Abstract
Uropathogenic Escherichia coli (UPEC) fall within a larger group of isolates producing extraintestinal disease. UPEC express type 1 pili as a critical virulence determinant mediating adherence to and invasion into urinary tract tissues. Type 1 pili expression is under regulation by a family of site-specific recombinases, including FimX, which is encoded from a genomic island called PAI-X for pathogenicity island of FimX. Using a new multiplex PCR, fimX and the additional PAI-X genes were found to be highly associated with UPEC (144/173 = 83.2 %), and more prevalent in UPEC of lower urinary tract origin (105/120 = 87.5 %) than upper urinary tract origin (39/53 = 74 %; P<0.05) or commensal isolates (28/78 = 36 %; P≤0.0001). The Fim-like recombinase gene fimX is the only family member that has a significant association with UPEC compared to commensal isolates. Our results indicate PAI-X genes, including the type 1 pili regulator gene fimX, are highly prevalent among UPEC isolates and have a strong positive correlation with genomic virulence factors, suggesting a potential role for PAI-X in the extraintestinal pathogenic E. coli lifestyle.
Introduction
Escherichia coli is a typical constituent of the enteric tract in many animals, including humans. Pathogenic E. coli strains produce a wide variety of intestinal and extra-intestinal diseases, such as diarrhoea, urinary tract infections, septicaemia and meningitis (Orskov & Orskov, 1992). Urinary tract infection (UTI) is a leading infection of children, women and the elderly (Foxman, 2010; Foxman et al., 2000), and uropathogenic E. coli (UPEC) is responsible for over 80 % of the approximately 10 million annual cases of community-acquired UTIs in the USA (Russo & Johnson, 2003). While the majority of UTIs involve bladder infection (cystitis), UPEC may also produce ascending infections to the kidneys, causing pyelonephritis or more advanced diseases such as urosepsis and meningitis among certain patient groups (Laupland et al., 2002; Rushton, 1997).
Molecular epidemiology to assign different unique genetic traits by strain group may be powerful. Defining distinguishing genetic features of commensal strains and pathogenic strains associated with different disease outcomes provides an opportunity to use this information for hypothesis generation, namely the identification of potential virulence loci, as well as for potential diagnostics. While specific pathotypes of E. coli are difficult to define given a high degree of genomic heterogeneity, certain genetic features can distinguish pathogenic strains from commensal strains (Groisman & Ochman, 1994). Phylogenetic analyses have shown that E. coli strains fall into four main phylogenetic groups (A, B1, B2 and D) (Herzer et al., 1990; Lecointre et al., 1998). Virulent extraintestinal pathogenic E. coli (ExPEC) belong mainly to group B2 and, to a lesser extent, to group D and encompass the UPEC strains as well (Boyd & Hartl, 1998; Picard et al., 1999). A large proportion of faecal-commensal, but a minority of UPEC strains, belong to group A (Bailey et al., 2010). However, even among the B2 and D groups, UPEC are genetically heterogeneous with an assortment of factors either directly demonstrated or epidemiologically linked to virulence, including adhesive factors like pili (P pili, type 1 pili and S pili), toxins (HlyA and CNF1), and invasion-promoting factors (Hek and IbeA) (Groisman & Ochman, 1994). Many of these factors are clustered together on horizontally acquired genomic islands termed pathogenicity-associated islands (Hacker, 1990).
Animal and human studies suggest that the type 1 pilus is a critical virulence determinant in the pathogenesis of UPEC cystitis, and bacteria lacking type 1 pili are attenuated, secondary to reduced adherence and invasion into superficial bladder epithelial cells (Connell et al., 1996; Mulvey et al., 1998; Wright et al., 2007). Corroborating the molecular pathogenesis studies of type 1 pili in UTI, vaccination with pili subcomponents in animal studies has been shown to reduce the severity and duration of infection (Palaszynski et al., 1998). Type 1 pili are encoded by the fim operon (fimAICDFGH) and their expression is controlled at multiple regulatory levels (Corcoran & Dorman, 2009; Kelly et al., 2006; McClain et al., 1991). The orientation of the invertible type 1 pili promoter region (fimS) (Abraham et al., 1985) is determined by the action of multiple tyrosine recombinases. UPEC strains are known to possess up to five Fim-like tyrosine recombinases (FimB, FimE, FimX, IpuA and IpuB) based on sequence homology, and all but IpuB are have been shown to invert fimS (Bryan et al., 2006; Gally et al., 1996; Hannan et al., 2008; Klemm, 1986; McClain et al., 1991; Xie et al., 2006) with differing biochemical activities: FimE inverts the promoter from ON to OFF, FimB catalyses bidirectional inversion, but favours OFF to ON, IpuA also has bidirectional activity, while FimX is able to invert fimS from OFF to ON.
While fimB and fimE are closely linked to the type 1 pili operon, fimX is located at an unlinked genetic locus we have termed PAI-X (Fig. 1a) for pathogenicity island of FimX. Among a small collection of E. coli isolates, we previously demonstrated that fimX was highly associated with UPEC isolates (Hannan et al., 2008). More recently, we showed that the recombinase FimX exclusively regulates the predicted LuxR-like response regulator HyxR that is encoded from the same pathogenicity island and is a negative regulator of the nitrosative stress response and intracellular macrophage survival (Bateman & Seed, 2012). Therefore, a goal of this study was to determine the prevalence of the type 1 pili recombinase regulator genes and PAI-X genes present in an extended, diverse collection of pathogenic and commensal E. coli isolates. Furthermore, by establishing the prevalence of the additional PAI-X genes among E. coli, we aimed to determine if PAI-X is associated with UPEC isolates from a particular UTI syndrome, indicating a possible role for the factors encoded by these genes in the UPEC pathogenic lifestyle.
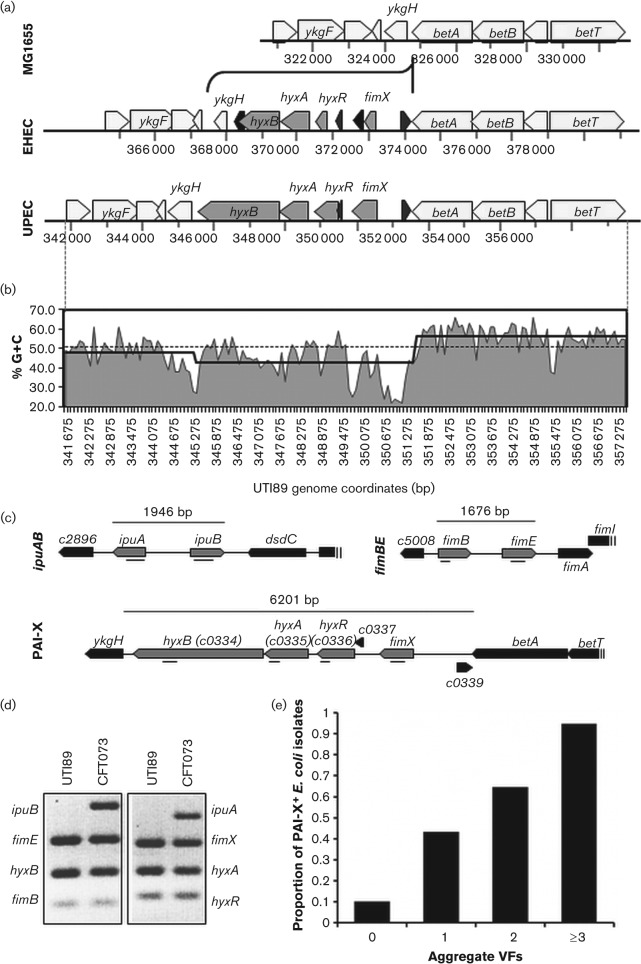
Fig. 1.
