Abstract
Transcriptional repression is a common approach to control gene expression in synthetic biology applications. Here, an engineered DNA binding protein based upon a transcription activator-like effector (TALE) scaffold was shown to outperform LacI in blocking transcription from a promoter and to repress expression of a downstream gene in an operon.
Synthetic biology is a field that leverages recombinant DNA technologies to build novel living systems useful for studying chemical production, biosensing, complex genetic circuits and many other applications.1 A typical synthetic biology design combines regulatory and function-encoding DNA molecules to produce a desired phenotype in a model organism. Optimal functionality of these synthetic systems often requires significant engineering to achieve appropriate levels of gene expression for the artificial construct and the native gene products with which it interacts. Currently, molecular tools are available for altering prokaryotic gene expression at the transcriptional (e.g. promoter replacement, artificial zinc fingers), posttranscriptional (e.g. anti-sense RNAs, RNA turnover), and translational (e.g. initiation signals, codon optimization) levels. However, many of these tools work in cis and are plagued by a limited number of compatible options, ease of implementation, and unintended effects on native physiology. In order to build increasingly complex genetic circuits and to optimize expression of native genes, new regulators, particularly those that function in trans, are needed.
A new option has recently gained significant attention in controlling eukaryotic gene expression. Virulence-effecting transcription activator-like effectors (TALEs) are proteins made by Xanthomonas bacterial species and injected into plants to increase susceptibility to infection.2,3 The utility of these virulence effectors lies in the one-to-one binding relationship between residues within the DNA-binding domain and a targeted DNA base pair.
TALEs are composed of three sections: (i) a central tandem repeat domain that controls binding to targeted sites, (ii) an N-terminal translocation signal, and (iii) a C-terminal region containing a transcriptional activation domain as well as a nuclear localization signal.4 Polymorphisms at specific locations within each repeat unit, referred to as repeat variable diresidues (RVDs), are responsible for DNA nucleotide recognition.4,5 The amino acid identity of the RVDs alone can be used to predict the corresponding DNA target sequence of a TALE, and custom TALEs can be programmed to bind a user-defined sequence by simply rearranging the order of the RVDs.6–8
Researchers are now harnessing the predictable one-to-one relationship between a repeat unit and its target DNA base pair to design TALEs that reliably bind to user-defined DNA sequences in yeast and mammalian cells,9,10 as well as nonnative sequences in plants,11 to modulate gene expression and introduce mutations. Surprisingly, the utility of TALEs as transcriptional regulators in bacteria has yet to be explored. In this manuscript we compare the performance of a synthetic TALE in repressing transcriptional initiation (Fig. 1a) and elongation (Fig. 1b) in E. coli relative to a native regulatory protein that targets the same DNA sequence. Our results provide evidence that TALEs can be used as novel repressors of gene expression in bacteria.
