Abstract
Background. We explored the concept of heterologous prime/boost vaccination using 2 therapeutic vaccines currently in clinical development aimed at treating chronically infected hepatitis C virus (HCV) patients: prime with a DNA-based vaccine expressing HCV genotype 1a NS3/4A proteins (ChronVac-C) and boost with a modified vaccinia virus Ankara vaccine expressing genotype 1b NS3/4/5B proteins (MVATG16643).
Methods. Two ChronVac-C immunizations 4 weeks apart were delivered intramuscularly in combination with in vivo electroporation and subsequently 5 or 12 weeks later boosted by 3 weekly subcutaneous injections of MVATG16643. Two mouse strains were used, and we evaluated quality, magnitude, and functionality of the T cells induced.
Results. DNA prime/MVA boost regimen induced significantly higher levels of interferon γ (IFN-γ) or interleukin 2 (IL-2) ELISpot responses compared with each vaccine alone, independent of the time of analysis and the time interval between vaccinations. Both CD8+ and CD4+ T-cell responses as well as the spectrum of epitopes recognized was improved. A significant increase in polyfunctional IFN-γ/tumor necrosis factor α (TNF-α)/CD107a+ CD8+ T cells was detected following ChronVac-C/MVATG16643 vaccination (from 3% to 25%), and prime/boost was the only regimen that activated quadrifunctional T cells (IFN-γ/TNF-α/CD107a/IL-2). In vivo functional protective capacity of DNA prime/MVA boost was demonstrated in a Listeria-NS3-1a challenge model.
Conclusions. We provide a proof-of-concept that immunogenicity of 2 HCV therapeutic vaccines can be improved using their combination, which merits further clinical development.
Keywords: HCV, prime/boost, therapeutic vaccine, electroporation
After acute hepatitis C virus (HCV) infection, 20% of individuals clear the virus, which is dependent on sustained Th1 CD4+ T lymphocyte–mediated responses together with polyfunctional CD8+ T cells [1–4]. HCV is highly efficient in establishing persistent infections, and the specific T-cell responses are impaired during chronic infection [5–8] due to appearance of escape mutants [4, 9], inhibitory effects exerted by viral proteins [10], or expression of coinhibitory receptors resulting in T-cell exhaustion [11, 12].
One way to prime T cells and/or reactivate impaired T cells is to express HCV antigens under a more “immunogenic” setting than natural infection where massive antigen expression appears in the tolerogenic liver environment. Therapeutic vaccination has been tested with some success in HCV-infected patients showing evidence of T-cell activation [13, 14]. Vectored vaccines tailored to generate robust T-cell immunity have recently reached the clinic. MVATG16643, a modified virus Ankara (MVA)–based vaccine [15], has shown in a phase I trial good safety and the capacity to induce interferon γ (IFN-γ)–producing T cells with significant although transient viral load decrease in chronically infected patients [16]. Used in combination with pegylated (PEG)–interferon α (IFN-α)/ribavirin in a phase II clinical trial, MVATG16643 resulted in a significant early viral response [17]. The DNA vaccine ChronVac-C [18] has also shown the capacity to induce T cells, which had transient effects on viral load in a phase I/IIa trial [19], and it has been suggested that it improves cure rates when given before PEG–IFN-α/ribavirin treatment [19]. The combined use of DNA vaccines with viral vectors in a prime/boost regimen has been proven useful for enhancing response levels in clinical studies [20–22]. Similarly, in vivo electroporation (EP)–mediated delivery of DNA vaccines either as stand-alone or in a prime/boost setting with viral vectors has also served to enhance the development of polyfunctional CD8+ T-cell responses [23, 24]. Here we have performed a proof-of-concept study to define the extent of improvement that could result from a prime/boost approach based on ChronVac-C and MVATG16643—2 individual vaccines currently in the clinic. These 2 vaccine regimens have been optimized extensively individually previously, and we wanted to investigate whether the combination of 2 regimens could confer additional benefits over the individual regimens. This study thus combines for the first time in the HCV setting approaches previously shown individually to enhance immunogenicity (ie, DNA vaccine delivery with in vivo EP and prime/boost using viral vectors). We performed an exhaustive evaluation of this concept in wild-type C57BL/6J and human leucocyte antigen (HLA)–A2 transgenic mice. This strategy was found very potent for the improvement of polyfunctional CD4+ and CD8+ HCV-specific responses and resulted in a significant increase of epitope recognition.
MATERIALS AND METHODS
Animals
C57BL/6J (H-2b) mice aged 8–12 weeks were maintained at Karolinska Institutet following the Ethical Committee regulations for animal research. HLA-A2.1 transgenic mice [25] aged 7–11 weeks were used following the requirements of CEE directive 86/609 and the French law.
Plasmid and Modified Vaccinia Virus Ankara Strain Vectors
ChronVac-C plasmid coNS3/4A-pVAX1 containing the full-length codon optimized NS3/4A gene of HCV genotype 1a has been described previously [26]. MVA encoding for HCV genotype 1b NS3, NS4A/B, and NS5B antigens [27] and MVATGN33.1 (empty MVA) have been described previously [15, 16]. Immunogens are referred to as DNA = ChronVac-C; ctr-DNA = empty pVAX1; MVA = MVATG16643; ctr-MVA = MVATGN33.1.
Recombinant Proteins and Synthetic Peptides
NS3 protein 1a (aa 1207–1612), NS3 helicase 1b (aa 1192–1457), NS5B 1b (aa 2420–2989), and Core 1b (negative control) were produced in Escherichia coli. Chicken egg albumin (OVA) and Concanavalin A (ConA) were purchased from Sigma-Aldrich.
SIINFEKL (OVA CTL) and ISQAVHAAHAEINEAGR (OVA Th) sequences originates from Ovalbumin. GAVQNEVTL and GAVQNEITL (GAV gt1a and 1b) sequences originates from NS3 (H2-Db; aa 1629–1637). EIPFYGKAIPLEAIK (E13K gt1a), and EIPFYGKAIPIEAIK (E13K gt1b) sequences originates from NS3 that contain a mouse class II epitope (aa 1372–1386). The following HLA-A2 peptides were used: GLLGCIITSL (GLL gt1a and 1b) (aa 1038–1047), CINGVCWTV (CIN) (genotype 1a), and CVNGVCWTV (CVN) (genotype 1b; aa 1073–1081) from NS3; and DLMGYIPLV (genotype 1b) from Core (negative control). Peptide libraries were used: 1/ for genotype 1b, 163 overlapping peptides covering NS3/4A (FPNS34 1b, 0.7 µM of each peptide), and 125 overlapping peptides covering NS5B (FPNS5B, 1 µM of each peptide); 2/ for genotype 1a, two pools of 53 peptides covering NS3/4A: FPNS34A 1a-1 and FPNS34A 1a-2 (0.8 µM of each peptide) [28].
Immunization Protocols
Fifty micrograms of DNA were injected intramuscularly in the tibialis cranialis muscle followed by in vivo electroporation using the Medpulser DNA Delivery System [18]. MVA (107 pfu per mouse) was administered subcutaneously at the tail base.