Overview of the genomic location, features, and organization of PAI-X among sequenced E. coli isolates. (a) Genomic organization and conservation of PAI-X among sequenced isolates from EHEC (EDL933) and UPEC (UTI89) pathotypes. MG1655 is used as a K-12, commensal reference. (b) Percentage G+C trace of PAI-XUTI89 and surrounding flanking regions. Percentage G+C is plotted as area under the curve for the entire region (betT through ykgE) using a 100 bp window. The average G+C across the entire UTI89 genome is plotted as a grey dotted line. The average % G+C content across PAI-X and flanking sequences is plotted as a black stepped line. (c) Grey arrows represent ORFs detected by multiplex PCR. Directionality of the ORFs, representing forward or reverse strand orientation, is shown. Amplicons are indicated as a black line (_). The genomic sequence for ipuA and ipuB is shown for CFT073 while fimB, fimE and PAI-X sequences are shown for the prototypic cystitis isolate UTI89. (d) Representative results of the multiplex PCR for the Fim-like recombinases and associated genomic islands using multiplex PCR. UTI89 and CFT073 are shown as prototypic UPEC strains to show visualization of all bands. (e) All E. coli isolates (n = 259), regardless of clinical syndrome, were scored based on the number of selective VFs present by PCR analysis and evaluated for the correlation between E. coli total VFs and the presence of PAI-X. Aggregate VFs refers to the number of total VFs present (out of the five assayed). Sample sizes varied between groups: 0 VFs (n = 30), 1 VF (n = 44), 2 VFs (n = 68), ≥3 VFs (n = 117).
Methods
Bacterial strains and cultivation.
All strains used in this study are listed in Table 1. A list of all individual clinical E. coli isolates with descriptive information and multiplex PCR results are listed in Table S1 (available in Microbiology online). Bacterial strains were routinely cultured in Luria–Bertani broth without antibiotics. Isolates were derived from four sources: (1) well-characterized, published representative pathogenic and commensal strains (n = 9); (2) isolates collected in previously published clinical studies in Seattle, representing various clinical syndromes of urinary tract infection (n = 155) (Czaja et al., 2009; Garofalo et al., 2007; Hooton et al., 1996; Johnson et al., 1987, 1988, 1991) as well as control faecal-commensal isolates (n = 20) (Stapleton et al., 1991); (3) strains from the E. coli reference (ECOR) collection (n = 68) (Ochman & Selander, 1984) and (4) clinical enterohaemorrhagic E. coli (EHEC) isolates (n = 7).
Table 1. Bacterial strains used in this study.
| Strains | Relevant features | Reference |
| UTI89 | E. coli cystitis isolate | Mulvey et al. (1998) |
| MG1655 | K-12 E. coli isolates | Blattner et al. (1997) |
| NU14 | E. coli cystitis isolate | Hultgren et al. (1986) |
| 536 | E. coli pyelonephritis isolate | Berger et al. (1982) |
| DS17 | E. coli pyelonephritis isolate | Tullus et al. (1984) |
| GR12 | E. coli pyelonephritis isolate | Svanborg Edén et al. (1983) |
| J96 | E. coli pyelonephritis isolate | Hull et al. (1981) |
| CFT073 | E. coli pyelonephritis/blood isolate | Mobley et al. (1990) |
| EDL933 | Enterohemhorrhagic E. coli isolate | Riley et al. (1983) |
| ECOR | E. coli reference set (n = 68) | Ochman & Selander (1984) |
| E. coli cystitis isolates | Single and recurrent clinical E. coli cystitis isolates (n = 68) | Czaja et al. (2009); Garofalo et al. (2007); Hooton et al. (1996) |
| E. coli asymptomatic isolates | Single and recurrent clinical E. coli ASB isolates (n = 45) | Garofalo et al. (2007); Hooton et al. (1996) |
| E. coli faecal isolates | Clinical E. coli faecal–commensal isolates (n = 20) | Stapleton et al. (1991) |
| E. coli pyelonephritis isolates | Clinical E. coli pyelonephritis isolates (n = 22) | Johnson et al. (1991) |
| E. coli urosepsis isolates | Clinical E. coli urosepsis isolates (n = 20) | Johnson et al. (1987); Johnson et al. (1988) |
The characterized representative strains included K-12 strain MG1655 (Blattner et al., 1997); cystitis isolates UTI89 (Mulvey et al., 1998) and NU14 (Hultgren et al., 1986); pyelonephritis isolates 536, DS17, GR12 and J96 (Berger et al., 1982; Hull et al., 1981; Svanborg Edén et al., 1983; Tullus et al., 1984); blood isolate CFT073 from a patient with concurrent pyelonephritis (Mobley et al., 1990) and EHEC isolates EDL933 (Riley et al., 1983).
For clinical isolates collected through studies carried out in Seattle, strains were archived in a specimen repository at the University of Washington UTI Research Laboratory and provided de-identified and anonymously through an Institutional Review Board-approved protocol. The study population for all studies was non-pregnant, outpatient women ages 18 to 49 seen in the Hall Health Primary Care Center (student health clinic) at the University of Washington (Czaja et al., 2009; Garofalo et al., 2007; Hooton et al., 1996) or Group Health Cooperative of Puget Sound (Garofalo et al., 2007; Hooton et al., 1996), except for isolates collected from in-patients hospitalized with pyelonephritis and/or urosepsis. Pyelonephritis isolates were collected from 20 women ages 18 to 45 hospitalized in one of four hospitals in Seattle (Johnson et al., 1991) and 2 were out-patients (Garofalo et al., 2007). Urosepsis isolates were collected from women ages 18 to 45 with bacteraemia arising from a urinary tract source, hospitalized in one of four hospitals in Seattle, WA (Johnson et al., 1987, 1988, 1991). Exclusion criteria for all studies of out-patients included known anatomical or functional abnormalities of the urinary tract, chronic illness requiring medical supervision, pregnancy or planned pregnancy during the next three months and, except for studies of pyelonephritis, symptoms or signs of acute pyelonephritis.
