1. Introduction
Post-translational modifications (PTM) of histones include acetylation, methylation, ubiquitination, phosphorylation, ADP-ribosylation and sumoylation, which play important roles in regulating transcription, chromatin assembly, DNA repair, recombination and DNA replication (1-2). Histone modifications also serve as epigenetic marks that can be inherited through cell division to maintain lineage specificity (3). Therefore, determination of PTM functions has been a central focus of the chromatin field for the past two decades (4).
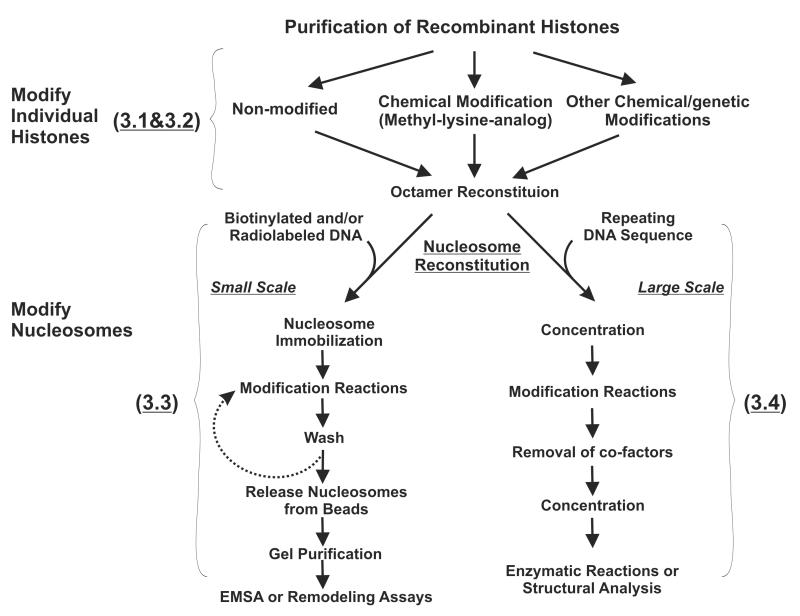
Chemically modified histone peptides are commonly utilized to identify PTM recognition modules or used as substrates for various enzymatic reactions in vitro. Although this type of assay has led to many major discoveries in the field, some intrinsic shortcomings limit their broad applications. First, short histone peptides may only cover a partial functional surface of histones; second, without DNA, free histone peptides may not recapitulate the native conformation of intact nucleosomes; third, technical limitations of peptide synthesis may prevent the desired combination of PTM when multiple histones are involved or a great distance between modifications is needed. Therefore, using chromatin templates that carry specific PTM becomes increasingly desirable for biochemical analysis of histone modifications. In this chapter, we describe two sets of protocols for reconstituting designer nucleosomes that contain specifically modified histones. We first present a small scale reconstitution method in which radiolabeled DNA templates are used and resulting nucleosomes are suitable for electro-mobility shift assays (EMSA) or chromatin remodeling reactions (Figure 1 and subheading 3.1, 3.2 and 3.3). We then discuss a generic method to prepare modified nucleosomes on a large scale for functional and structural studies. Histone modifications can be introduced either at the level of individual histones through chemical approaches (Figure 1, subheading 3.2) (5-7) or at the level of nucleosomes via a broad range of site specific histone modifying enzymes (Figure 1, subheading 3.3 and 3.4) (8-9). With proper pairings of these two strategies, one can expect to generate a single nucleosome containing various combinations of histone modifications for studying cross-talk between PTMs. Due to space limitation, in the cases where procedures described here were adapted from previously established protocols (5, 10-12), we will primarily emphasize the modifications which we have made and the critical parameters that are important for successful subsequent steps.
Figure 1.
A schematic diagram of our strategies to modify nucleosomes for studying PTM cross-talk. Topics covered in each subtitle were summarized with a bracket.
2. Materials
2.1. Preparation of recombinant histone octamers
Recombinant histone H3, H4, H2A and H2B
Spectra/Por Dialysis membrane (3500 MWCO with a flat width of 18mm, Spectrum)
Amicon ultra concentrator (5000 MWCO, Millipore)
Unfolding buffer: 6M guanidinium chloride, 20mM Tris-HCl (pH 7.5), with freshly added 5mM DTT.
Refolding buffer: 2M NaCl, 10mM Tris.HCl (pH 7.5), 1mM EDTA, 5mM 2-Mercaptoethanol
Superdex 200 column (GE Healthcare)
SDS running buffer: 25mM Tris, 250mM glycine, pH 8.3, 0.1% SDS
3× SDS loading buffer: 40% glycerol, 3% SDS, 0.8M 2-Mercaptoethanol, 15μM Bromophenol Blue
Coomassie Blue staining buffer: 3mM Coomassie Blue R250, 50% methanol, 10% acetic acid
Destaining Buffer: 50% methanol, 10% acetic acid
2.2 Installing MLA (Methyl-Lysine-analog) onto histones
Recombinant histones
50°C incubator
Alkylating buffer (made freshly): 2M HEPES pH 7.8, 8M Guanidine-HCl, 0.2M methionine
1M DTT (all DTT buffers in this session are made freshly from powder)
Alkylating agents: Agent1 (2-N-Methylaminoethyl chloride HCl for mono-methylation); Agent2 (2-Chloro-N,N-dimethylethylamine hydrochloride) for di-methylation; Agent3 ((2-Bromoethyl) trimethylammonium bromide) for tri-methylation.