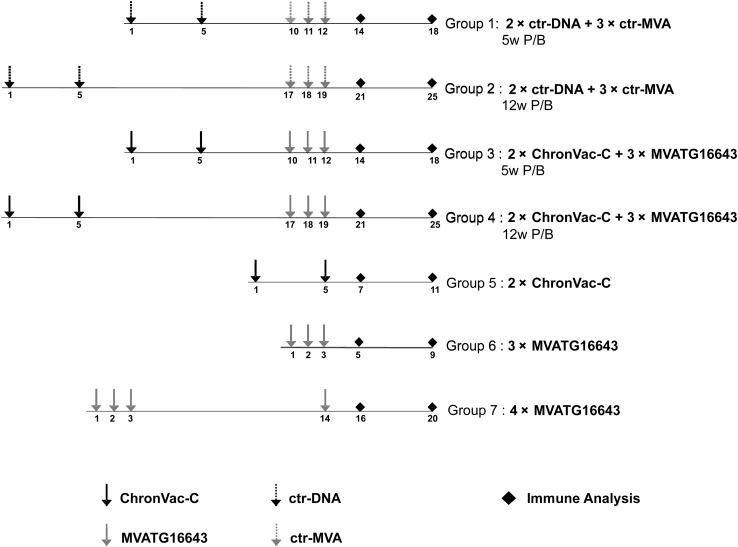
The study was conducted similarly in both wild-type (n = 5 per group) and HLA-A2 transgenic (n = 3–4 per group) mice (Figure 1).
Figure 1.
Study design in C57BL/6J and HLA-A2 mouse strains. Mice groups 1–4 received 2 injections of ChronVac-C or empty vector pVAX1 4 weeks apart (weeks 1 and 5) followed by 3 injections of MVATG16643 or MVATGN33.1 1 week apart (weeks 10, 11, and 12 or weeks 17, 18, and 19). Time interval between DNA prime/modified virus Ankara (MVA) boost was 5 weeks (groups 1 and 3) or 12 weeks (groups 2 and 4). Group 5 received 2 ChronVac-C injections 4 weeks apart (weeks 1 and 5). Group 6 received 3 MVATG16643 injections 1 week apart (weeks 1, 2, and 3). Group 7 received 3 injections of MVATG16643 1 week apart (weeks 1, 2, and 3) followed by a fourth injection of MVATG16643 (week 14). Hepatitis C virus (HCV)–specific T-cell responses were analyzed either 2 weeks or 6 weeks after last immunization.
Abbreviations: ctr-DNA, pVAX1; ctr-MVA, MVATGN33.1; P/B, prime/boost.
ELISpot Assay
Two assays were employed, each optimized for each animal strain. For C57BL/6J mice, pooled splenocytes were tested using anti-IFN-γ or anti–interleukin 2 (IL-2) Enzyme-Linked ImmunoSpot (ELISpot) assay according to manufacturers protocol [9, 18], in presence of peptides/proteins for 24 hours (IL-2) and 48 hours (IFN-γ). Spot counts were calculated as mean number of spot forming cells (SFCs)/106 cells using the AID ELISpot reader system version 2.6. A mean number of IFN-γ– or IL-2–producing cells >50 SFC/106 cells was considered as positive. ELISpot assay for HLA-A2 mice is described in the Supplementary Materials.
Determination of NS3-Specific CD8+ T Cell Frequencies
The frequency of NS3-specific CD8+ T cells was analyzed by direct ex vivo staining of splenocytes using the NS3 GAVQNEVTL 1a Pro5 pentamer exactly as described previously [29].
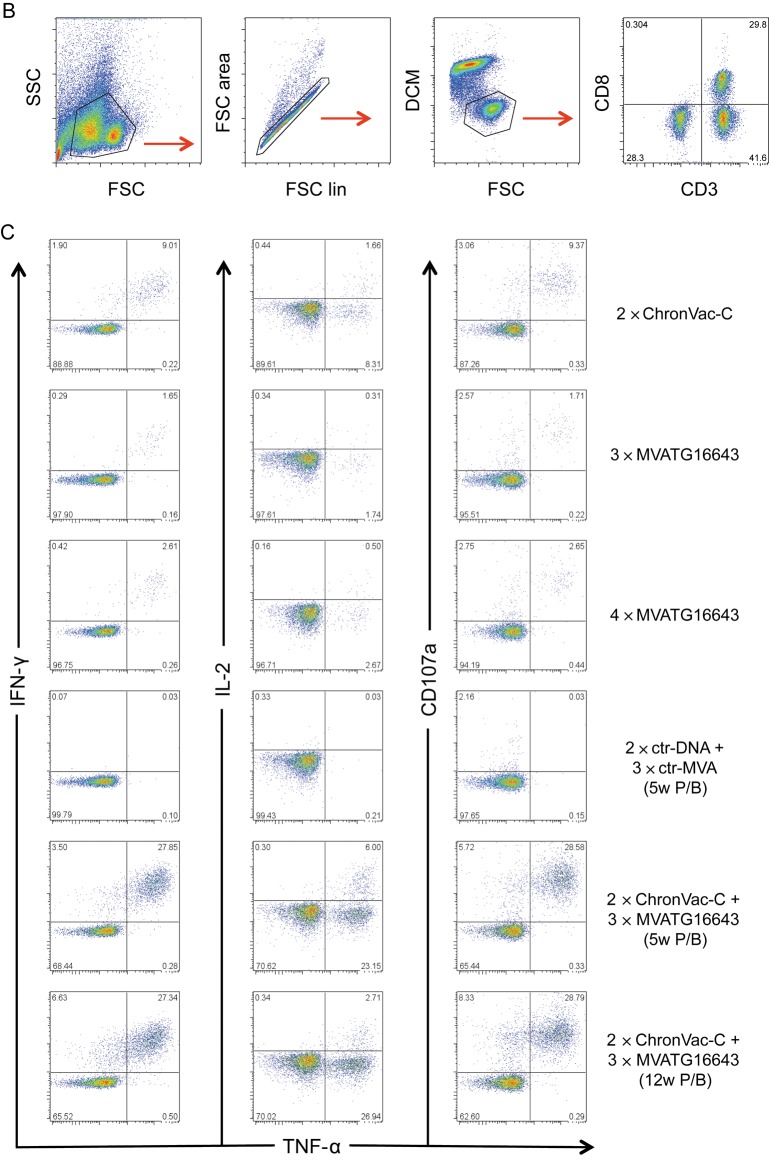
Characterization of Polyfunctional T Cells in C57BL/6J Mice by Flow Cytometry
Five × 105 splenocytes were restimulated for 12 hours in the presence of GolgiPlug with NS3/4A (1a (0.8 µM/each peptide) and 1b (0.7 µM/each peptide)) and NS5B 1b (1 µM/each peptide) peptide libraries or GAV epitopes 1a and 1b (5 µg/mL). Staining was done using Pacific Blue rat antimouse CD3 APC-Cy7 rat antimouse CD8a, PE rat antimouse CD107a, APC rat antimouse IL-2, FITC rat antimouse IFN-γ, and PE-Cy7 rat antimouse TNF-α. LIVE/DEAD Fixable Aqua Dead Cell Stain Kit was used as a dead cell marker. Permeabilization of cells was performed using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit. Cells were acquired on a CyAn ADP flow cytometer and analyzed using FlowJo 8.8.6 software. Results were given as individual cytokines produced alone and/or in combination as percent of total CD8+ T cells. The pie charts represent the proportion of cytokine-secreting CD8+ T cells that produce 1, 2, 3, or 4 cytokines/surface markers.