Clinical UTI syndromes were defined using the same criteria in all studies. Syndromes were defined as follows: acute, uncomplicated cystitis (n = 68 isolates) was defined as the presence of typical symptoms (dysuria, frequency and/or urgency), and a midstream urine culture containing ≥102 c.f.u. bacteria ml−1 (Czaja et al., 2009; Garofalo et al., 2007; Hooton et al., 1996); asymptomatic bacteriuria (ASB; n = 45) was defined as a midstream urine culture containing >105 c.f.u. bacteria ml−1 documented twice at least 24 h apart in a subject with no typical symptoms of UTI (Garofalo et al., 2007; Hooton et al., 1996); acute uncomplicated pyelonephritis (n = 22) was defined as costovertebral angle pain and/or tenderness, pyuria and bacteriuria with ≥104 c.f.u. ml−1 (Garofalo et al., 2007; Johnson et al., 1991) and urosepsis (n = 20) was defined as bacteraemia arising from a urinary tract source (Johnson et al., 1987, 1988). Commensal isolates (n = 20) were defined as faecal isolates (prospectively collected between 1981 and 1987) from healthy, non-pregnant women with no cystitis symptoms, no history of UTI in the prior year and no antibiotic use in the previous month (Stapleton et al., 1991).
Sixty-eight E. coli isolates from the ECOR collection (Ochman & Selander, 1984) representing phylogenetic groups A, B1, B2, and D human and non-human faecal-commensal isolates (n = 61) and human urinary tract isolates (ASB, n = 1; pyelonephritis, n = 6; cystitis, n = 4) were used as a well-characterized strain references. ASB, cystitis and pyelonephritis isolates from the ECOR collection were included as UPEC isolates for analysis.
An additional seven previously unpublished, clinical isolates of EHEC were also included for analysis. These isolates were provided de-identified and anonymously through an Institutional Review Board-approved protocol, and represent Shiga-toxin positive, diarrhoeagenic E. coli from a diverse patient population at Washington University Medical Center in St. Louis.
Multiplex PCR analysis.
One microlitre of bacterial culture suspension was used as template for multiplex PCR. All strains were analysed by multiplex PCR to determine the presence of the Fim-like recombinases and PAI-X genes. Primers were designed to conserved regions from gene alignments based on sequence data from available E. coli genomes (NCBI Genomes). The primer specificity was predicted by performing a blast analysis of the primer sequence to the UTI89 reference genome (Altschul et al., 1997). Up to four primer pairs were used per reaction (for primer sequences see Table 1). Primer groups were as follows: Group 1 (ipuB, fimE, hyxB, fimB), 94 °C (5 min), then 30 cycles of 94 °C (30 s), 51 °C (30 s), 72 °C (30 s), followed by 72 °C (7 min) and 4 °C hold; Group 2 (ipuA, fimX, hyxA, hyxR), 94 °C (5 min), then 30 cycles of 94 °C (30 s), 52 °C (30 s), 72 °C (30 s), followed by 72 °C (7 min) and 4 °C hold.
Since UPEC also colonize the enteric tract, we assayed the ECOR reference strains (n = 68) and faecal isolates (n = 20) for the presence of additional virulence factors (VFs), using the multiplex primers listed in Table 2, to determine if any of the faecal isolates represent potential extraintestinal pathogens. The ECOR strain set has been previously analysed by multiplex PCR for various VFs (Johnson et al., 2001). Primers and programs were grouped as follows: Group 3 (kpsMT II, kpsMT III, ibeA), 94 °C (5 min), then 30 cycles of 94 °C (30 s), 55 °C (30 s), 72 °C (35 s), followed by 72 °C (7 min) and 4 °C hold; Group 4 (sfa/foc, traT, papA), 94 °C (5 min), then 30 cycles of 94 °C (30 s), 55 °C (30 s), 72 °C (50 s), followed by 72 °C (7 min) and 4 °C hold; Group 5 (cnf1, hlyA, fyuA), 94 °C (5 min), then 30 cycles of 94 °C (30 s), 55 °C (30 s), 72 °C (70 s), followed by 72 °C (7 min) and 4 °C hold; and Clonal Groups (chuA, yjaA, TSPE4.C2), 94 °C (5 min), then 30 cycles of 94 °C (30 s), 55 °C (30 s), 72 °C (30 s), followed by 72 °C (7 min) and 4 °C hold.
Table 2. Primers used in this study.
Multiplex PCR primers and assays for additional VFs: fyuA (Johnson & Stell, 2000), traT (Johnson & Stell, 2000), kpsMT II (Johnson & Stell, 2000), kpsMT III (Johnson & Stell, 2000), papAH (Johnson & Stell, 2000), sfa/focDE (Le Bouguenec et al., 1992), ibeA (Huang et al., 1995), hlyA (Johnson & Stell, 2000), cnf1 (Yamamoto et al., 1995), and clonal groups (Clermont et al., 2000) were performed as described. Primer groupings are listed in Methods.
| Primers (forward/reverse) | Target | Sequence (5′→3′) | Size | Reference |
| fimE MPX 1/fimE MPX2 | fimE | CTAACTGGAAAGGCGCTGAC/GAATATTTCGATGCCCGAGA | 225 | This study |
| fimB MPX 1/fimB MPX2 | fimB | GCCTCATGCTGCACGTAAT/CAATCGACAAATTTCACTCG | 79 | This study |
| fimX MPX 1/fimX MPX 2 | fimX | CCAGAGCATGTCCTTTCCTG/TTCCTCTGCTTAAGCCACAAC | 216 | This study |
| ipuA MPX 1/ipuA MPX 2 | ipuA | GCGATGTTTGCATGATTTTA/TTTTACCCGCAGCAGAAACT | 303 | This study |
| ipuB MPX 1/ipuB MPX 2 | ipuB | TGCGCAAATTTATTACTCATAGTG/TGTCTCGAGATTTTATTTCCTTGA | 334 | This study |
| hyxR MPX 1/hyxR MPX 2 | hyxR | TCGATGAGCGGAATGTTGTC/GGCTGCTCTATACGGGATGC | 89 | This study |
| hyxA MPX 1/hyxA MPX 2 | hyxA | GCATTTCCATCACCGTGAAA/GTGCGCAGTTTCTCAAAACG | 142 | This study |
| hyxB MPX 1/hyxB MPX 2 | hyxB | GGGTATCACCACCCAGCATT/CAGGATGCTGTCCGTCTGAG | 139 | This study |
All PCRs were performed using APEX Taq polymerase and accompanying buffers (Genesse Scientific). Amplicons were visualized on a 2 % TBE gel stained with ethidium bromide and photographed with a UV transilluminator (Bio-Rad). Each reaction was performed a minimum of three times, as biological replicates.
Alignment of PAI-X coding sequences from O157 : H7 EHEC strains.
The coding sequences for fimX, hyxR, hyxA and hyxB were gathered for UTI89 (EMBL coding sequence: ABE05839) and O157 : H7 strain ELD933 (EMBL coding sequence: AAG54651) from the EMBL-EBI Integr8 web portal (Kersey et al., 2005). The resulting sequences were aligned using the clustal w multiple alignment algorithm (Thompson et al., 1994) in the BioEdit Sequence Alignment Editor (Hall, 1999), using the ‘Toggle’ feature to translate the coding sequence. Mutations in EHEC, relative to UTI89, that resulted in a premature stop codon were noted.
Sensitivity and specificity calculations.