5mM 2-Mercaptoethanol /H2O: 5mM 2-Mercaptoethanol in H2O
PD-10 Column (GE Healthcare)
2.3. Preparation of radioactive labeled nucleosomes for EMSA and sliding
Primers for PCR amplification of probe DNA: Primer 774 (Bio-601L5′): 5′-BiO -cgAGGCCTcagctgGATATCacaggatgtatatatctgacacgtgcc (Note: the underlined sequence contains multiple restriction enzyme digestion sites which can be used to release the DNA/nucleosomes from the beads. EcoRV is our primary choice.) Primer 773 (601L-216-3′): 5′-TGACCAAGGAAAGCATGATTCTTCACAC
The 601 positioning sequence containing plasmid (pBL386-601R)
γ-32P-ATP, 1mCi/7μl
T4 Polynucleotide Kinase (T4 PNK)
LA-Taq™ DNA polymerase (TaKaRa)
ProbeQuant G-50 mini column (GE Healthcare)
DNA size standards
Gel extraction kit (e.g. QIAquick® gel extraction kit, Qiagen)
Recombinant histone octamers (modified and unmodified) as prepared via subheading 3.1.
Initial Buffer: 50mM HEPES (pH 7.5), 1mM EDTA, 0.1% Ipegel CA-630, 20% glycerol, 5mM DTT, 0.5mM PMSF, 100μg/ml BSA
Final Buffer: 10mM Tris-HCl(pH 7.5), 1mM EDTA, 0.1% Ipegel CA-830, 20% glycerol, 5mM DTT, 0.5mM PMSF, 100μg/ml BSA
10×TBE buffer: 890mM Tris base, 890mM boric acid, 20mM EDTA
IF100 Buffer: a mixture of 1 volume of Initial Buffer, 1 volume of Final Buffer and 0.04 volume of 5M NaCl
Dynabeads® M-280 Streptavidin coated magnetic beads (Invitrogen)
Buffer H600: 25mM HEPES pH7.6, 0.5mM EDTA, 0.1mM EGTA, 2.5mM MgCl2, 10% Glycerol, 0.02% Ipegel CA-830, 1mM DTT, 0.1mg/ml BSA and 600mM KCl.
Histone modifying complexes that are active on nucleosome templates (See Note7)
100μM Acetyl-CoA
200μM S-Adenosyl methionine (SAM)
5×HAT buffer: 250mM Tris-HCl pH 8.0, 250mM KCl, 5mM DTT, 25% glycerol, 0.03% Ipegel CA-830
5×HMT buffer: 250mM Tris-HCl pH 8.0, 250mM NaCl, 5mM MgCl2, 10mM DTT, 25% glycerol, 0.03% Ipegel CA-830
RE Digestion Buffer: 50mM HEPES pH 7.9, 100mM NaCl, 2mM MgCl2, 0.04% Ipegel CA-830, 0.1mg/ml BSA and 5mM DTT
Gel Elution Buffer: 10mM Tris pH7.4, 100mM NaCl, 1mM EDTA, 5mM DTT, 0.5mM PMSF, 0.1mg/ml BSA
5× Gel Final Buffer: 10mM Tris pH7.4, 100mM NaCl, 1mM EDTA, 5mM DTT, 0.5mM PMSF, 0.1mg/ml BSA, 50% glycerol, 0.25% Ipegel CA-830
2.4. Preparation of modified nucleosome in a large scale
EcoR V (100,000 U/ml)
Ice-cold 100% Ethanol
Isopropanol
5M NaCl, autoclaved
3M Sodium acetate (pH 5.2), autoclaved
TAE buffer; 0.04 M Tris-acetate, 1mM EDTA (pH 8.0), 3.3mM glacial acetic acid
10× DNA loading dye
40% PEG8000, autoclaved
Spectra/Por Dialysis membrane (3500 MWCO with a flat width of 18mm, Spectrum)
GelRed™ Nucleic Acid Gel Stain; 10,000× in water (Biotium)
10×TEB: 100mM Tris-HCl pH7.5, 10mM EDTA, 10mM 2-Mercaptoethanol
TEBS (Dialysis buffer): 1× TEB supplemented with different concentrations of NaCl as follows: TEBS2.0, TEBS1.2, TEBS1.0, TEBS0.8 and TEBS0.6 contain 2M, 1.2M, 1M, 0.8M and 0.6M NaCl respectively.
Native PAGE gel and electrophoresis equipments
10×TBE buffer: 890mM Tris base, 890mM boric acid, 20mM EDTA
Acetyl Coenzyme A, [Acetyl-3H] and non-labeled Acetyl-CoA
Sephadex G-50 purification column
P81 Phosphocellulose membrane
1× HAT Wash buffer: 50mM NaHCO3/Na2CO3 pH9.2
Acetone
Scintillation fluid for filters
TE 10/0.1: 10mM Tris.HCl 8.0; 0.1mM EDTA
2.5 Histone deacetylase (HDAC) assays
1× HDAC Buffer: 20mM Tris-HCl pH8.0, 5mM 2-Mercaptoethanol, 5% glycerol.