Surrogate Challenge Assay With Listeria-HCV NS3
Challenge assay based on recombinant Listeria monocytogenes expressing NS3 protein from genotype 1a HCV-1 isolate (TC-LNS3) was performed as previously described [15, 30].
Statistical Analysis
For C57BL/6J–based experiments, comparisons were performed using GraphPad InStat 3, Macintosh and Microsoft Excel 2008, Macintosh. Kinetic of measurements was compared using the area under the curve (Excel). Parametrical data were compared using the analysis of variance and nonparametrical data with Mann–Whitney U test. For HLA-A2.1–based experiments, analyses of immunogenicity were conducted using Statistica 9 software (Kruskal–Wallis test followed by a Mann–Whitney U test). For Listeria-NS3 challenge, analysis was conducted using a Tukey parametric test after checking variances homogeneity.
RESULTS
Quantification of IFN-γ– or IL-2–Producing T Cells in Wild-Type Mice
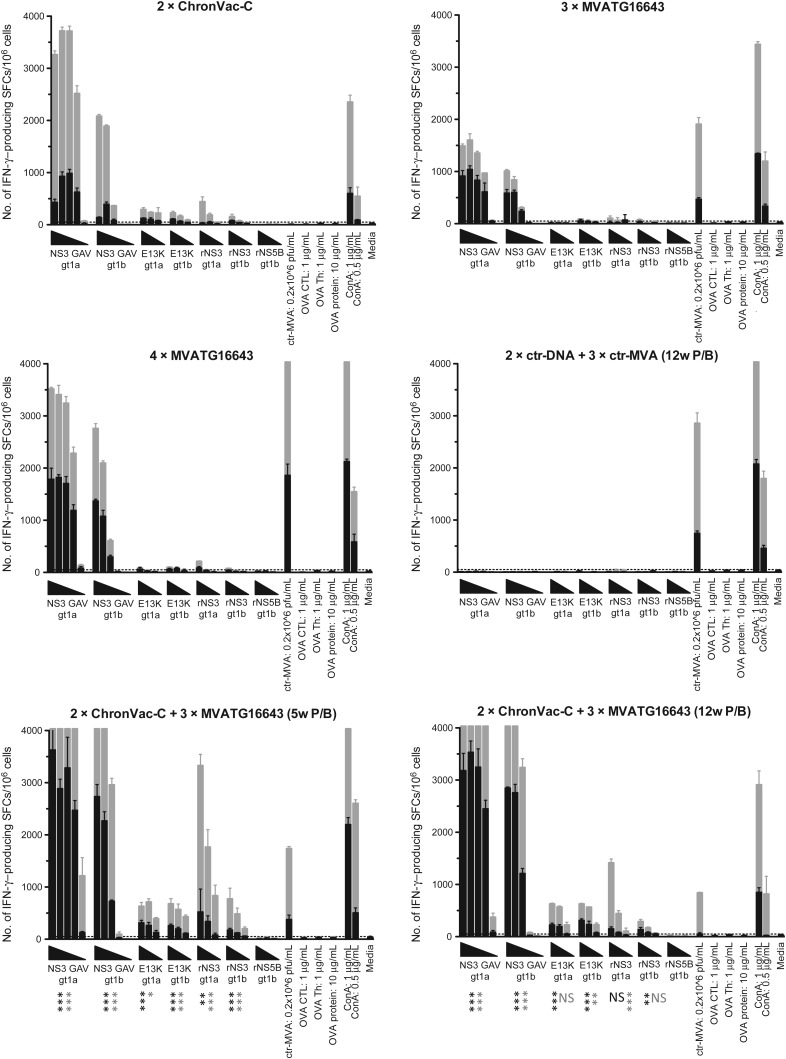
C57BL/6J mice immunized according to Figure 1 were compared for their ability to activate HCV-specific T-cell responses using IFN-γ ELISpot assays (Figure 2). Two weeks after last immunization, vaccination with the vaccines alone (2 injections of ChronVac-C and 3 of MVATG16643) induced equivalent number of IFN-γ–producing T cells (range for all antigen dilutions, 982–18 and 1038–43 SFCs/106 splenocytes specific of GAV 1a epitope) (Figure 2, black bars). For MVATG16643, these were enhanced following a fourth injection inducing 1818–78 SFCs/106 splenocytes. When heterologous prime/boost was applied, number of spots was greatly enhanced for all stimulations, reaching up to 3627 and 2845 SFC/106 splenocytes, for example, for the GAV 1a and 1b epitopes (P < .001; Figure 2, black bars). Interestingly, responses of the ChronVac-C/MVATG16643 injected groups were significantly improved between the 1st and 2nd time point of analysis (week 6 from last immunization; Figure 2, gray bars): responses >4000 (upper detection limit) and 2808 SFCs/106 splenocytes were detected against the GAV 1a and 1b epitopes and rNS3 1a protein, respectively. At that time point, the heterologous prime/boost groups were superior to the ChronVac-C and MVATG16643 groups. The activated T-cell response was cross-reactive between genotype 1a and 1b epitopes, although the response to the 1a epitope was consistently stronger. The IL-2 splenocyte-driven production was also investigated (Supplementary Figure 1). The weak production observed for this cytokine after stimulation with rNS3 1a was also improved by the DNA prime/MVA boost approach to a level of 327–67 SFCs/106 splenocytes (P < .05). Both IFN-γ and IL-2 responses to NS5B 1b were undetectable in all groups (Figure 2, black and gray bars; Supplementary Figure 1).
Figure 2.
Interferon γ (IFN-γ) ELISpot responses in C57BL/6J mice. IFN-γ ELISpot was performed on 5 mice per group 2 (black bars) and 6 (gray bars) weeks after last immunization. Results are given as mean spot-forming cells (SFCs)/106 (+ standard deviation) splenocytes values after stimulation with deescalating doses (GAV 1a and 1b: 0.25, 12.5, 0.625 ng/mL, 31.2 and 1.56 pg/mL; E13K 1a and 1b: 10, 1, 0.1 µg/mL; rNS3 1a and 1b and rNS5B 1b: 10, 2 and 0.4 µg/mL) of peptides/proteins as represented by triangles (each bar represents a specific concentration). Cutoff was set to 50 SFCs/106 splenocytes. Statistical difference between prime/boost (P/B) groups (eg, 2 × ChronVac-C + 3 × MVATG16643 (5-week P/B) or (12-week P/B) compared with each vaccine alone or control groups (eg, ChronVac-C, MVATG16643, and 2 × ctr-DNA + 3 × ctr-MVA) is indicated as follows: *P < .05, **P < .01, and ***P < .001 using area under the curve and analysis of variance. The P value shown is the least significant value between the P/B group and the vaccines alone or the control groups.