Specificity and sensitivity metrics were calculated as previously described (Altman & Bland, 1994) and were used in this study as a common metric to compare the performance of detecting different E. coli genes to classify strains as UPEC. Calculations were based on data derived from Table 3. The formulas used for calculating sensitivity and specificity are shown below:
Table 3. Molecular epidemiology of the Fim-like recombinases and PAI-X factors in commensal and pathogenic E. coli isolates.
P values (Fisher’s exact) were calculated using two-tailed tests. Symbols indicating significance for group comparisons are as follows: human versus non-human commensals: *P≤0.05; lower UTI versus upper UTI: *P≤0.05; **P≤0.01; ***P≤0.001. Commensal total versus UPEC total: ***P≤0.001; ****P≤0.0001.
| Commensal† | UPEC | |||||||||||||
| Human | Non-human | Total | Lower UTI‡ | Upper UTI‡ | Total | EHEC | ||||||||
| (n = 48) | (n = 30) | (n = 78) | (n = 120) | (n = 53) | (n = 173) | (n = 8) | ||||||||
| Target | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| fimB | 46 | 96 | 30 | 100 | 76 | 97 | 119 | 99.2 | 53 | 100 | 172 | 99.4 | 8 | 100 |
| fimE | 48 | 100 | 30 | 100 | 78 | 100 | 120 | 100.0 | 53 | 100 | 173 | 100.0 | 8 | 100 |
| ipuA | 9 | 19 | 3 | 10 | 12 | 15 | 28 | 23.3 | 13 | 25 | 41 | 23.7 | 1 | 13 |
| ipuB | 9 | 19 | 3 | 10 | 12 | 15 | 28 | 23.3 | 13 | 25 | 41 | 23.7 | 1 | 13 |
| fimX | 17 | 35 | 11 | 37 | 28 | 36 | 105 | 87.5 | 39 | 74* | 144 | 83.2**** | 5 | 63 |
| hyxR | 17 | 35 | 11 | 37 | 28 | 36 | 105 | 87.5 | 39 | 74* | 144 | 83.2**** | 5 | 63 |
| hyxA | 17 | 35 | 11 | 37 | 28 | 36 | 105 | 87.5 | 39 | 74* | 144 | 83.2**** | 5 | 63 |
| hyxB | 17 | 35 | 11 | 37 | 28 | 36 | 105 | 87.5 | 39 | 74* | 144 | 83.2**** | 5 | 63 |
| ibeA | 6 | 13 | 1 | 3 | 7 | 9 | 56 | 46.7 | 11 | 21** | 67 | 38.7**** | 2 | 25 |
| traT | 18 | 38 | 9 | 30 | 27 | 35 | 94 | 78.3 | 43 | 81 | 137 | 79.2**** | 5 | 63 |
| fyuA | 27 | 56 | 9 | 30|| | 36 | 46 | 109 | 90.8 | 48 | 91 | 157 | 90.8**** | 5 | 63 |
| hlyA | 10 | 21 | 1 | 3|| | 11 | 14 | 45 | 37.5 | 20 | 38 | 61 | 37.6**** | 2 | 25 |
| cnf1 | 7 | 15 | 1 | 3 | 8 | 10 | 69 | 57.5 | 16 | 30*** | 85 | 49.1**** | 4 | 50 |
Commensal strains represent the ECOR reference set as well as faecal isolates from healthy women. Only clonal groups A, B1, B2 and D were included. ECOR isolates that were human ASB, CY or PY isolates were included in the UPEC group for analysis.
Lower UTI isolates represent single and recurrent ASB and CY isolates. Upper UTI isolates represent PY and urosepsis isolates.
Sensitivity = number true positive/(number true positive+number false negative)
Specificity = number true negative/(number true negative+number false positive)
Statistical analysis.
Statistical analyses using the Fisher’s exact test were performed using GraphPad Prism (GraphPad Software) where indicated in figure legends. Two-tails were used in the determination of statistical significance, which was defined by attaining P≤0.05. Statistical determinations were not weighted for epistatic relationships. Logistic regression was performed in R using the MASS, arm, aod and car packages. Model refinement was performed using a forward step-wise approach.
Results
Organization of the fimX-associated island, PAI-X and association with VFs
Three recombinases FimB, FimE and FimX regulate the phase variation of type 1 pili in the prototypic cystitis strain UTI89 (Hannan et al., 2008). fimB and fimE are located immediately upstream of the type 1 pili operon, while fimX is located at an unlinked genetic locus we have termed PAI-X for Pathogenicity-associated Island of FimX (located at approximately 4.2 min on the UTI89 chromosome), adjacent to the betABIT choline-glycine betaine locus (Fig. 1a). The ~6.2 kb region contains four ORFs (>100 aa): fimX, hyxR, hyxA and hyxB for Hypotheticals of PAI-X (Fig. 1a). The fimX genomic locus (PAI-X) meets most, if not all, criteria for a pathogenicity island as defined by Hacker & Kaper (2000). For instance, PAI-X has disparate G+C content compared to the rest of the UTI89 genome: PAI-X has 42.9 % G+C content relative to 50.6 % G+C content for UTI89 generally (Fig. 1b) (Chen et al., 2006; EnCor Biotechnology, http://www.encorbio.com/protocols/Nuc-MW.htm). As shown later in the results, PAI-X is more prevalent among pathogenic E. coli strains than commensal strains, indicating PAI-X is associated with virulence (Table 3). Although prior work demonstrated that FimX plays a role in the regulation of type 1 pili during acute cystitis (Hannan et al., 2008), the role of the hyx genes during infection remains unknown.
To determine the prevalence and complement of Fim-like recombinase genes present in the genomes of a range of E. coli isolates as well as the association of PAI-X genes with known and epidemiologically linked VFs, a multiplex PCR approach was developed to detect the Fim-like recombinases (fimB, fimE, fimX, ipuA, ipuB) and hyx genes (hyxR, hyxA, hyxB) (Fig. 1c, d) in a variety of pathogenic strains isolated from women with a variety of clinical syndromes (see Methods for a detailed description of E. coli strains and patient populations). For comparison, the assay was performed on non-human and the human commensal isolates from the ECOR reference set (Ochman & Selander, 1984), an independent set of faecal isolates from healthy women and a small group of EHEC isolates. Additionally, isolates were screened by PCR for the presence of five known or epidemiologically associated VF genes including cnf1, hlyA, fyuA, traT and ibeA. The proportion of E. coli isolates carrying PAI-X increased with the total number of virulence factor genes (Fig. 1e), demonstrating that PAI-X acquisition has a strong, positive correlation with known VFs.
The type 1 pili regulator gene fimX and associated pathogenicity island PAI-X are highly prevalent among UPEC
As expected, fimB and fimE are ubiquitous among both pathogenic and non-pathogenic E. coli as both fimB and fimE were present in >98 % of UPEC and commensal strains (Table 3). We found that ipuA and ipuB were always present together, consistent with previous findings (Bryan et al., 2006), and that ipuA and ipuB were present in commensal (15 %) and UPEC (23.7 %) isolates in statistically equal abundance (Table 3). In contrast, fimX was highly prevalent among UPEC (83.2 %), but not commensal isolates (36 %; P<0.0001), and was more prevalent in lower (87.5 %) than upper UTI isolates (74 %; P = 0.03) (Table 3). ipuA and ipuB, on the other hand, were equally distributed among upper (25 %) and lower UTI isolates (23.3 %). There were no significant differences in the prevalence of fimX or ipuAB among the various individual UTI syndromes (data not shown). The Fim-like recombinase gene fimX is the only family member to have a specific, significant association with UPEC isolates.