1M HCl/0.4M acetic acid
Ethyl acetate
Scintillation cocktail (e.g. ScintiSafe 30% Cocktail (Scintanalyzed*, Fisher))
3. Methods
3.1 Preparation of recombinant histone octamers
1. Lyophilize 0.15 μmol of recombinant histone H3, H4, H2A and H2B (6 hrs to overnight) (See Note 1)
Dissolve each histone pellet in 1 ml of Unfolding Buffer for 1 hour at room temperature on an orbital mixer. Determine the concentration of histones by measuring OD276 and using the calculated extinction coefficients (See Note 2).
Mix the four histones at an equimolar ratio and adjust the final concentration to 1mg/ml with Unfolding Buffer. Place the mixture at RT for 30 minutes then dialyze (3500 MWCO) against pre-chilled Refolding Buffer at 4°C as follows: 800 ml for 1 hr; another 800 ml for 1hr; then overnight in remaining 2.4 L.
Centrifuge at 26940.0 ×g (SS-34 rotor) for 10 mins at 4°C to remove any aggregates, then concentrate the supernatant to a final volume of 1ml using an Amicon-Ultra concentrator (5000 MWCO). Centrifuge the sample at 21,000×g for 5 mins at 4°C to eliminate precipitates and then load it onto a Superdex 200 column (CV=120ml) pre-equilibrated with Refolding Buffer. The flow rate is set at 1ml/min and 1 ml fractions are collected after 30 ml.
Run peak fractions on a 16% SDS-PAGE gel to check the stoichiometry of histone octamers. Wild type histone octamers elute around 65-68 ml, and tailless histone octamers comes out at 70-74 ml.
Pool all fractions containing histone octamers and concentrate to about 1 ml (producing solution of about 3-8 mg/ml). To increase the consistency of nucleosome reconstitution, we determine the concentration of histone octamers through a gel quantification method, in which a standard curve is created using a sample of histone at known concentration. When Xenopus, Drosophila or human histones are used, the yield of octamers is about 80% of total histone proteins. Recombinant yeast histones are reconstituted less efficiently, with an average yield of about 20%.
3.2 Installing MLA (Methyl-Lysine-analog) to histones
The protocol in this session is based on a previous method with minor modifications (5).
Lyophilize 0.45 μmol (three 0.15 μmol aliquots) of genetically engineered Kc (the lysine of interest replaced by a cysteine) mutant histones (See Note 3).
Resuspend entire pellets in 1ml of freshly prepared alkylating buffer supplemented with 20μl of 1M DTT. Place the tube on a roller mixer at RT for 30 minutes until histones are completely dissolved, then transfer the tube to 37°C for 1hr to ensure cysteine residues are completely reduced by DTT. Wrap the tubes with foil to avoid direct light exposure in subsequent steps.
- Chemical reactions:
- Mono-methylation:
- Add 100 μl of 1M freshly made Agent1 (2-N-Methylaminoethyl chloride HCl), incubate at RT for 4hrs.
- Add 10 μl of 1M DTT and incubate at RT for another 10 hrs
- Di-methylation:
- Add 50 μl of 1 M freshly made Agent2 (2-Chloro-N,N-dimethylethylamine hydrochloride), and incubate at RT for 2 hrs.
- Add 10 μl of 1 M DTT and incubate at RT for 30 mins
- Replenish 50 μl of 1 M Agent2, and incubate at RT for 2 hrs.
- Tri-methylation:
- Directly add 100 μg of Agent3 (2-Bromoethyl trimethylammonium bromide) powder into the tube, and incubate at 50°C for 2.5 hrs with occasional stirring at the beginning to dissolve the powder.
- Add 10 μl of 1 M DTT and incubate at 50°C for another 2.5 hrs
Chemical reactions are stopped by addition of 50 μl of 2-Mercaptoethanol (14M)
Discard the upper storage buffer in PD-10 columns and pre-equilibrate the column four times with 5 ml of 5mM 2-Mercaptoethanol/H2O .
Load the reaction mixtures onto the PD-10 columns. Once samples are completely absorbed, add 1.5 ml of 5 mM 2-Mercaptoethanol/H2O but do NOT collect the flow through.
Transfer the PD-10 column to a fresh 50ml conical tube. Add 3.5 ml of 5mM 2-Mercaptoethanol/H2O and collect eluates, which contain purified chemically modified histones.
Determine the concentration of histones by measuring OD276 and using deducted extinction coefficients. Aliquot 0.15 μmol of modified histones in each tube, and store them at −80°C.
To reconstitute these MLA histones into octamers, proceed to Step 1 in section 3.1.
3.3. Preparation of radioactive nucleosomes for EMSA and sliding assays
End-label biotinylated DNA probe containing the 601 positioning sequence (See Note 4)
-
1.