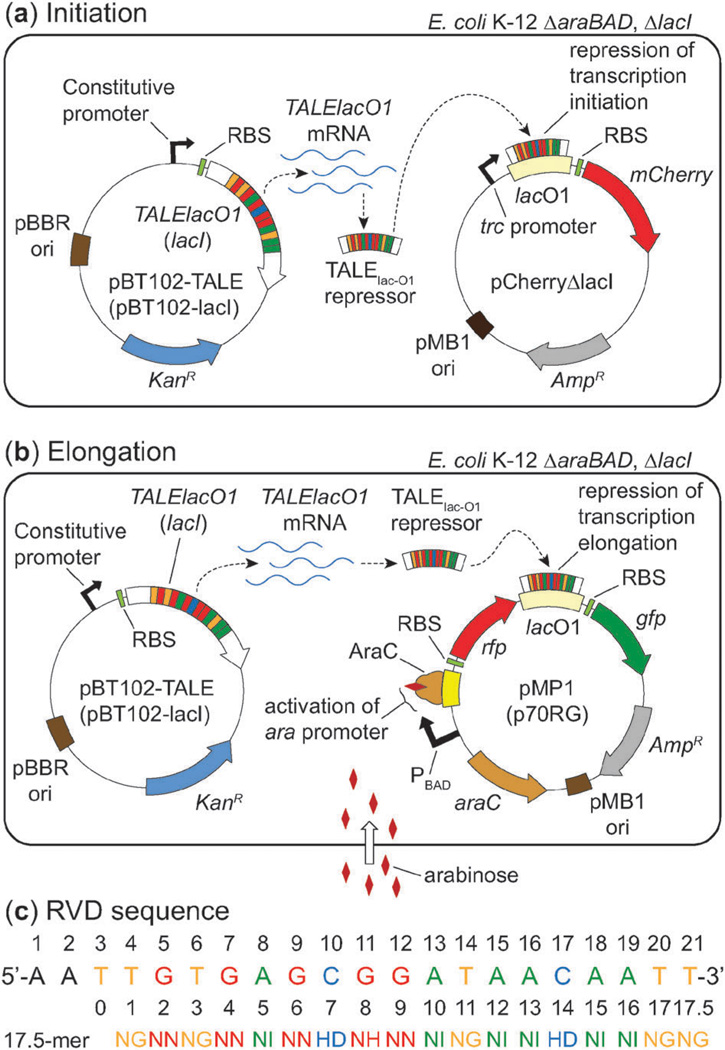
Fig. 1.
Cartoons of the systems used to investigate designer TALEs as repressors of E. coli transcription. (a) A model for examining TALE-mediated repression of transcription initiation. (b) A model for studying TALE-mediated repression of transcription elongation. Abbreviations: AmpR = ampicillin resistance gene (β-lactamase), KanR = kanamcyin resistance gene (neomycin phosphotransferase II), ori = origin of replication, RBS = ribosome binding site. (c) An alignment of the 17.5-mer RVD sequence of the TALE construct (TALElacO1) to the 21-bp lac operator. Amino acids for each RVD are in single letter format.
A common method of repressing gene expression in prokaryotes involves the binding of a protein to a DNA sequence (a.k.a. operator) near or overlapping its promoter. This interaction occludes recruitment of RNA polymerase and prevents transcription initiation, thereby repressing gene expression. The reversible interaction between the LacI repressor protein and the lac operator is the paradigm for transcriptional repression.12,13 We developed a simple platform to compare the regulation using our designer TALE (referred to subsequently as TALElacO1) relative to the natural LacI repressor. The RVD sequence was changed such that it would bind 18 base pairs of the lac O1 operator (repeat position 0 binds the requisite 5′ thymine base pair) (Fig. 1c). Our platform uses two plasmids for studying TALE-mediated gene repression: an expression vector for production of the TALElacO1 or LacI repressor, and a reporter plasmid incorporating the lac operator to repress transcription initiation or elongation of a fluorescent reporter protein (Fig. 1a and b).