Like fimX, hyxRAB was also more significantly associated with pathogenic (83.2 %) versus commensal isolates (36 %; Table 3). In UPEC isolates, the hyx genes were always present with fimX, and thus, were also more significantly associated with UPEC isolates, demonstrating that, when present, the major genetic features of PAI-XUTI89 are highly conserved among other UPEC isolates of urinary tract and blood.
Distribution of the Fim-like recombinase and PAI-X genes by E. coli phylogenetic group
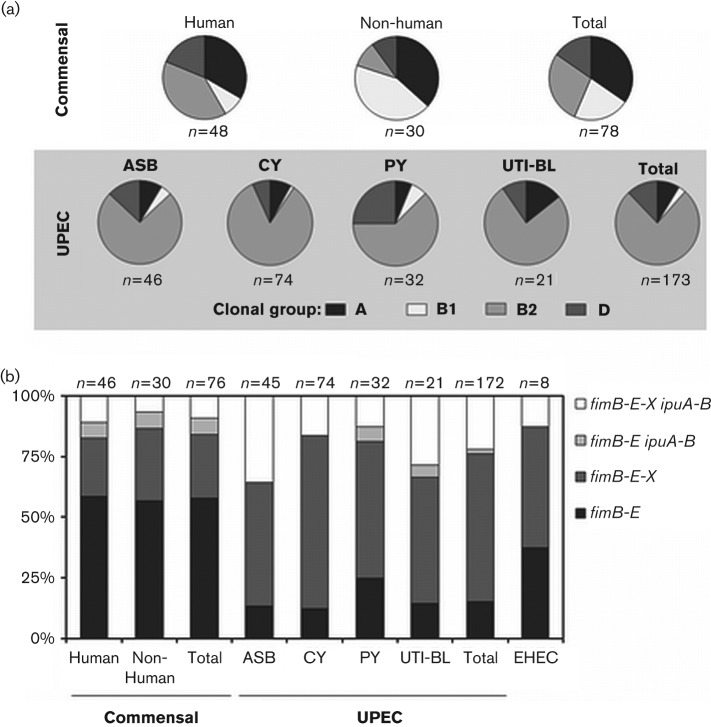
Phylogenetic group analysis was performed as previously described (Clermont et al., 2000; Gordon et al., 2008). Consistent with previous results (Carlos et al., 2010), commensal E. coli of non-human origin contained a larger percentage of group B1 strains (46.2 %) than those of human origin (11.8 %; P = 0.004) while, not surprisingly, faecal isolates of human origin contained a larger percentage of group B2 strains (26.5 %; P = 0.03 compared with faecal isolates of non-human origin) (Fig. 2a). Consistent with other published clinical strain sets, the majority of UPEC isolates were group B2 (76.3 %) and to a lesser extent group D (Fig. 2a). Group D strains were at least 2.5-fold more highly associated with pyelonephritis isolates (25.0 %), than isolates from other syndromes (13/141 = 9.2 %; P = 0.03). The distribution of the isolates among the phylogenetic groups is consistent with previously published clinical sets.
Fig. 2.
Clonal group variation and Fim-like recombinase prevalence by isolate source and syndrome. (a) Phylogenetic associations among commensal and pathogenic isolates. The percentage of isolates in each of the four clonal groups is shown. Clonal groups were assigned based on a previously published multiplex PCR for chuA, yjaA, and genomic fragment TSPE4.C2 (Clermont et al., 2000; Gordon et al., 2008). ECOR strain clonal groups were determined previously (Johnson et al., 2001) and are in accordance with our experimental results. CY, cystitis; PY, pyelonephritis; UTI-BL, urosepsis. (b) The percentage of isolates that encode one of four recombinase profiles observed, grouped as follows: fimB-E only (similar to MG1655); fimB-E-X only (similar to UTI89); fimB-E ipuA-B (rarely observed); and all five recombinases present, fimB-E-X ipuA-B (similar to CFT073). Two human commensal isolate and one ASB isolate were excluded from analysis as they did not carry the fimB gene by PCR analysis.
Analysis of the Fim-like recombinases and PAI-X association with phylogenetic group revealed that fimB and fimE were equally distributed among all groups (Table 4), consistent with the ubiquitous presence of these genes. One the other hand, the genes ipuA and ipuB were frequently identified in group B2 (28.6 %) or group D (27.3 %) strains, but were rarely found in group A strains (2.4 %) and were absent in group B1 strains (0 %; Table 4). PAI-X was found in the vast majority of group B2 isolates (95.5 %) but few group A strains (9.5 %; P<0.0001), consistent with group A strains making up the largest proportion of commensal isolates (Fig. 2a). PAI-X showed a much lower association with group D strains (33.3 %), and all 11 of these isolates were from the human UPEC group (Table 4), which is consistent with PAI-X being a phylotype B2 and D, human UPEC-associated marker. PAI-X was also present in 45.5 % of B1 isolates (Table 4), but the majority (8/10 = 80.0 %) were from non-human commensals.
Table 4. Phylogenetic distribution of the Fim-like recombinases and PAI-X among all E. coli isolates.
All E. coli isolates (n = 251), excluding eight EHEC isolates, were analysed for clonal group by PCR. PAI-X factors were assayed separately but grouped together for analysis. P values (Fisher’s exact) were calculated using two-tailed tests. Statistical significance is indicated in the table as: *P≤0.05; **P≤0.01; ***P≤0.001; ****P≤0.0001. Significance was calculated for indicated group versus all other strains. Parentheses indicate a negative association.
| A | B1 | B2 | D | |||||
| n = 42 | n = 22 | n = 154 | n = 33 | |||||
| Target | No. | % | No. | % | No. | % | No. | % |
| fimB | 42 | 100 | 22 | 100 | 151 | 98.1 | 33 | 100 |
| fimE | 42 | 100 | 22 | 100 | 154 | 100.0 | 33 | 100 |
| ipuA | 1 | 2(***) | 0 | 0(**) | 44 | 28.6*** | 9 | 27 |
| ipuB | 1 | 2(***) | 0 | 0(**) | 44 | 28.6*** | 9 | 27 |
| PAI-X | 4 | 10(****) | 10 | 45(*) | 147 | 95.5**** | 11 | 33(****) |
UPEC have acquired additional Fim-like recombinase genes
There were four main recombinase profiles observed among isolates: strains that encode fimB fimE only, strains that encode fimB fimE in addition to fimX, strains that encode ipuA ipuB in addition to fimB fimE, and strains that encode all five recombinase genes, fimB fimE fimX ipuA ipuB (Fig. 2b). The majority of commensal strains (both human and animal origin) only carried the genes for fimB and fimE (58 %; Fig. 2b). In both commensal (7 %) and UPEC isolates (1.7 %), addition of ipuA ipuB in the absence of fimX was infrequent. Interestingly, UPEC strains generally contained an expanded repertiore of recombinase genes. For instance, 84.8 % of UPEC contained at least one added Fim-recombinase gene in addition to fimB and fimE, compared with 42 % of commensal isolates (P<0.0001) (Fig. 2b). Therefore, UPEC isolates are twofold more likely to have acquired recombinases in addition to ‘core genome’ family members fimB and fimE, suggesting that pathogenic E. coli have undergone adaptation of the regulatory programme controlling type 1 pili.