End-label one primer by setting up the following reaction: 1μl of 20μM Primer 773 (601L-216-3′), 1 μl of 10× Kinase Buffer (T4 PNK NEB), 0.5 μl of γ-32P -ATP, 6.5 μl of H2O, 1 μl of T4 PNK. Incubate at 37°C for 60 mins then heat inactivate PNK at 70°C for 10 mins.
-
2.
PCR amplify the biotinylated DNA probe: mix 10 μl of 10×LA Taq buffer, 8 μl of 2.5 mM dNTPs, 10 μl of the entire kinase reaction from Step1, 1 μl of 20 μM Primer 774 (Bio-601-5′), 1 μl of pBL386-601R plasmid (10 ng/μl), 67 μl of H2O and 0.5 μl of LA Taq. 28-30 cycles of standard PCR conditions should result in adequate amount of probes.
-
3.
Pass the entire PCR reaction above through a ProbeQuant G-50 mini-column to remove unincorporated γ-32P ATP as suggested by manufacturers. The eluates are then directly loaded onto a 2.0% agarose gel in 1.5× TAE Buffer. The DNA band with the correct size is excised and purified using standard Qiagen Gel extraction purification Kits. We elute DNA with 20 μl of pre-warmed (42°C) 0.1× TE. Another 10 μl elution is performed to increase yield. Eluates are combined before the final quantification (See Note 5).
Nucleosome reconstitution via a serial salt-dilution method
-
4.
Nucleosome reconstitution is started by sequentially adding 5 M NaCl, 2 mg/ml BSA, H2O, 1 pmol of radio-labeled DNA and the proper amount of histone octamers (see Note 6) into a 10 μl reaction so that the final concentration of NaCl and BSA is 2 M and 0.1 mg/ml, respectively.
-
5.
The salt concentration is gradually reduced by adding 3.3 μl, 6.7 μl, 5 μl, 3.6 μl, 4.7 μl, 6.7 μl, 10 μl, 30 μl and 20 μl of Initial Buffer with a 15 minute interval between additions while samples are incubated at 30°C.
-
6.
The reaction is brought to 0.1 M NaCl by adding 100 μl of Final Buffer and incubated for another 15 minutes at 30°C. The efficiency of nucleosomes reconstitution is examined by running 2 μl of each sample on a 5% native PAGE gel in 0.3× TBE Buffer.
Immobilization of radio-labeled nucleosomes on magnetic beads
-
7.
Transfer 20 μl of streptavidin-coated magnetic beads slurry (10 mg/ml) to a screw cap tube, and wash beads twice with 200 μl of IF100 Buffer using a magnetic stand. Withdraw the supernatant after the last wash.
-
8.
Add the reconstitution mixture from Step 6 to the tube containing pre-washed magnetic beads and incubate overnight at 4°C on a rotating mixer to allow biotinylated nucleosomes binding to the streptavidin-coated magnetic beads.
-
9.
Nucleosome-bound beads are washed twice with 1ml of Buffer H600 to eliminate non-specific binding proteins. The beads are finally resuspended in 200μl of IF100 buffer and store in 4°C or directly proceed to the next step. This method typically results in a 90-95% of immobilization efficiency as estimated by radioactivity (cpm) recovery.
Histone modification reaction catalyzed by chromatin modifying enzymes
-
10.
Spin briefly the screw cap tube containing immobilized nucleosomes from Step 9, and then let the beads settle on a magnetic stand for 2 mins. Withdraw the supernatant carefully with a pipet, then wash the beads twice using 300μl corresponding reaction buffer supplemented with 0.1mg/ml BSA (in our case here, either 1× HAT (BSA) or 1× HMT (BSA) is used)
-
11.At this point, the beads can be directly subjected to various histone modification reactions (See Note 7). We typically set up the reactions in a separate tube. The last wash of 1× reaction buffer in Step 10 is removed right before the reaction mixtures are applied to the beads.
-
HAT Reaction (100μl)Mix 20 μl of 5× HAT buffer, 1 μl of 10 mg/ml BSA, 2.5 μl of 100 μM Acetyl-CoA and the proper amount of histone acetylases complexes (See Note 8) and bring the final volume to 100 μl with H2O.
-
HMT Reaction (100μl)Mix 20 μl of 5× HMT buffer, 1 μl of 10 mg/ml BSA, 2.5 μl of 200 μM SAM and the proper amount of histone methyltransferase and bring the final volume to 100 μl with H2O.
-
-
12.
Gently invert the tubes for a few times, then place them on a rotating mixer at 30°C for more than 4 hours
-
13.
Collect beads on a magnetic stand. Wash them three times with 500 μl of Buffer H600 to remove cofactors and enzymes.
-
14.
(Optional) Repeat Step 10 and 13 for additional modification reactions.
Restriction enzyme digestion to release nucleosomes from magnetic beads
-
15.
The beads from the above procedures (Step 13 or 14, containing ~1 pmol DNA) are washed with 200 μl of RE digestion buffer twice.
-
16.