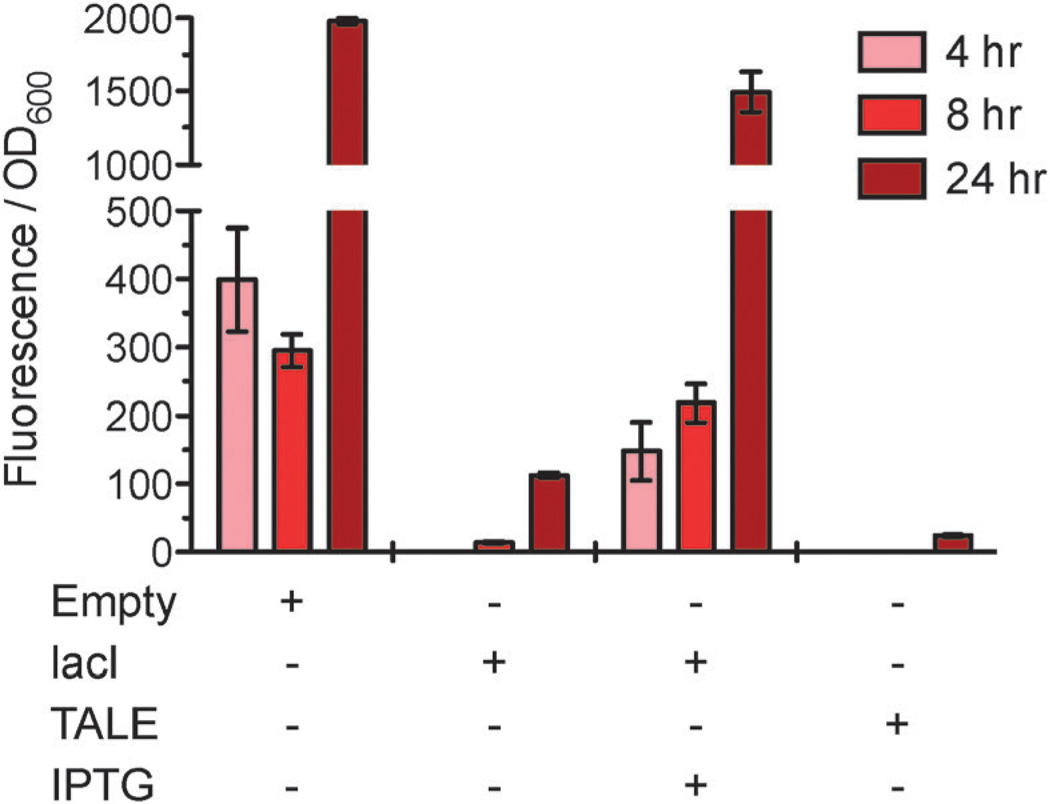
To test repression of transcription initiation, a reporter plasmid (pCherΔlacI) was constructed with mCherry cloned downstream of the LacI-regulated trc promoter. Cultures (6 mL) of E. coli harboring the appropriate combinations of plasmids were grown in culture tubes at 37 °C and sampled over time to measure optical density and red fluorescence. In the absence of LacI (pBT102 or “Empty”), a strong red fluorescent signal was observed (Fig. 2) consistent with derepression of the trc promoter and high-level expression of mCherry. As expected, the presence of LacI resulted in a significant loss of red fluorescence consistent with repression of the trc promoter by LacI. Similarly, expression of TALElacO1 nearly abolished the red fluorescent signal, suggesting that the designer TALE was acting as intended; presumably binding of the TALE to the lac operator blocks promoter recognition and prevents transcription initiation. Similar results were obtained when cultures were spotted onto solid media containing isopropyl-β-d-1-thiogalactopyranoside (IPTG), a non-metabolized mimic of allolactose. Consistent with culture tube results, cells expressing lacI in the presence of IPTG appeared red, whereas colonies expressing TALElacO1 did not show red fluorescence in the presence or absence of IPTG (Fig. S1, ESI‡).
Fig. 2.
TALElacO1 represses expression of mCherry via inhibition of transcription initiation. Expression of TALElacO1 or lacI results in a reduction of red fluorescence intensity relative to the empty vector over time.
Collectively, these data suggest that the designer TALE was able to repress transcription initiation in a manner analogous to the LacI protein. By 24 hours, repression of mCherry by TALElacO1 reduced red fluorescence by approximately two orders of magnitude. This reduction was significantly greater than the corresponding reduction caused by LacI (24 ± 2 vs. 113 ± 3 a.u.), suggesting that TALE mediated repression of transcription initiation was more potent with this experimental setup.