Abbreviations: ctr-DNA, pVAX1; ctr-MVA, MVATGN33.1; ELISpot, Enzyme-Linked ImmunoSpot; IFN-γ, interferon γ; NS, not significant; P/B, prime/boost; SFC, spot-forming cell.
The response pattern was reiterated in HLA-A2 transgenic mice at 2 and 6 weeks after the last immunization (data not shown and Supplementary Figure 2, respectively). Six weeks after the last injection, the heterologous prime/boost induced higher IFN-γ–producing T-cell frequencies than was observed with the ChronVac-C alone after stimulation with E13K 1b, NS3 1b (P < .05), GLL 1a, and NS3 1a (trend). It also resulted in higher IFN-γ–producing T-cell frequencies than MVATG16643 alone after stimulation with GLL, E13K, NS3 1b (P < .05), CVN, CIN, and NS3 1a (trend). In comparison with 4 MVATG16643 injections, a trend in improvement in favor of prime/boost was seen overall. The only significant improvement was seen for the CVN epitope (P < .05). NS5B-specific responses remained very weak with all immunization schedules.
Overall, the heterologous ChronVac-C prime/MVATG16643 boost results in higher frequencies of HCV-specific IFN-γ– and IL-2–producing T cells than that obtained with the separate vaccines. In a very convincing manner, this regimen also enlarged the spectrum of epitopes recognized in HLA-A2 mice when compared with the single vaccines. Both T-cell frequency and epitope scope increases most likely result from additive responses induced by each vaccine but also possibly from potential synergy of the responses. This is suggested by results seen after FPNS34A 1b, NS3 1b, NS3 1a, and E13K stimulations, for which the SFC levels were far higher than anticipated from the simple cumulative SFC levels obtained with single vaccines. Finally, the heterologous prime/boost approach was beneficial without significant differences between the 2 prime/boost time intervals (5 or 12 weeks).
Expansion of T Cells in Wild-Type Mice
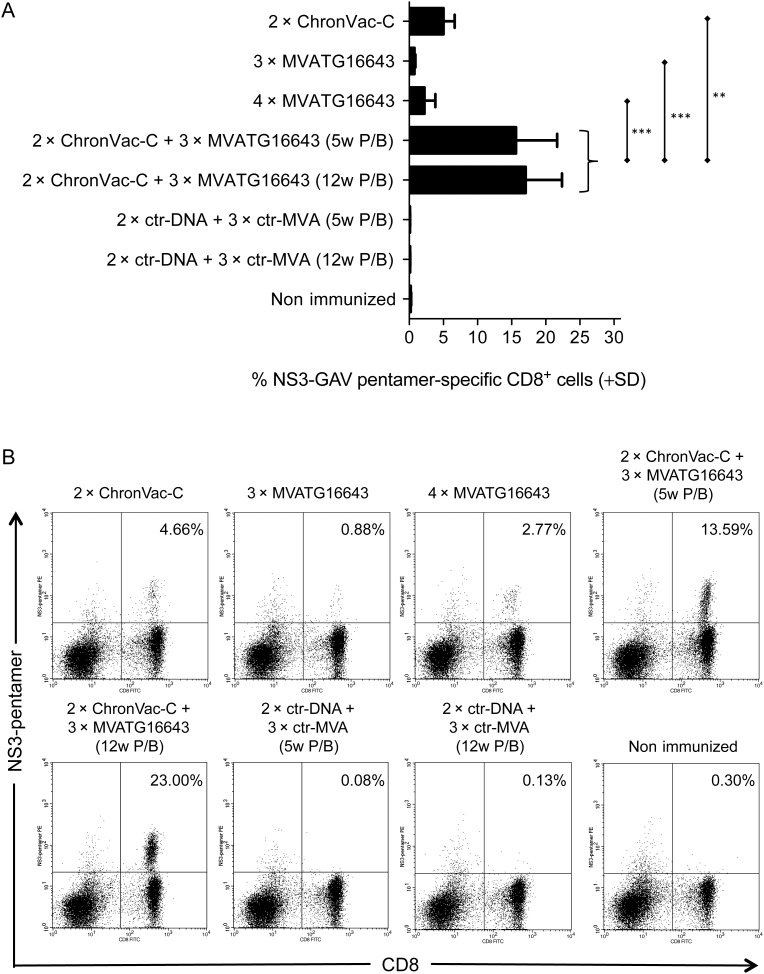
Expansion of NS3-specific CD8+ T cells was determined 6 weeks after the last immunization by direct ex vivo pentamer staining. CD8+ T cells specific of the GAV 1a epitope primed by ChronVac-C or MVATG16643 represented <7.7% of the total CD8+ T-cell population (Figure 3). The ChronVac-C prime/MVATG16643 boost protocol greatly improved T-cell expansion up to 23% of total CD8+ cells. Because this observation is limited to a single epitope, this is a quite striking T-cell expansion (P < .01 as compared with ChronVac-C alone; P < .001 as compared with schedules based on MVATG16643 alone).
Figure 3.
Expansion of NS3-specific CD8+ T cells in C57BL/6J mice. Mice (n = 5) were immunized as outlined in Figure 1 and sacrificed 6 weeks after last immunization. A, GAV 1a epitope-specific CD8+ T cells precursor frequency was determined by direct ex vivo pentamer staining with data given as the percentage of GAV pentamer-positive CD8+ T cells (+ standard deviation). Statistical difference between prime/boost (P/B) groups (eg, 2 × ChronVac-C + 3 × MVATG16643 (5-week + 12-week P/B) compared with ChronVac-C and MVATG16643 alone) is indicated as follows: **P < .01, and ***P < .001 using Mann–Whitney U test. B, Representative flow cytometric plots showing the pattern of GAV pentamer-positive CD8+ T cells.
Abbreviations: ctr-DNA, pVAX1; ctr-MVA, MVATGN33.1; P/B, prime/boost; SD, standard deviation.