Add 50 units of EcoRV to 50 μl of RE digestion Buffer then apply the mixture to pre-washed beads from Step 15. Incubate the tubes on a rotating mixer at 37°C for 4-8 hrs. Transfer the supernatant to another tube; monitor and compare the radioactivity of supernatant with the beads using a Geiger survey meter. A typical recovery rate is about 70-80%. The resulting nucleosomes can be stored at 4°C for up to two months.
Gel-purification of all nucleosomes at the final stage
-
17.
If the volume of the final elution is well above 50 μl, use Amicon ultra concentrator (10000 MWCO) to bring it down to about 50 μl.
-
18.
Load samples on a 5% PAGE gel in 0.3×TBE. Run at 11 volts/cm at 4°C for 5 hrs.
-
19
. Cover the wet gel with plastic wrap and expose directly to an X-ray film for 30 mins. Use a black sharpie to mark down the gel position so that the bands corresponding to the desired nucleosomes can be precisely excised when the developed film is used as a reference.
-
20
. Smash the excised gel slice into small pieces. Add about 1.5 gel-volume of Gel elution buffer (~200-400 μl), and elute the nucleosomes overnight at 4°C on a roller mixer.
-
21
. Centrifuge at 21,000 × g for 5 mins at 4°C. Use barrel tips to withdraw the liquid from the gel then transfer to a 0.5ml eppendorf tube. Add 1/4 volume of 5× Gel Final buffer to the eluates and store them at 4°C.
These nucleosomes can be directly used as substrates in EMSA assays or chromatin remodeling assays as described in another chapter by Tom Owen-Hughes.
3.4. A Large Scale Preparation of Modified Nucleosomes
-
1.
Construct vectors that contain multiple copies of certain nucleosome positioning sequences (such as the 601 sequence and 5S) or other designer sequences. Our strategy to construct repeating 601 sequences in pBlueScript vector is illustrated in Figure 2, which is adopted from a previously established method (10) (See Note 9)
-
2.
Prepare milligram quantities of plasmids that contain the repeating sequence (see Note 10)
-
3.
Digest 5 mg of plasmids using 500 units of EcoRV (16 copies of 216 bp fragments) in a 10 ml reaction at 37°C for more than 16 hrs. Check the completeness of digestion by running 1 μl of the sample on a 1.0% agarose gel. If undigested products remain, add more EcoRV and incubate for another 4 hrs.
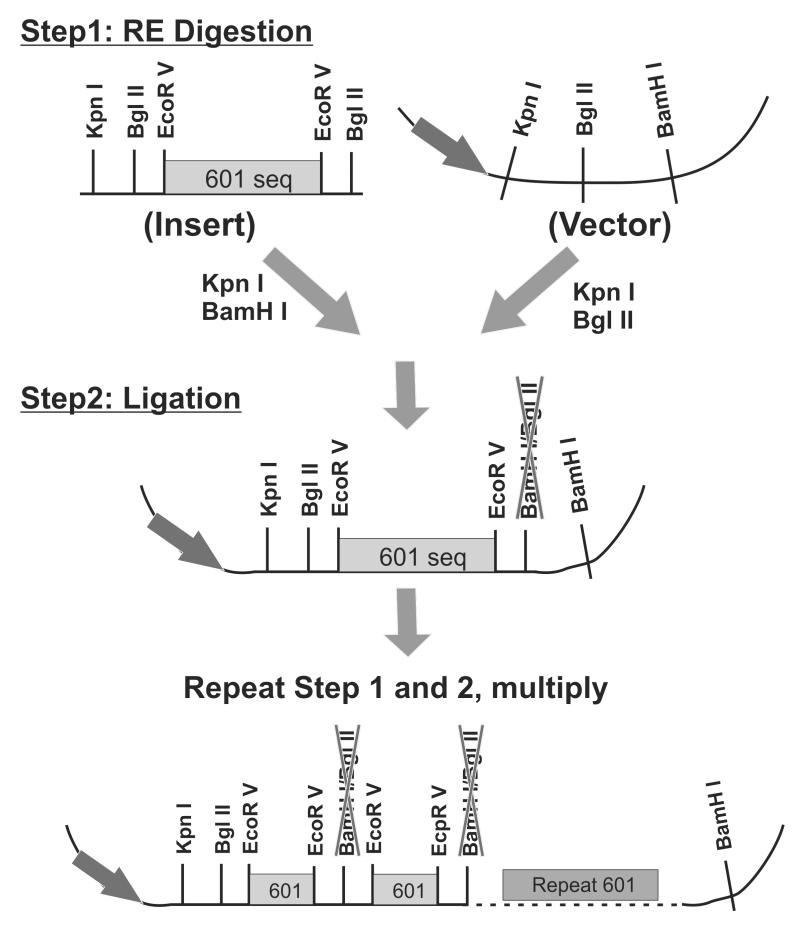
Figure 2.
Construction of vectors that contains multiple copies of nucleosome positioning sequence. A plasmid containing a single copy of 601 sequence was digested with two sets of restriction enzymes (Step 1) and ligated to produce 2× copies of 601 (Step 2).
Optimize the conditions for PEG purification of small DNA fragments
-
4.