In E. coli, there are over 600 predicted operons in which multiple genes are co-transcribed from the same promoter.14 Tools to differentially regulate expression of genes within an operon can be used to control the ratio of proteins while maintaining on-off regulation from an inducible promoter.15 It is not known if E. coli regulates gene expression via binding of proteins to sequences within structural genes or in intergenic spaces of operons. This strategy is however used by Saccharomyces cerevisiae wherein the DNA-binding protein Rap1p blocks elongation by causing RNAPII to prematurely terminate transcription.16 Analogously, we hypothesized that artificial transcription factors, such as TALEs, could be designed to bind within ORFs or intergenic regions to fine tune gene expression in synthetic biology applications. Previous research suggests that this strategy could work in prokaryotes, as RNAP is known to stall at roadblocks formed by protein-bound DNA.17
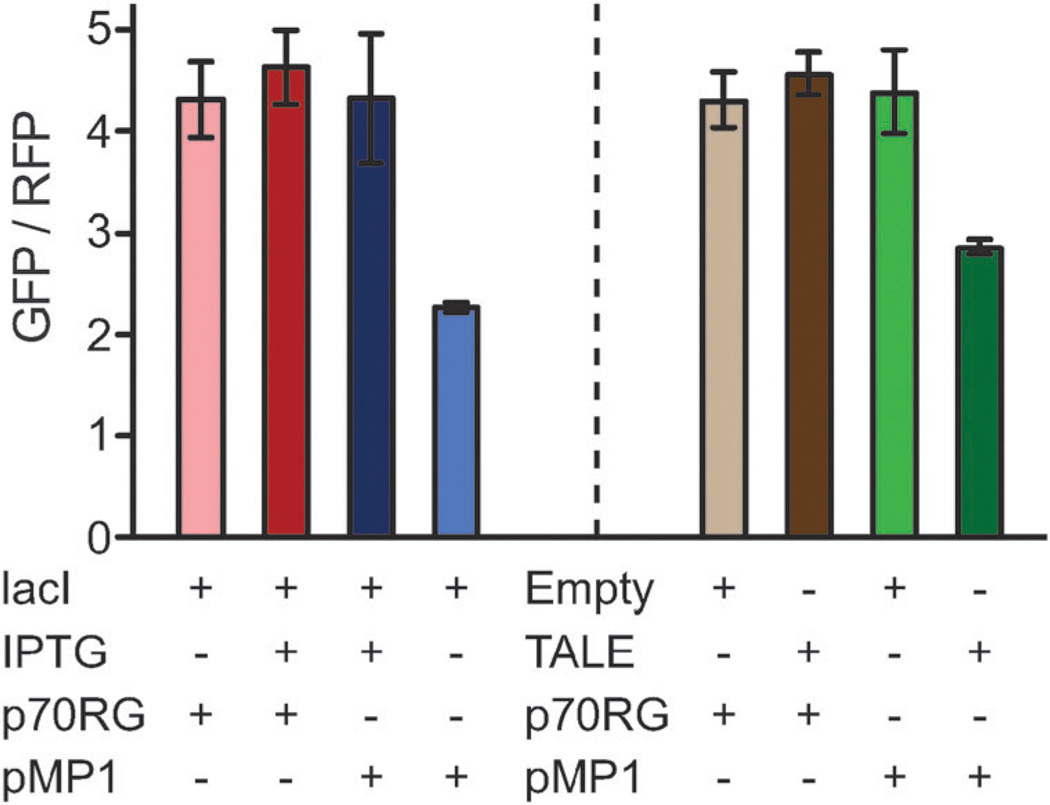
To test our hypothesis, a reporter plasmid, pMP1, was constructed to express a synthetic operon of genes encoding red (dsRed, 5′) and green (gfpuv, 3′) fluorescent proteins with an intergenic lac operator (Fig. 1b). A plasmid without the intergenic lac O1 (p70RG)15 served as a control. We hypothesized that expression of LacI or TALElacO1 would reduce the ratio of GFP to RFP fluorescence relative to the controls (p70RG or de-repression of LacI by IPTG).