PAI-X is a genetic marker for human UPEC
To elucidate whether specific genetic features of E. coli were predictive of isolates being urinary tract-associated UPEC, we evaluated the sensitivity and specificity of VF genes for predicting or excluding an E. coli isolate as UPEC (see Methods section for a detailed explanation). These served as common metrics to compare the likelihood that specific E. coli genes were predictive of an isolate being part of the UPEC group. Applying this analysis to the data from Table 3, several genes have high sensitivity (equivalent to prevalence) as a positive indicator for an isolate being UPEC. For, instance fyuA and PAI-X have the two highest sensitivities at 90.8 and 83.2 %, respectively, indicating that these genomic markers are highly associated with UPEC (Table 5). Conversely, a high specificity would indicate those genes that, when absent, predict an isolate is not UPEC. Genes like cnf1 and hlyA, which are almost never found in commensal isolates, have specificities ≥97 %, indicating that negative tests for these genomic markers are useful in excluding an isolate from the UPEC group. Although hlyA and cnf1 have high specificities, their low prevalence in the UPEC population yields a low sensitivity value, making their presence less useful in predicting if an isolate is UPEC. Of the VFs tested, PAI-X had the best combination of sensitivity and specificity, at 83.2 and 65 %, respectively (Table 5).
Table 5. Sensitivity and specificity of VFs predictive of human UPEC.
Sensitivity and specificity are described in Methods. Calculations are based on data derived from Table 3.
| Sensitivity | Specificity | |
| ibeA | 67/173 = 38.7 % | 42/48 = 88 % |
| traT | 137/173 = 79.2 % | 30/48 = 63 % |
| fyuA | 157/173 = 90.8 % | 21/48 = 44 % |
| hlyA | 61/173 = 37.6 % | 38/48 = 79 % |
| cnf1 | 85/173 = 49.1 % | 41/48 = 85 % |
| PAI-X | 144/173 = 83.2 % | 31/48 = 65 % |
Based on the potential for multiple associations between VFs and sources/syndrome, multivariate logistic regression analysis was used to simultaneously assess bacterial VFs and phylogenetic group as predictors of UTI in general, upper tract disease (pyelonephritis with/without bloodstream involvement) and ASB (Table 6). Logistic regression modelling first included phylogenetic group and the VF genes ibeA, traT, fyuA, ipuA, hlyA, cnf1 and fimX. Subsequently a forward step approach was used to refine the models. From an analysis of all isolates against candidate predictors of UTI, phylogenentic group B2 and VF genes ibeA, traT, fyuA and cnf1 emerged as significant predictors of UTI (Wald χ2, P = 1.6×10−8). The models for distinguishing upper tract isolates from lower tract isolates (cystitis and ASB) were not as robust. In the two best models, ibeA and cnf1 had odds ratios (OR) of <1 (Wald χ2, P = 0.025), suggesting they are modest negative predictors of upper urinary tract infections. Significant positive predictors, however, did not emerge, although ibeA and ipuA had moderate to strong OR (4.1 and 4.5 respectively; Wald χ2, P = 0.0007) as positive predictors of ASB.
Table 6. Stepwise multivariate logistic regression analysis for predictors of UTI, upper tract infection, and ASB.
Phylogenetic groups (Group) are A, B1, B2 and D. The Group variable was set as B2 versus all other groups. All stepwise models initially included model A values of Group, ibeA, traT, fyuA, hylA, cnf1, ipuA and fimX. The genes ipuB and (hyxR, hyxA and hyxB) were not included because they were found to have 100 % correlation with ipuA and fimX, respectively. Forward stepwise refinement of model A was performed in each prediction scenario; model B, Group+traT+fyuA+cnf1; model C, hlyA+cnf1; model D, Group+ibeA+ipuA. CI, confidence interval; DF, degrees of freedom; OR, odds ratio.
| Candidate predictor values | OR (95 % CI) | P value | Wald χ2 (DF, P value) | Model |
| Predictors of UTI | ||||
| Group | 3.6 (1.4–9.7) | 2.7×10−15 | 42.1 (4, 1.6×10−8) | A |
| ibeA | 1.4 (0.52–3.8) | 0.004 | ||
| traT | 6.7 (3.2–15.0) | 9.4×10−9 | ||
| fyuA | 4.3 (1.9–10.2) | 0.0003 | ||
| Group | 4.4 (2.0–10.1) | 2.7×10−15 | 54.3 (4, 4.7×10−11) | B |
| traT | 7.7 (3.8–16.5) | 5.3×10−10 | ||
| fyuA | 4.2 (1.9–9.8) | 0.0002 | ||
| cnf1 | 2.5 (1.1–6.2) | 0.03 | ||
| Predictors of upper tract infection | ||||
| ibeA | 0.4 (0.17–0.94) | 0.003 | 7.4 (2, 0.025) | A |
| cnf1 | 0.3 (0.11–0.69) | 0.007 | ||
| cnf1 | 0.19 (0.07–0.45) | 0.0001 | 12.5 (2, 0.002) | C |
| Predictors of ASB | ||||
| ibeA | 3.8 (1.5–10.4) | 0.003 | 12.1 (2, 0.002) | A |
| ipuA | 4.3 (1.6–12.8) | 0.004 | ||
| ibeA | 4.1 (1.7–10.7) | 0.003 | 14.6 (2, 0.0007) | D |
| ipuA | 4.5 (1.7–12.6) | 0.002 | ||
Discussion
Pathogenicity-associated islands are common among ExPEC (Blum et al., 1995; Chen et al., 2006; Lloyd et al., 2007) and have some characteristic features, including: (1) disparate G+C content from the core genome; (2) association with pathogenic organisms, but not commensal counterparts; (3) contain virulence-associated genes; (4) insertion sites or flanking direct repeats; (5) a high frequency of insertion at tRNA sites and (6) often mobility (pseudo)genes (Dobrindt et al., 2004; Gal-Mor & Finlay, 2006; Hacker & Kaper, 2000). PAI-X was associated with E. coli isolated from patients experiencing a variety of clinical syndromes, including asymptomatic bacteriuria, cystitis, pyelonephritis and urosepsis. Prior studies have also clearly demonstrated that FimX-mediated control of type 1 pili expression is activated in vivo and that independent expression of type 1 pili by FimX in vivo is sufficient, but not necessary, to produce acute cystitis (Hannan et al., 2008). PAI-X is not associated with a tRNA and direct repeats or insertion elements were not immediately apparent. Many islands are also associated with phage genes and transposases and, while FimX is part of the phage integrase superfamily, it has never been observed to function as an integrase or excisionase, rather mediating site-specific recombination or inversion.