Set up five testing conditions each containing 1 μg of digested plasmid (2 μl of the reaction mixture in Step 3), and 0.5 M NaCl. A different amount of 40% PEG8000 is then added to each condition so that the final concentration of PEG is 6%,7%, 8%, 9%, or 10%, respectively. Bring the final volume of each reaction to 100 μl using 1× Buffer3 (NEB) and mix well.
-
5.
Incubate these tubes on ice for 1hr, then centrifuge at 21,000×g at 4°C for 20 mins. Carefully transfer the supernatant, which contains the EcoRV digested small fragments, to a new eppendorf tube and resuspend the pellets (vector backbone) in 20 μl TE 10/0.1.
-
6.
Add 250 μl (or 2.5 volumes) of pre-chilled ethanol to each tube, and incubate at −20°C for 30 mins. Spin down the fragments at 21,000×g at 4°C for 20min. Dissolve the pellets in 50 μl of TE 10/0.1.
-
7.
Load 2μl of samples from Step 5 and Step 6 on a 1% agarose gel (Figure 3). Select the condition which results in the highest yield of small fragment without any backbone contamination.
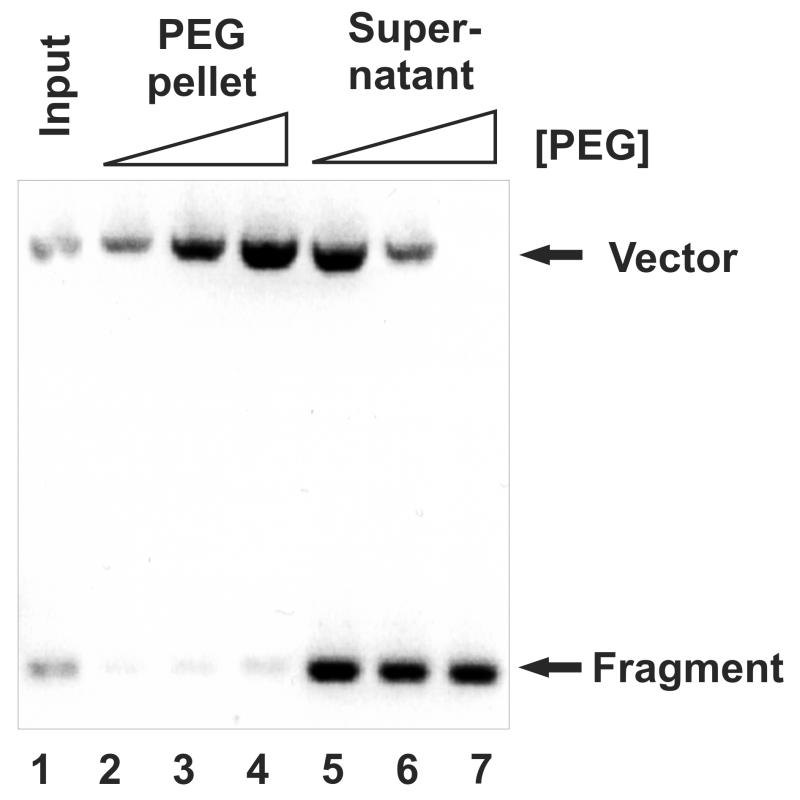
Figure 3.
Optimized PEG purification of small DNA templates. 1 μg of plasmid DNA was digested with EcoRV and precipitated with 7% (lane 2 and 5), 8% (lane 3 and 6) and 9% of PEG (lane 4 and 7). DNA purified from both pellet (Lane 2,3,4) and supernatant (Lane 5,6,7) after PEG precipitation was run on 1% agarose gel and the ratio of small fragment (fragments) and backbone (vector) was determined. 0.5 μg of DNA directly purified after digestion was loaded in Lane 1 (Input).
Purification of small DNA fragments for nucleosome restitution
-
8.
Add 2 ml of 5 M NaCl to the 10ml RE digestion mixture (Step 3) and the proper amount of 40% PEG8000 as determined above (Steps 4-7). Bring the final volume to 20 ml using 1× Buffer3 before mixing the solution and incubate on ice for 1hr.
-
9.
Centrifuge at 26940.0 xg (SS-34) at 4°C for 20mins. Transfer the supernatant to another Oak Ridge tube. Add 20 ml of Isopropanol and incubate at RT for 15 mins.
-
10.
Centrifuge at 26940.0 xg (SS-34) at 4°C for 20mins. Rinse the pellets with 10 ml of 70% EtOH. Dissolve the air-dried pellets in 1 ml of TE 10/0.1, and determine the concentration of DNA fragments by comparing to known concentration DNA marker on a 2% agarose gel.
Reconstitution of Nucleosome Core Particles
-
11.
