograde movement of huntingtin and HAP 1 in axons. Neuroreport. 1997;8:2247–2251. doi: 10.1097/00001756-199707070-00031. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis Elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E, Nassir J, Gutekunst CA, et al. Targeted disruption of Huntingtin-associated protein-1 (Hap1) results in postnatal death due to depressed feeding behavior. Human Molecular Genetics. 2002;11:945–959. doi: 10.1093/hmg/11.8.945. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr., Frokjaer-Jensen , Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Colomer V, Engelender S, Sharp A, et al. Huntingtin-associated protein 1 (HAP1) binds to a Trio-like polypeptide, with a rac1 guanine nucleotide exchange factor domain. Hum Mol Genet. 1997;6:1519–1525. doi: 10.1093/hmg/6.9.1519. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Dietrich P, Zeitlin S. Expression of the Huntingtin-associated protein 1 gene in the developing and adult mouse. Neurosci Lett. 2000;282:37–40. doi: 10.1016/s0304-3940(00)00872-7. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Zeitlin S, Dietrich P. Huntingtin-associated protein 1 (Hap1) mutant mice bypassing the early postnatal lethality are neuroanatomically normal and fertile but display growth retardation. Hum Mol Genet. 2004;13:3115–3125. doi: 10.1093/hmg/ddh328. [DOI] [PubMed] [Google Scholar]
- Engelender S, Sharp A, Colomer V, et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150(Glued) subunit of dynactin. Human Molecular Genetics. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci U S A. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Garriga G. Genes necessary for C. elegans cell and growth cone migrations. Development. 1997;124:1831–1843. doi: 10.1242/dev.124.9.1831. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos , Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Ishihara T, Katsura I. Ligand-gated chloride channel is necessary for correct thermotaxis. Worm Breeder’s Gazette. 1996;14:69. [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages , et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Gorska-Andrzejak , Stowers RS, Borycz J, Kostyleva R, Schwarz TL, Meinertzhagen IA. Mitochondria are redistributed in Drosophila photoreceptors lacking Milton, a kinesin-associated protein. J Comp Neurol. 2003;463:372–388. doi: 10.1002/cne.10750. [DOI] [PubMed] [Google Scholar]
- Gutekunst C, Li S, Yi H, Ferrante R, Li X, Hersch S. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): Comparison with huntingtin in rat and human. J of Neuroscience. 1998;18:7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst CA, Levey AI, Heilman CJ, et al. Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc Natl Acad Sci U S A. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- HDCRG T. H. s. D. C. R. G. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J Biol Chem. 2003;278:5399–5409. doi: 10.1074/jbc.M209384200. [DOI] [PubMed] [Google Scholar]
- Kittler JT, Thomas P, Tretter V, et al. Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating {gamma}-aminobutyric acid type A receptor membrane trafficking. Proc Natl Acad Sci U S A. 2004;101:12736–12741. doi: 10.1073/pnas.0401860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Gutekunst CA, Hersch SM, Li XJ. Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci. 1998a;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Hosseini SH, Gutekunst CA, Hersch SM, Ferrante RJ, Li XJ. A human HAP1 homologue. Cloning, expression, and interaction with huntingtin. J Biol Chem. 1998b;273:19220–19227. doi: 10.1074/jbc.273.30.19220. published erratum appears in J Biol Chem 1999 Apr 2;274(14):9906. [DOI] [PubMed] [Google Scholar]
- Li SH, Li H, Torre ER, Li XJ. Expression of huntingtin-associated protein-1 in neuronal cells implicates a role in neuritic growth. Mol Cell Neurosci. 2000;16:168–183. doi: 10.1006/mcne.2000.0858. [DOI] [PubMed] [Google Scholar]
- Li SH, Yu ZX, Li CL, et al. Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington’s disease. J Neurosci. 2003;23:6956–6964. doi: 10.1523/JNEUROSCI.23-17-06956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li S, Sharp A, et al. A Huntingtin-Associated Protein Enriched In Brain With Implications for Pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- Li X, Sharp A, Li S, Dawson T, Snyder S, Ross C. Huntingtin-associated protein (HAP1): Discrete neuronal localizations in the brain resemble those of neuronal nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:4839–4844. doi: 10.1073/pnas.93.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chin LS, Levey AI, Li L. Huntingtin-associated protein 1 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate and functions in endosomal trafficking. J Biol Chem. 2002;277:28212–28221. doi: 10.1074/jbc.M111612200. [DOI] [PubMed] [Google Scholar]
- Liu KS, Sternberg PW. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron. 1995;14:79–89. doi: 10.1016/0896-6273(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Marcora E, Gowan K, Lee JE. Stimulation of NeuroD activity by huntingtin and huntingtin-associated proteins HAP1 and MLK2. Proc Natl Acad Sci U S A. 2003;100:9578–9583. doi: 10.1073/pnas.1133382100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Kim M, Velier J, et al. Analysis of Huntingtin-associated protein 1 in mouse brain and immortalized striatal neurons. Journal of Comparative Neurology. 1999;403:421–430. [PubMed] [Google Scholar]
- Metzger S, Rong J, Nguyen HP, et al. Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum Mol Genet. 2008;17:1137–1146. doi: 10.1093/hmg/ddn003. [DOI] [PubMed] [Google Scholar]
- Nasir J, Duan K, Nichol K, et al. Gene structure and map location of the murine homolog of the Huntington-associated protein, Hap1. Mammalian Genome. 1998;9:565–570. doi: 10.1007/s003359900819. [DOI] [PubMed] [Google Scholar]
- Nasir J, Maclean A, Engelender S, et al. Chromosomal localization of the Huntingtin associated protein (HAP-1) gene in mouse and humans with radiation hybrid and interspecific backcross mapping. Mamm Genome. 1999;10:397–398. doi: 10.1007/s003359901009. [DOI] [PubMed] [Google Scholar]
- Page K, Potter L, Aronni S, Everitt B, Dunnett S. The expression of Huntingtin-associated protein (HAP1) mRNA in developing, adult and ageing rat CNS: implications for Huntington’s disease neuropathology. European Journal of Neuroscience. 1998;10:1835–1845. doi: 10.1046/j.1460-9568.1998.00185.x. [DOI] [PubMed] [Google Scholar]
- Parker JA, Connolly JB, Wellington C, Hayden M, Dausset J, Neri C. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc Natl Acad Sci U S A. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Kambampati V, Mains RE, Eipper BA. Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J Neurosci. 2001;21:8426–8434. doi: 10.1523/JNEUROSCI.21-21-08426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Faumont S, Gaston MR, Pearson BJ, Lockery SR. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature. 2001;410:694–698. doi: 10.1038/35070575. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Strange K. From genes to integrative physiology: ion channel and transporter biology in Caenorhabditis elegans. Physiol Rev. 2003;83:377–415. doi: 10.1152/physrev.00025.2002. [DOI] [PubMed] [Google Scholar]
- Tang TS, Tu H, Chan EY, et al. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39:227–239. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L. Endogenous inhibitors of RNA interference in Caenorhabditis elegans. Bioessays. 2004;26:715–718. doi: 10.1002/bies.20078. [DOI] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]