Our molecular epidemiology data provide strong evidence that this locus is widespread among a diverse group of UPEC clinical isolates of urinary tract origin but is present in a minority of commensal E. coli isolates. UPEC are true commensal-pathogens, meaning that these potential pathogens cycle through the commensal reservoir of the gastrointestinal tract without causing symptoms. However, phylotype B2 E. coli, especially those encoding UTI-associated VFs, likely represent potential pathogens even if they are isolated from a commensal niche. This suggests that the prevalence of PAI-X and other VFs is likely to be overrepresented among E. coli strains defined as commensals based solely on host site and not taking into account genomic virulence potential. Our molecular epidemiology data also support the idea that fimX is the only Fim-recombinase gene specifically associated with UPEC isolates, suggesting that strains carry fimB fimE as part of the ‘core genome’ then can acquire ipuA ipuB subsequent to fimX acquisition in pathogenic isolates. Our findings are consistent with the notion that UPEC strains have acquired additional regulatory inputs to control and possibly fine-tune the expression of a major VF, type 1 pili. Alternatively, the high prevalence of fimX among UPEC may be conserved due to its role regulating hyxR and the subsequent effects on UPEC intracellular survival within macrophages and tolerance to reactive nitrogen intermediates. Future work will be needed to ascertain the evolutionary benefit of FimX coordinated regulation of type 1 pili and hyxR during in vivo infection.
Among ExPEC in general, fimX was always found associated with hyxR, hyxA and hyxB, suggesting that when this locus is present, it is broadly structurally conserved. Although sequence data from model strains suggest these genes reside together at the same locus, hybridization and/or sequencing studies will be necessary to ascertain if fimX and hyxRAB are always linked. Our molecular epidemiology data also suggest that this locus is less prevalent among pyelonephritis and urosepsis isolates than lower UTI isolates (ASB, cystitis); although, the majority of these strains still carry PAI-X, perhaps accounting for why fimX (representing PAI-X) did not emerge as a predictive factor for any particular syndrome in multivariate logistic regression. Future studies will be needed to determine if there is a biological significance to this statistical difference. Many isolates that cause urosepsis are genetically related to strains known to cause cystitis and pyelonephritis, suggesting that the simple presence or absence of any genetic factors is insufficient to define urosepsis isolates as a group.
As illustrated by several of the sequenced EHEC isolates, the simple presence or absence of PAI-X by PCR analysis is unlikely to predict the functional state of the factors. FimX, HyxR, HyxA and HyxB are highly conserved at the amino acid level with greater than 98 % identity, respectively, among the prototypic UPEC isolates UTI89, 536 and CFT073. However, sequence analysis of PAI-X in EHEC 0157:H7strains EDL933 and EC4115 indicated the presence of a premature nonsense mutation in fimX (Tyr128→Stop), hyxB (Ser370→Stop) and hyxR (Ser56→Stop) (Fig. 1a). In EHEC strains, there is also a potential in-frame secondary translational start site at residue 107 of hyxR, which would create a significant N-terminal truncation. Thus, it is likely that fimX, hyxR and hyxB do not yield functional products in these EHEC strains. These data indicate that PAI-X is likely widely distributed among UPEC, although sequencing efforts will reveal if these strains carry polymorphisms producing coding sequence disruptions as found in many EHEC isolates. In contrast, the high identity at the nucleotide level for all of the PAI-X genes between sequenced EHEC isolate EDL933 and prototypic UPEC isolate UTI89 suggests that PAI-X may have a common provenance. PAI-X, including fimX, was highly conserved in UPEC isolates but disrupted among sequenced EHEC isolates, suggesting a potential evolutionary benefit of this locus specific to the colonization, transmission or pathogenesis of UPEC isolates generally.
A role for PAI-X in virulence is supported by our molecular epidemiology data and the strong, positive correlation between the number of VFs present in the genome of an isolate and the presence of PAI-X. Previous studies have shown that FimX is capable of regulating virulence during experimental cystitis (Hannan et al., 2008). FimX is sufficient but not necessary to mediate type 1 pili expression during cystitis, thereby promoting UPEC bladder epithelial invasion. Translation and blast analysis of hyxR suggest it is a putative helix–turn–helix, LuxR-like response regulator. Recently, our laboratory has also shown that FimX exclusively regulates the PAI-X gene hyxR through bidirectional promoter inversion, and that HyxR regulates the intracellular survival of E. coli during macrophage infection (Bateman & Seed, 2012). The additional two conserved ORFs found in this locus appear to code for a putative outer membrane autotransporter (HyxB) and a conserved hypothetical protein (HyxA), which has homologues only in the genomes of E. coli and S. enterica (protein blast E-score <5×10−5). Further work will be needed to discern the individual contributions of the other PAI-X genes in colonization and extraintestinal disease, but it is clear that PAI-X constituents alter UPEC virulence. The high prevalence of PAI-X genes distributed among UPEC may suggest that PAI-X encoded factors like FimX and HyxR have frequent and conserved roles in host interactions, warranting future studies to assess their full biological functions.
Acknowledgements
This work was supported by the National Institutes of Health grant K08DK07444301to P. C. S; a Duke Children’s Miracle Network award to P. C. S; the National Institute of Diabetes and Digestive and Kidney Diseases grant P01 DK053369 to A. E. S., T. M. H. and W. E. S.; and the Specialized Centers of Research from the National Institute of Diabetes and Digestive and Kidney Diseases and the Office of Research on Women’s Health, National Institutes of Health grant P50 DK064540 to P. C. S., A. E. S., T. M. H. and W. E. S. We would like to thank Ravi Jhaveri for critical review of this manuscript. We thank Scott Hultgren for his directorship of the SCOR collaborative. We thank Marsha Cox for her management and preparation of the clinical isolates. The authors do not declare any conflicts of interest and have no financial disclosures. This work has been presented, in part, at the 46th Annual Meeting of the Infectious Diseases Society of America, Philadelphia, 29 October–1 November 2009 (oral abstract 798).
Abbreviations:
- ASB
asymptomatic bacteriuria
- EHEC
enterohaemorrhagic E. coli
- ExPEC
extraintestinal pathogenic E. coli
- UPEC
uropathogenic E. coli
- UTI
urinary tract infection
- VF,
virulence factor
Footnotes
One supplementary table is available with the online version of this paper.