Polyfunctionality of Activated T Cells in Wild-Type and HLA-A2 Mice
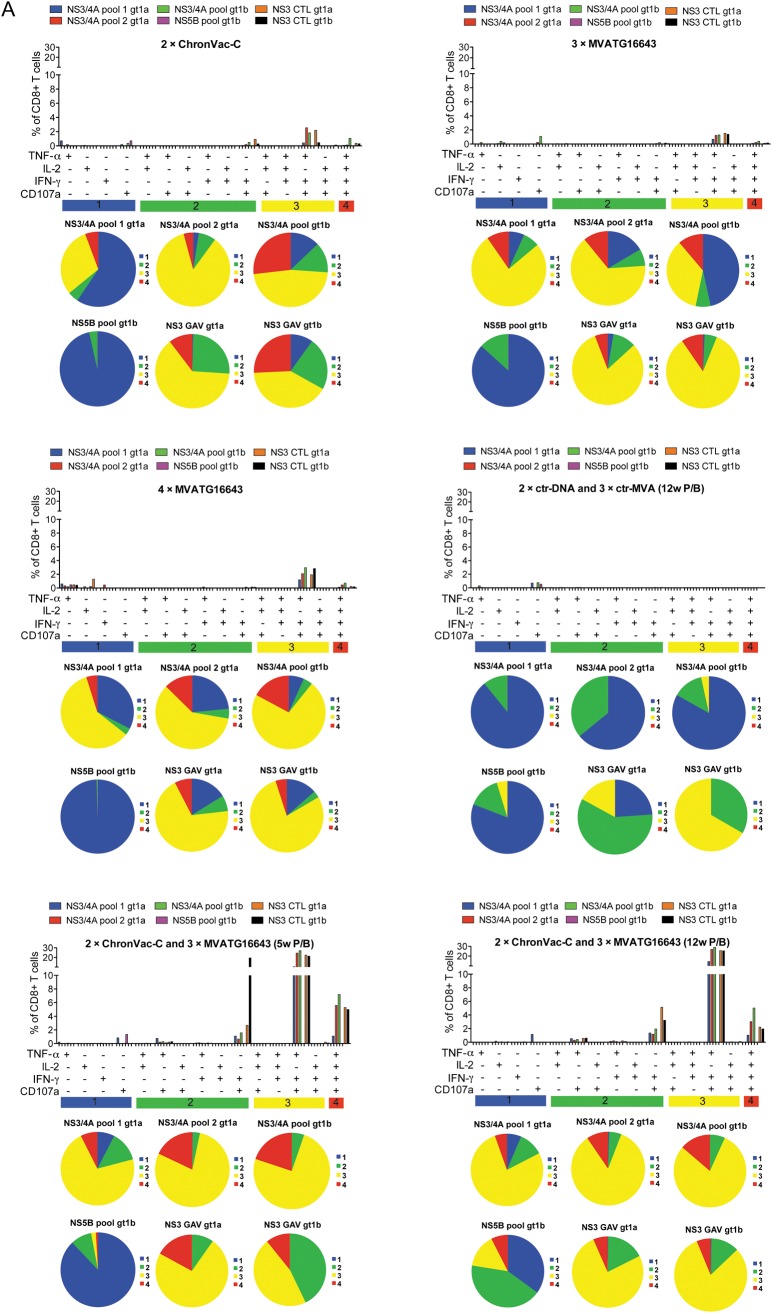
Polyfunctionality of the T-cell responses was determined in C57BL/6J mice 6 weeks after last immunization by a multicolor flowcytometric assay [31] (Figure 4). Each separate vaccine mainly primed trifunctional T cells expressing IFN-γ, tumor necrosis factor α (TNF-α), and CD107a specific for NS3/4A peptide pools 1a and 1b and GAV 1a epitope, although to a level <3%. The heterologous prime/boost activated the same type of trifunctional T cells, but a dramatic increase in the cell percentage was detected after stimulation with both GAV epitopes and both NS3/4A peptide pools, reaching up to 29% of total CD8+ T cells. In addition, a significant amount of quadrifunctional T cells also producing IL-2 was activated, with significant percentages reaching 7.2% of total CD8+ T cells. Only weak responses against NS5B could be detected in this assay. Thus, the ChronVac-C prime/MVATG16643 boost strategy was highly potent at improving the polyfunctionality of the NS3-specific T-cell response.
Figure 4.
Polyfunctionality of the NS3-specific T-cell response in C57BL/6J mice. Six weeks after last immunization, spleens from 5 mice per group were pooled. A, Bars illustrate the percentage of CD8+CD107a+ tumor necrosis factor a (TNF-α)–interleukin 2 (IL-2)–interferon γ (IFN-γ)–producing cells after 12 hours of antigen stimulation. Splenocytes were stimulated with overlapping peptide pools (NS3/4A gt 1a and 1b), and NS5B (gt 1b) or the specific peptide NS3-GAV (gt 1a and 1b). See Methods section for details. Distribution of single-, double-, triple-, and quadri-cytokine–producing CD8+ T cells is shown as various colors in pie chart diagrams. Each pie chart represents the response in pooled groups to the 6 different hepatitis C virus (HCV) stimulations. The responses are grouped by number of functions, matched to the colored bars numbered 1 (blue), 2 (green), 3 (yellow), and 4 (red). Notable is that >70% of the average responses to each of the 6 antigens express fewer than 4 functions. B, Gating schema for identification of polyfunctional CD8+ T cell responses. Shown are representative data on GAV epitope 1a-specific responses in a pool of mice immunized with 2 × ChronVac-C. C, Shown are representative data on GAV epitope 1a–specific responses in pools of mice immunized with: 2 × ChronVac-C, 3 × MVATG16643, 4 × MVATG16643, 2 × ctr-DNA + 3 × ctr-MVA, 2 × ChronVac-C + 3 × MVATG16643 (5-week prime/boost [P/B]) or (12-week P/B). Flow cytometric plots show the pattern of CD8+ T-cell TNF-α secretion along with IFN-γ, IL-2, and upregulation of CD107a. Abbreviations: ctr-DNA, pVAX1; ctr-MVA, MVATGN33.1; DCM, dead cell marker; FSC, forward scatter; IFN-γ, interferon γ; IL, interleukin; P/B, prime/boost; SSC, side scatter; TNF-α, tumor necrosis factor α.
Abbreviations: ctr-DNA, pVAX1; ctr-MVA, MVATGN33.1; DCM, dead cell marker; FSC, forward scatter; IFN-γ, interferon γ; IL, interleukin; P/B, prime/boost; SSC, side scatter; TNF-α, tumor necrosis factor α.
We also analyzed polyfunctionality of CD8+ and CD4+ T-cell responses in HLA-A2 mice by IFN-γ/TNF-α intracellular staining 6 weeks after last immunization (Supplementary Figure 3). The major population was IFN-γ–/TNF-α–producing T cells. The ChronVac-C prime/MVATG16643 boost tended to induce higher IFN-γ/TNF-α + CD8+ T-cell percentages than ChronVac-C alone (Supplementary Figure 3A) for GLL and FPNS34 1b (28% vs 12% and 3.8% vs 0.39, respectively) and higher IFN-γ/TNF-α + CD4+ T-cell percentages (Supplementary Figure 3B) specific for FPNS34A 1a-2 (0.26% vs 0.04%; P < .05), E13K 1b, and FPNS34A 1b (trend). In comparison with MVATG16643 alone, the ChronVac-C prime/MVATG16643 boost was more efficient at inducing IFN-γ/TNF-α + CD8+ T cells specific for GLL (28% vs 3.9%; P < .05) and FPNS34A 1b (trend). Higher IFN-γ/TNF-α + CD4+ T-cell percentages were also observed with ChronVac-C prime/MVATG16643 boost after stimulation with FPNS34A 1a-1, FPNS34A 1a-2 (0.22% vs 0.04% and 0.26% vs 0.03%; P < .05), E13K 1b, and FPNS34A 1b (trend). The heterologous prime/boost compared with the homologous MVATG16643 prime/boost induced higher percentages of IFN-γ/TNF-α + CD8+ T cells (GLL and FPNS34A 1b) and of IFN-γ/TNFα + CD4+ T cells (E13K 1b, FPNS34A 1b, FPNS34A 1a-1 and -2) although without significant differences (except for FPNS34A 1a-1).