Plasmid combinations were co-transformed into MG1655 ΔlacI ΔaraBAD and 6 mL cultures were grown in tubes at 37 °C. The optical density and fluorescence were measured at 6, 8, and 10 hours after inoculation. The ratio of GFP to RFP fluorescence observed at the 8-hour time point was representative of all measurements (Fig. 3 and Fig. S2, ESI‡). Expression of lacI or TALElacO1 in strains containing the pMP1 reporter plasmid resulted in a reduction of the GFP to RFP ratio relative to controls (~1.5- to 2-fold). Conversely, the ratio was unchanged in strains harboring p70RG. The GFP to RFP fluorescence ratio was restored in the presence of IPTG for strains expressing lacI and pMP1. As expected, the presence of both LacI and TALElacO1 reduced green fluorescence. Unexpectedly, the presence of LacI (but not TALElacO1) also increased the level of red fluorescence by an unknown mechanism (Fig. S2, ESI‡). Together, these data confirm that relative protein expression can be altered (reversibly with LacI) by DNA binding proteins.
Fig. 3.
TALElacO1 and LacI inhibit transcription elongation from an operon, reducing expression of gfpuv (GFP) relative to dsRed (RFP). Data shown corresponds to 8 hours following inoculation.
While the small decrease in relative expression was statistically significant, the data confirms that repression of transcription elongation by protein roadblocks is not absolute and that transcriptional read-through does occur (Fig. S2, ESI‡). This sharply contrasts with the repression of initiation strategy, where expression of TALElacO1 strongly reduces red fluorescence by ~300-fold relative to the de-repressed promoter. This difference in potency is not surprising as RNA polymerase has been shown to by-pass protein roadblocks in vitro.17 Further exploration of the variables in this system (e.g. # of operator sites, TALE affinity, inducible binding, RNA turnover) could lead to the development of design rules for controlling ratios of gene products.
TALEs are likely to become increasingly popular tools for synthetic biologists due to their specificity, predictable code, and modular design. While this study employed a simplistic plasmid-based system for examining TALE-mediated gene repression, the true potential of TALEs lies in their ability to bind any DNA sequence, enabling custom TALEs to target any chromosomal sequence and evoke changes in gene expression. This proof-of-concept study sets the stage for a more in depth examination of TALEs as artificial transcription factors in prokaryotes. Varying degrees of transcription repression could be achieved by altering the affinity of the TALE for its target sequence by: (1) reducing the number of repeat domains;6 (2) scrambling the RVD sequence to reduce specificity; and (3) introducing multiple TALEs that bind adjacent DNA sequences. Perhaps the most intriguing and powerful use of TALEs lies in the opportunity to engineer these proteins to not only repress, but also activate gene expression in bacteria. In combination with TALE repressors, a user may be able to turn a series of genes “on” and “off” in a predictable fashion. In conclusion, the demonstration of site-specific and modular transcriptional repression by TALEs opens the door to use this platform to make more complex genetic circuits for the fields of bacterial genetics, physiology, and synthetic biology.
Supplementary Material
Acknowledgments
This work was funded by the Wisconsin Alumni Research Foundation, the National Science Foundation (EFRI-1240268), and the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Sciences DE-FC02-07ER64494). M.C.P. was supported as a trainee in the Biotechnology Training Program (NIH).
Footnotes
This article is part of the ChemComm ‘Emerging Investigators 2013’ themed issue.
Electronic supplementary information (ESI) available: Detailed experimental procedures and Fig. S1, S2. See DOI: 10.1039/c2cc37107c
Notes and references
- 1.Khalil AS, Collins JJ. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Nucleic Acids Res. 2011;39:82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller J, Leung E, Meng X, Zhang L, Rebar E, Gregory P, Urnov F, Meyer BJ. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholze H, Boch J. Curr. Opin. Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanove AJ, Voytas DF. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 6.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 7.Moscou MJ, Bogdanove AJ. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 8.Streubel J, Blücher C, Landgraf A, Boch J. Nat. Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 9.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morbitzer R, Römer P, Boch J, Lahaye T. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis M. C. R. Biol. 2005;328:521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Schlax PJ, Capp MW, Record MT. J. Mol. Biol. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 14.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Nat. Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 16.Yarrington RM, Richardson SM, Huang CRL, Boeke JD. Genetics. 2012;190:523–U382. doi: 10.1534/genetics.111.136648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavco PA, Steege DA. J. Biol. Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.