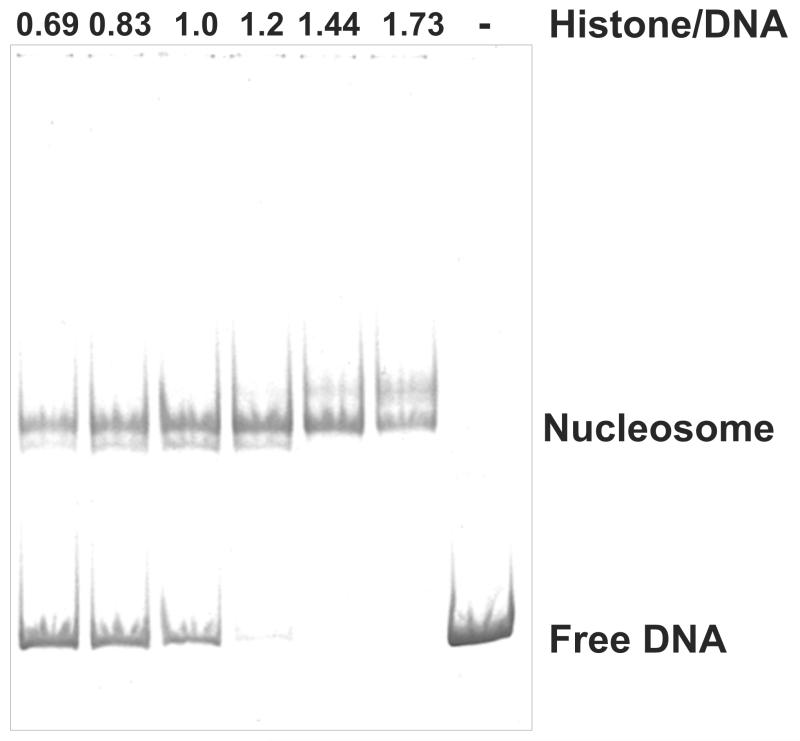
Empirically determine the optimal ratio of histone/DNA for the large scale nucleosome reconstitution. Set up nucleosome reconstitution reactions as described in subheading 3.3 (Steps 4-7). Use 2 μg of DNA fragment for each reaction and titrate histone octamers in 1.25 fold increments. Load 20 μl of the final mixtures on a 4.5% native PAGE gel and run it at 11 volts/cm for 1.5 hrs at RT. Stain the gel with the GelRed dye for 20 mins, visualize DNA (nucleosomes) under UV light (Figure 4).
-
12.
Mix 500 μg of DNA fragments (from Step 10), 4 ml of 5 M NaCl, 100 μl of 10× TEB buffer. Use H2O to bring the final reconstitution mixture to 1 ml (including histone octamers). Gently invert the tube several times then add the proper amount of histone octamers as determined above. Incubate the tube at 30°C for 30min, and then transfer the mixture to a dialysis bag (3500 MWCO).
-
13.Dialyze against 800ml of each buffer listed below at RT with indicated duration:
TEBS2.0 (2M NaCl) 1hr TEBS1.0 2hrs TEBS0.8 2hrs TEBS0.6 2hrs 1×TEB overnight (at 4°C) -
14.
Transfer the reconstitution mixture to an eppendorf tube and incubate at 37°C for 30mins. Remove aggregates by centrifuging at 21,000×g for 10 mins at 4°C. Adjust the final concentration of nucleosomes to 1-2 mg/ml using Amicon ultra concentrators.
Figure 4.
Determination of the Histone/DNA ratio for large scale nucleosome reconstitution. Nucleosome core particles were reconstituted with 2 μg of DNA templates and histone octamers with 1.25 fold increments. 10% resulting nucleosomes were run on a 4.5% native PAGE gel and visualized by GelRed staining under UV light. Numbers on top of each lane indicate the ratio of histone to DNA.
Histone modification reactions (acetylation)
-
15.
Mix 20 μg of reconstituted nucleosomes, 30 μl of 5× HAT buffer, 4 μl of 20 μM 3H-acetyl CoA and the proper amount of histone acetyltransferases, and adjust the final volume to 150 μl with H2O. Incubate at 30°C for 20 hrs.
-
16.
Carefully load the entire reaction onto a G-50 mini column prewashed with 1× HAT buffer. Centrifuge at 700×g for 2 mins to collect modified nucleosomes. Un-incorporated co-factors (acetyl-CoA in this case) should remain in the column (See Note 11).
-
17.
(Optional) Repeat Step 15 and 16 to install other modifications enzymatically.
-
18
. The overall acetylation level can be determined by the standard filter-binding assay (See Note 12) using a scintillation counter. To check the acetylation specificity, 0.5 μg of purified acetylated nucleosomes is subject to western blotting using antibodies against specifically acetylated histones.
3.5 Histone Deacetylase (HDAC) Assays Using Nucleosomal Templates
Mix 0.5 μg of acetylated nucleosome substrates (3H labeled) with a proper amount of histone deacetylase. Bring the final volume to 25 μl using 1× HDAC buffer.
Incubate at 30°C for 1 hr.
Add 36 μl of 1 M HCl/0.4 M acetic acid, and briefly vortex the tubes to stop the reactions.
Add 800 μl of ethyl acetate and vigorously vortex for 5 seconds.
Centrifuge at 12,000×g for 10 minutes at 4°C.
Transfer supernatant (~720 μl) to a scintillation vial containing 4 ml of scintillation fluid for liquid counting.
Vortex each vial vigorously for 5 seconds, then measure the radioactivity using a scintillation counter.