In accordance with ELISpot data, intracellular cytokine staining (ICS) analysis led to the conclusion that ChronVac-C prime/MVATG16643 boost induced higher HCV-specific CD8+ and CD4+ T-cell responses than ChronVac-C or MVATG16643 alone or the MVATG16643 prime/boost.
Efficacy of the ChronVac-C Prime/MVATG16643 Boost in an HCV Surrogate Challenge Assay
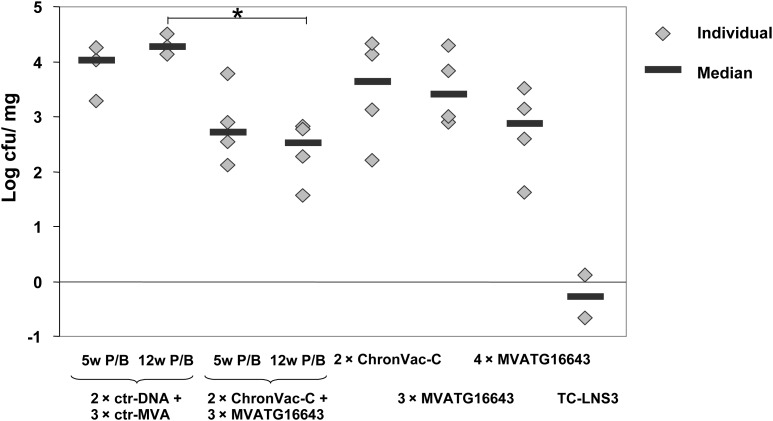
We used a recombinant Listeria-NS3 1a (TC-LNS3) as infectious challenge to evaluate functionality of vaccine-educated responses. The challenge was given 2 weeks after the last vaccination, and the number of viable bacteria in the spleen of mice was determined. Figure 5 shows that the prime/boost schedules compared (ChronVac-C/MVATG16643 with a 5-week or a 12-week interval and MVATG16643/MVATG16643) are the most efficient at reducing Listeria titers. Significant protection was observed in the ChronVac-C/MVATG16643 vaccinated group with the 12-week time interval (mean bacterial load of 2.52 log CFU/mg vs 4.27 log CFU/mg for the negative control group). These data underlined that the immune responses induced by the heterologous vaccine regimen display functionality in vivo, conferring significant control on the replication of a Listeria expressing an HCV-NS3 protein.
Figure 5.
Protection of HLA-A2 mice against Listeria-NS3 challenge. Mice immunized according to Figure 1 (3 mice for ctr-groups, 4 mice for group 3–7) and 2 mice immunized with a TC-LNS3 low dose (positive control, 0.1 LD50 of TC-LNS3) were included. All mice were challenged intravenously with a TC-LNS3 high dose 2 weeks after last immunization and killed 2 days later. Spleens were removed, homogenized, and serially diluted in phosphate-buffered saline 1X 0.1% Triton. Dilutions were plated out on brain heart infusion agar. Results are shown as the number of viable bacteria in the spleen (log CFU/mg organ) of each mouse (gray squares). Statistical analysis was conducted using a Tukey parametric test after checking variances homogeneity. * indicates significant differences between groups (P < .05).
Abbreviations: ctr-DNA, pVAX1; ctr-MVA, MVATGN33.1; P/B, prime/boost.
DISCUSSION
Treatment of HCV infection has recently benefited from the addition of protease inhibitors used in combination with PEG–IFN-α/ribavirin, leading to improvement in cure rate reaching up to 70%–80% for genotype 1–infected patients in clinical trials. However, problems such as increased cost, increased side effects, and difficulties to treat specific populations still remain. Therapeutic vaccines represent a novel class of compounds displaying a mechanism of action complementary to those of direct acting antivirals [32, 33]. The most advanced HCV therapeutic vaccines to date are in phase I/IIb clinical development, and, although it is expected that their primary positioning will be in combination with antivirals, it remains interesting to develop an improved vaccine regimen that may cure patients without addition of drugs. The threshold of immunity required to reach such result will likely have to be greatly enhanced. Prime/boost strategies based on DNA vaccines and viral vectors have emerged as a powerful approach in various infectious models to improve the number of antigen-specific T cells and to increase efficacy in challenge assays [34, 35]. In the HCV field, prime/boost approaches have been developed with some success in animal models using DNA, adenovirus, or polypeptides as primer and canarypox viruses, DNA, adenovirus or alphaviral particles as booster vaccines [36–38]. However, the only prime/boost study so far to reach the clinic in the HCV field is the human adenovirus 6 (Ad6) prime and chimpanzee adenovirus 3 (Ad3) boost approach that activated T cells specific for multiple HCV proteins with secretion of Th1-type cytokines [39].
In this proof-of-concept study, we compared established vaccine schedules shown earlier to result in potent CD8+ and CD4+ T-cell immunity both for MVATG16643 (3 injections 1 week apart) and for ChronVac-C (2 injections 4 weeks apart) [15, 18], with the combined use of these 2 vaccines in prime/boost settings. The DNA vaccine was first administered, followed by boosting with the MVA vaccine (with a 5- or a 12-week time interval). To provide an exhaustive analysis, we used both C57BL/6J mice and “humanized” HLA-A2 transgenic mice. Immune responses induced were analyzed by the use of CD8+-restricted epitopes, peptide libraries, and proteins derived from all 3 NS3, NS4A/B, and NS5B antigens from both genotype 1a and genotype 1b vaccine sequences.
A superiority of the ChronVac-C prime/MVATG16643 boost over either of the vaccine used alone was suggested at different levels. First, pentamer staining of GAV-specific responses in C57BL/6J demonstrated a considerable expansion of CD8+ T cells up to 23% following the prime/boost vs 7.7% at best for single vaccine–based strategies. This improvement did translate to enhancement at the functional level. ELISpot assay showed, for example, that the heterologous prime/boost induces in C57BL/6J mice a GAV-specific response 3.5-fold higher than that induced by ChronVac-C or MVATG16643 alone. In HLA-A2 mice, IFN-γ–producing CD4+ T cells specific for FPNS34A 1a-2 epitope were 6.5- to 8.7-fold higher than responses developed after ChronVac-C or MVATG16643 immunization. This is a key feature in the development of an immune-based therapy against chronic HCV infection where both CD4+ and CD8+ arms of the immune response have been found to be altered.
Numerous studies have shown that the quality even more than the level of T-cell responses is crucial for determining the outcome of various infections [40, 41]. For HCV, it was even reported that resolution obtained under early interferon therapy correlated with the development of polyfunctional IFN-γ/IL-2/CD107α+ virus-specific CD8+ T cells [42]. Whereas ChronVac-C or MVATG16643 alone and homologous MVATG16643 prime/boost were able to induce 2%–3.5% of trifunctional IFN-γ/TNF-α/CD107α+ CD8+ T cells, ChronVac-C prime/MVATG16643 boost resulted in the striking detection of this population up to 29% of CD8+ T-cells. In addition, the heterologous prime/boost induced up to 7.2% of quadrifunctional T cells (producing also IL-2). These results reenforced the superiority of the heterologous prime/boost regimen not only in terms of magnitude of induced T-cell responses but also in terms of their polyfunctionality.