Footnotes
We prepare each individual recombinant histone based on the general protocol described by the Luger lab (10). We typically use 3-liter culture for wild type histones and 1.5 liter for mutants, which yield about 100-500mg of histones. We found that applying the solubilized inclusion bodies through a single ion-exchange column (5ml Hi-Trap SP (GE)) results in adequate purity for the procedures described here.
The concentrations of histones are determined by OD276 using the theoretical extinction coefficients calculated on the ExPASy Proteomics Server. For mutant histones, concentrations are further confirmed by running on a SDS-PAGE gel and comparing to a standard curve of a histone with known concentration.
For histones containing native cysteines, these residues should first be mutated to alanine. The resulting “wild type” histones should be tested in available in vitro and/or in vivo functional assays to ensure that the C-to-A mutation itself introduces minimal interference within the intended experiments. The commonly used histone H3C110A does not result in any noticeable phenotype in all tested assays thus far. Your favorite lysine(s) can then be substituted with cysteine(s) in this “wild type” background. Multiple cysteines can be introduced in one histone (such as H3Kc9 and H3Kc36); however, only one type of methylation analog can be installed at these residues using the current protocol. We have tested various combinations of double lysine mutant histones, and found that the recipes described here are sufficient for completion of such double reactions.
If chemical approach can generate all intended histone modification combinations, non-biotinylated DNA templates can be used. Proceed to nucleosome reconstitution (Step 4-6). In this case, these nucleosomes can be directly gel-purified starting from Step 17 in subheading 3.3. 32P also can be incorporated into DNA using a body-labeling method in which PCR reactions using the same primer sets are carried out in the presence of 32P-labeled nucleotides.
We measure radioactivity incorporation (cpm) by counting 1μl of purified DNA on a scintillation counter. We then determine the DNA concentration (ng/μl) by running 1 μl of DNA on a 2% Agarose gel with known concentration DNA markers. This allows us to deduce the ratio of cpm/ng for the labeled DNA and estimate the amount of DNA during subsequent steps by monitoring the radioactivity (cpm).
Theoretically histone and DNA should be in a 1:1 molar ratio of nucleosomes binding sites to octamers or a 1:1.3 mass ratio of histone octamers to DNA. We typically perform a titration test with 1.25 fold increments. Once the optimal histone/DNA ratio for any given histone octamer is determined, it can be consistently used for the same preparation with different DNA probes.
The histone modifying enzymes that are suitable for the assays described in this chapter should be highly active and possess nucleosomal activity. Preliminary tests are normally performed to determine the necessary amount of enzymes and the completeness of the modification reactions. If 80-90% of a target population is modified, these nucleosomal templates are generally satisfactory for most other assays.
The generic histone acetyltransferase complexes used here are purified from yeast using the TAP method (13). We typically use Ada2-TAP, use of which yields a mixture of three HAT complexes: SAGA, SLIK and ADA, to provide the histone H3 specific HAT, and the NuA4 (Epl1-TAP) complex to provide the histone H4 HAT.
A pBlueScript vector carrying a single copy 601sequence flanked by several restriction sites (Figure 2, top left panel) is digested with KpnI and BamHI, the small fragment is ligated to the same vector digested with KpnI and BglII to produce the 2× 601 plasmid. Since the junction site of BamHI and BglII can no longer be cut by either enzyme, the two steps in Figure 2 can be multiplied several times to generate more repeats. We found that for the single sequence that is less than 300bp in length, a 16× version of construct can be routinely obtained.
A previous established procedure for a large scale plasmid preparation (10) is adopted with minor modifications. We normally start with 2 liters of 2×TY culture. After 18-22 hrs incubation at 37°C, about 20 grams of cells can be harvested, which yield about 10 mg of plasmid DNA.
The simple procedure described here only removes free co-factors. Most modifying enzymes remain in the final nucleosome fraction, which should be factored into the data interpretation as appropriate.
The following procedures are based on a previous protocol (12). Spot 5 μl of acetylated nucleosomes on a piece of negatively charged P81 phosphocellulose filter. Air dry filters for 5mins. Wash filters with 50 ml of HAT wash buffer at RT for 5 mins and then repeat two more times. Filters are briefly rinsed with 30 ml acetone and then air dried for 5 min. Place the filters into scintillation vials containing 4 ml of scintillation fluid (for filters) and measure the radioactivity using a scintillation counter.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Henikoff S, McKittrick E, Ahmad K. Epigenetics, histone H3 variants, and the inheritance of chromatin states. Cold Spring Harb Symp Quant Biol. 2004;69:235–243. doi: 10.1101/sqb.2004.69.235. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 9.Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. discussion 75-76. [DOI] [PubMed] [Google Scholar]
- 10.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 11.Owen-Hughes T, Utley RT, Steger DJ, West JM, John S, Cote J, Havas KM, Workman JL. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol Biol. 1999;119:319–331. doi: 10.1385/1-59259-681-9:319. [DOI] [PubMed] [Google Scholar]
- 12.Grant PA, Berger SL, Workman JL. Identification and analysis of native nucleosomal histone acetyltransferase complexes. Methods Mol Biol. 1999;119:311–317. doi: 10.1385/1-59259-681-9:311. [DOI] [PubMed] [Google Scholar]
- 13.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]