The spectrum of epitopes recognized was also improved by the ChronVac-C/MVATG16643 vaccination in HLA-A2 mice. This was due first to ChronVac-C–specific induction of anti–CVN/CIN responses or MVATG16643-specific induction of anti-NS5B responses; thus expected additive effects were observed. More remarkable was that potential synergy was observed for the CD4+-driven responses beyond the simple addition of values expected with each vaccine alone (see IFN-γ ELISpot and IFN-γ/TNF-α ICS data with E13K, NS3, and FPNS34A 1b and 1a stimulations). Several features of the DNA and the MVA vectors may underlie their synergic activities. DNA vaccines introduce encoded proteins into MHC class I and II antigen-processing pathways [18], whereas its nonreplicative nature focuses immune responses on the encoded antigens [34]. In contrast, MVAs express higher levels of encoded antigens, and T-cell responses induced seem to be dominated by cross-priming in vivo, despite the ability of the virus to infect antigen-presenting cells [43, 44].
Finally, beyond extensive quantitative and qualitative characterization, it was also important to address the in vivo functionality of the generated immune responses. For this purpose we utilized the Listeria surrogate challenge model, which allow us to determine migration of vaccine-primed T cells to the Listeria-infected organs, thereby monitoring eradication of antigen-expressing cells [15, 30]. We showed that the heterologous prime/boost regimen conferred significant protection against a challenge with a Listeria-expressing HCV-NS3 protein, whereas none of the single vaccine–based regimens succeeded in a significant control of the bacteria replication. Notable is the variation in bacterial load within groups of mice, although it is in a similar range as seen in previous studies [15, 31]. Collectively, all immunological data in this study favor a correlation between the strength of the induced T-cell response and protection against Listeria-NS3 challenge.
In conclusion, this study demonstrated the clear interest of the ChronVac-C prime/MVATG16643 boost strategy in agreement with reports arguing in favor of heterologous vaccine approaches. But results also go beyond: we used for the first time in the HCV field 2 vaccines already in clinical development. These vaccines express heterologous sequences derived from the 2 viral subtypes 1a and 1b belonging to the worldwide most prominent and hardest to treat genotype 1. We showed that the number of IFN-γ– and IL-2–producing CD4+ and CD8+ T cells, the epitope polyspecificity, and the polyfunctionality of induced CD8+ T cells were all greatly improved. In addition, vaccine-educated responses displayed protective function in vivo. These preclinical data support further evaluation of this vaccine combination in the clinic.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http:/jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We kindly thank ChronTech Pharma AB, Huddinge, Sweden, for providing synthetic peptides.
Financial support. This work was supported by grants from the Swedish Research Council (M. S., L. F. [K2012-99X-22017-01-3]); Swedish Cancer Society (M. S.): Stockholm County Council (M. S.); the Swedish Society of Medical Research (L. F.); the Swedish Society of Medicine (L. F.); Goljes Memorial Fund (L. F., G. A.): the Åke Wiberg Foundation (L. F.); the Royal Swedish Academy of Sciences (L. F., G. A.); Karolinska Institutet (L. F., M. S.); Lars Hiertas Memorial Fund (G. A.); Magnus Bergvalls Foundation (G. A.); and Karolinska Institutet / Södertörns University (postdoctoral grant, G. A.).
Potential conflicts of interest. M. S. is founder, paid consultant, and board member of ChronTech Pharma AB. L. F. and G. A. are paid consultants of ChronTech Pharma AB. A. F., E. J., E. G., J. Y. B., and G. I. are employed by Transgene SA. K. E. B. and N. Y. S. are employed by Inovio Pharmaceuticals. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Thimme R, Oldach D, Chang KM, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoukry NH, Grakoui A, Houghton M, et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med. 2003;197:1645–55. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauer GM, Barnes E, Lucas M, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127:924–36. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Cox AL, Mosbruger T, Lauer GM, et al. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–12. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–52. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 6.Grakoui A, Shoukry NH, Woollard DJ, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–62. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 7.Wedemeyer H, He XS, Nascimbeni M, et al. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol. 2002;169:3447–58. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 8.Gruener NH, Lechner F, Jung MC, et al. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J Virol. 2001;75:5550–8. doi: 10.1128/JVI.75.12.5550-5558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Söderholm J, Sällberg M. A complete mutational fitness map of the hepatitis C virus nonstructural 3 protease: relation to recognition by cytotoxic T lymphocytes. J Infect Dis. 2006;194:1724–8. doi: 10.1086/509513. [DOI] [PubMed] [Google Scholar]
- 10.Meylan E, Curran J, Hofmann K, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 11.Radziewicz H, Hanson HL, Ahmed R, et al. Unraveling the role of PD-1/PD-L interactions in persistent hepatotropic infections: potential for therapeutic application? Gastroenterology. 2008;134:2168–71. doi: 10.1053/j.gastro.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Urbani S, Amadei B, Tola D, et al. Restoration of HCV-specific T cell functions by PD-1/PD-L1 blockade in HCV infection: effect of viremia levels and antiviral treatment. J Hepatol. 2008;48:548–58. doi: 10.1016/j.jhep.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Klade CS, Wedemeyer H, Berg T, et al. Therapeutic vaccination of chronic hepatitis C nonresponder patients with the peptide vaccine IC41. Gastroenterology. 2008;134:1385–95. doi: 10.1053/j.gastro.2008.02.058. [DOI] [PubMed] [Google Scholar]
- 14.Eisenstein M. Vaccines: a moving target. Nature. 2011;474:S16–7. doi: 10.1038/474S16a. [DOI] [PubMed] [Google Scholar]
- 15.Fournillier A, Gerossier E, Evlashev A, et al. An accelerated vaccine schedule with a poly-antigenic hepatitis C virus MVA-based candidate vaccine induces potent, long lasting and in vivo cross-reactive T cell responses. Vaccine. 2007;25:7339–53. doi: 10.1016/j.vaccine.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 16.Habersetzer F, Honnet G, Bain C, et al. A poxvirus vaccine is safe, induces t-cell responses, and decreases viral load in patients with chronic hepatitis c. Gastroenterology. 2011;141:890–9. doi: 10.1053/j.gastro.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Wedemeyer H. Hepatology. 2011;54(Suppl. 4):989A–90A. doi: 10.1016/j.jhep.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Ahlén G, Söderholm J, Tjelle T, et al. In vivo electroporation enhances the immunogenicity of hepatitis C virus nonstructural 3/4A DNA by increased local DNA uptake, protein expression, inflammation, and infiltration of CD3+ T cells. J Immunol. 2007;179:4741–53. doi: 10.4049/jimmunol.179.7.4741. [DOI] [PubMed] [Google Scholar]
- 19.Weiland O, Ahlén G, Diepolder H, Jung MC, Levander S, Fons M, Mathiesen I, Sardesai NY, Vahlne A, Frelin L. Sällberg M.Therapeutic DNA Vaccination Using In Vivo Electroporation Followed by Standard of Care Therapy in Patients With Genotype 1 Chronic Hepatitis C. Mol Ther. 2013 doi: 10.1038/mt.2013.119. doi:10.1038/mt.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winstone N, Guimaraes-Walker A, Roberts J, et al. Increased detection of proliferating, polyfunctional, HIV-1-specific T cells in DNA-modified vaccinia virus Ankara-vaccinated human volunteers by cultured IFN-gamma ELISPOT assay. Eur J Immunol. 2009;39:975–85. doi: 10.1002/eji.200839167. [DOI] [PubMed] [Google Scholar]
- 21.Dunachie SJ, Walther M, Epstein JE, et al. A DNA prime-modified vaccinia virus ankara boost vaccine encoding thrombospondin-related adhesion protein but not circumsporozoite protein partially protects healthy malaria-naive adults against Plasmodium falciparum sporozoite challenge. Infect Immun. 2006;74:5933–42. doi: 10.1128/IAI.00590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConkey SJ, Reece WH, Moorthy VS, et al. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat Med. 2003;9:729–35. doi: 10.1038/nm881. [DOI] [PubMed] [Google Scholar]
- 23.Lang Kuhs KA, Ginsberg AA, Yan J, et al. Hepatitis C virus NS3/NS4A DNA vaccine induces multiepitope T cell responses in rhesus macaques mimicking human immune responses. Mol Ther. 2012;20:669–78. doi: 10.1038/mt.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutnick NA, Myles DJ, Hirao L, et al. An optimized SIV DNA vaccine can serve as a boost for Ad5 and provide partial protection from a high-dose SIVmac251 challenge. Vaccine. 2012;30:3202–8. doi: 10.1016/j.vaccine.2012.02.069. [DOI] [PubMed] [Google Scholar]
- 25.Pascolo S, Bervas N, Ure JM, et al. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2 m) HLA-A2.1 monochain transgenic H-2Db beta2 m double knockout mice. J Exp Med. 1997;185:2043–51. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frelin L, Ahlen G, Alheim M, et al. Codon optimization and mRNA amplification effectively enhances the immunogenicity of the hepatitis C virus nonstructural 3/4A gene. Gene Ther. 2004;11:522–33. doi: 10.1038/sj.gt.3302184. [DOI] [PubMed] [Google Scholar]
- 27.Kato N, Hijikata M, Ootsuyama Y, et al. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990;87:9524–8. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sällberg M, Ruden U, Magnius LO, et al. Rapid “tea-bag” peptide synthesis using 9-fluorenylmethoxycarbonyl (Fmoc) protected amino acids applied for antigenic mapping of viral proteins. Immunol Lett. 1991;30:59–68. doi: 10.1016/0165-2478(91)90090-w. [DOI] [PubMed] [Google Scholar]
- 29.Chen A, Ahlén G, Brass A, et al. Heterologous T cells can help restore function in dysfunctional hepatitis C virus nonstructural 3/4A-specific T cells during therapeutic vaccination. J Immunol. 2011;186:5107–18. doi: 10.4049/jimmunol.1001790. [DOI] [PubMed] [Google Scholar]
- 30.Simon BE, Cornell KA, Clark TR, et al. DNA vaccination protects mice against challenge with Listeria monocytogenes expressing the hepatitis C virus NS3 protein. Infect Immun. 2003;71:6372–80. doi: 10.1128/IAI.71.11.6372-6380.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyström J, Chen A, Frelin L, et al. Improving on the ability of endogenous hepatitis B core antigen to prime cytotoxic T lymphocytes. J Infect Dis. 2010;201:1867–79. doi: 10.1086/652808. [DOI] [PubMed] [Google Scholar]
- 32.Torresi J, Johnson D, Wedemeyer H. Progress in the development of preventive and therapeutic vaccines for hepatitis C virus. J Hepatol. 2011;54:1273–85. doi: 10.1016/j.jhep.2010.09.040. [DOI] [PubMed] [Google Scholar]
- 33.Halliday J, Klenerman P, Barnes E. Vaccination for hepatitis C virus: closing in on an evasive target. Expert Rev Vaccines. 2011;10:659–72. doi: 10.1586/erv.11.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estcourt MJ, Ramsay AJ, Brooks A, et al. Prime-boost immunization generates a high frequency, high-avidity CD8+ cytotoxic T lymphocyte population. Int Immunol. 2002;14:31–7. doi: 10.1093/intimm/14.1.31. [DOI] [PubMed] [Google Scholar]
- 35.Woodland DL. Jump-starting the immune system: prime-boosting comes of age. Trends Immunol. 2004;25:98–104. doi: 10.1016/j.it.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Pancholi P, Perkus M, Tricoche N, et al. DNA immunization with hepatitis C virus (HCV) polycistronic genes or immunization by HCV DNA priming-recombinant canarypox virus boosting induces immune responses and protection from recombinant HCV-vaccinia virus infection in HLA-A2.1-transgenic mice. J Virol. 2003;77:382–90. doi: 10.1128/JVI.77.1.382-390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folgori A, Capone S, Ruggeri L, et al. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nat Med. 2006;12:190–7. doi: 10.1038/nm1353. [DOI] [PubMed] [Google Scholar]
- 38.Lin Y, Kwon T, Polo J, et al. Induction of broad CD4+ and CD8+ T-cell responses and cross-neutralizing antibodies against hepatitis C virus by vaccination with Th1-adjuvanted polypeptides followed by defective alphaviral particles expressing envelope glycoproteins gpE1 and gpE2 and nonstructural proteins 3, 4, and 5. J Virol. 2008;82:7492–503. doi: 10.1128/JVI.02743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnes E, Folgori A, Capone S, et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med. 2012;4:115ra1. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–58. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 41.Cellerai C, Harari A, Stauss H, et al. Early and prolonged antiretroviral therapy is associated with an HIV-1-specific T-cell profile comparable to that of long-term non-progressors. PLoS One. 2011;6:e18164. doi: 10.1371/journal.pone.0018164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badr G, Bedard N, Abdel-Hakeem MS, et al. Early interferon therapy for hepatitis C virus infection rescues polyfunctional, long-lived CD8+ memory T cells. J Virol. 2008;82:10017–31. doi: 10.1128/JVI.01083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kastenmuller W, Drexler I, Ludwig H, et al. Infection of human dendritic cells with recombinant vaccinia virus MVA reveals general persistence of viral early transcription but distinct maturation-dependent cytopathogenicity. Virology. 2006;350:276–88. doi: 10.1016/j.virol.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 44.Gasteiger G, Kastenmuller W, Ljapoci R, et al. Cross-priming of cytotoxic T cells dictates antigen requisites for modified vaccinia virus Ankara vector vaccines. J Virol. 2007;81:11925–36. doi: 10.1128/JVI.00903-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.