References
- Abraham J. M., Freitag C. S., Clements J. R., Eisenstein B. I. (1985). An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A 82, 5724–5727. 10.1073/pnas.82.17.5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D. G., Bland J. M. (1994). Diagnostic tests. 1: Sensitivity and specificity. BMJ 308, 1552. 10.1136/bmj.308.6943.1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. K., Pinyon J. L., Anantham S., Hall R. M. (2010). Distribution of human commensal Escherichia coli phylogenetic groups. J Clin Microbiol 48, 3455–3456. 10.1128/JCM.00760-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman S. L., Seed P. C. (2012). Epigenetic regulation of the nitrosative stress response and intracellular macrophage survival by extraintestinal pathogenic Escherichia coli. Mol Microbiol, 83, 908–925. 10.1111/j.1365-2958.2012.07977.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger H., Hacker J., Juarez A., Hughes C., Goebel W. (1982). Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J Bacteriol 152, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Plunkett G., III, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K. & other authors (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- Blum G., Falbo V., Caprioli A., Hacker J. (1995). Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett 126, 189–195. [DOI] [PubMed] [Google Scholar]
- Boyd E. F., Hartl D. L. (1998). Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol 180, 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan A., Roesch P., Davis L., Moritz R., Pellett S., Welch R. A. (2006). Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect Immun 74, 1072–1083. 10.1128/IAI.74.2.1072-1083.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos C., Pires M. M., Stoppe N. C., Hachich E. M., Sato M. I., Gomes T. A., Amaral L. A., Ottoboni L. M. (2010). Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol 10, 161. 10.1186/1471-2180-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L., Hung C. S., Xu J., Reigstad C. S., Magrini V., Sabo A., Blasiar D., Bieri T., Meyer R. R. & other authors (2006). Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103, 5977–5982. 10.1073/pnas.0600938103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66, 4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell I., Agace W., Klemm P., Schembri M., Mărild S., Svanborg C. (1996). Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93, 9827–9832. 10.1073/pnas.93.18.9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran C. P., Dorman C. J. (2009). DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol 74, 1071–1082. 10.1111/j.1365-2958.2009.06919.x [DOI] [PubMed] [Google Scholar]
- Czaja C. A., Stamm W. E., Stapleton A. E., Roberts P. L., Hawn T. R., Scholes D., Samadpour M., Hultgren S. J., Hooton T. M. (2009). Prospective cohort study of microbial and inflammatory events immediately preceding Escherichia coli recurrent urinary tract infection in women. J Infect Dis 200, 528–536. 10.1086/600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U., Hacker J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2, 414–424. 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
- Foxman B. (2010). The epidemiology of urinary tract infection. Nat Rev Urol 7, 653–660. 10.1038/nrurol.2010.190 [DOI] [PubMed] [Google Scholar]
- Foxman B., Barlow R., D’Arcy H., Gillespie B., Sobel J. D. (2000). Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol 10, 509–515. 10.1016/S1047-2797(00)00072-7 [DOI] [PubMed] [Google Scholar]
- Gal-Mor O., Finlay B. B. (2006). Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol 8, 1707–1719. 10.1111/j.1462-5822.2006.00794.x [DOI] [PubMed] [Google Scholar]
- Gally D. L., Leathart J., Blomfield I. C. (1996). Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol 21, 725–738. 10.1046/j.1365-2958.1996.311388.x [DOI] [PubMed] [Google Scholar]
- Garofalo C. K., Hooton T. M., Martin S. M., Stamm W. E., Palermo J. J., Gordon J. I., Hultgren S. J. (2007). Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun 75, 52–60. 10.1128/IAI.01123-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., Clermont O., Tolley H., Denamur E. (2008). Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol 10, 2484–2496. 10.1111/j.1462-2920.2008.01669.x [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Ochman H. (1994). How to become a pathogen. Trends Microbiol 2, 289–294. 10.1016/0966-842X(94)90006-X [DOI] [PubMed] [Google Scholar]
- Hacker J. (1990). Genetic determinants coding for fimbriae and adhesins of extraintestinal Escherichia coli. Curr Top Microbiol Immunol 151, 1–27. 10.1007/978-3-642-74703-8_1 [DOI] [PubMed] [Google Scholar]
- Hacker J., Kaper J. B. (2000). Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54, 641–679. 10.1146/annurev.micro.54.1.641 [DOI] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41, 95–98. [Google Scholar]
- Hannan T. J., Mysorekar I. U., Chen S. L., Walker J. N., Jones J. M., Pinkner J. S., Hultgren S. J., Seed P. C. (2008). LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol Microbiol 67, 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzer P. J., Inouye S., Inouye M., Whittam T. S. (1990). Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol 172, 6175–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton T. M., Scholes D., Hughes J. P., Winter C., Roberts P. L., Stapleton A. E., Stergachis A., Stamm W. E. (1996). A prospective study of risk factors for symptomatic urinary tract infection in young women. N Engl J Med 335, 468–474. 10.1056/NEJM199608153350703 [DOI] [PubMed] [Google Scholar]
- Huang S. H., Wass C., Fu Q., Prasadarao N. V., Stins M., Kim K. S. (1995). Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun 63, 4470–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. A., Gill R. E., Hsu P., Minshew B. H., Falkow S. (1981). Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun 33, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren S. J., Schwan W. R., Schaeffer A. J., Duncan J. L. (1986). Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun 54, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Stell A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181, 261–272. 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Roberts P. L., Stamm W. E. (1987). P fimbriae and other virulence factors in Escherichia coli urosepsis: association with patients’ characteristics. J Infect Dis 156, 225–229. 10.1093/infdis/156.1.225 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Moseley S. L., Roberts P. L., Stamm W. E. (1988). Aerobactin and other virulence factor genes among strains of Escherichia coli causing urosepsis: association with patient characteristics. Infect Immun 56, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Lyons M. F., II, Pearce W., Gorman P., Roberts P. L., White N., Brust P., Olsen R., Gnann J. W., Jr, Stamm W. E. (1991). Therapy for women hospitalized with acute pyelonephritis: a randomized trial of ampicillin versus trimethoprim-sulfamethoxazole for 14 days. J Infect Dis 163, 325–330. 10.1093/infdis/163.2.325 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Delavari P., Kuskowski M., Stell A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183, 78–88. 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- Kelly A., Conway C., O Cróinín T., Smith S. G., Dorman C. J. (2006). DNA supercoiling and the Lrp protein determine the directionality of fim switch DNA inversion in Escherichia coli K-12. J Bacteriol 188, 5356–5363. 10.1128/JB.00344-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P., Bower L., Morris L., Horne A., Petryszak R., Kanz C., Kanapin A., Das U., Michoud K. & other authors (2005). Integr8 and genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res 33 (Database issue), D297–D302. 10.1093/nar/gki039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. (1986). Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J 5, 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laupland K. B., Zygun D. A., Davies H. D., Church D. L., Louie T. J., Doig C. J. (2002). Incidence and risk factors for acquiring nosocomial urinary tract infection in the critically ill. J Crit Care 17, 50–57. 10.1053/jcrc.2002.33029 [DOI] [PubMed] [Google Scholar]
- Le Bouguenec C., Archambaud M., Labigne A. (1992). Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 30, 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointre G., Rachdi L., Darlu P., Denamur E. (1998). Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol 15, 1685–1695. 10.1093/oxfordjournals.molbev.a025895 [DOI] [PubMed] [Google Scholar]
- Lloyd A. L., Rasko D. A., Mobley H. L. (2007). Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol 189, 3532–3546. 10.1128/JB.01744-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain M. S., Blomfield I. C., Eisenstein B. I. (1991). Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol 173, 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L., Green D. M., Trifillis A. L., Johnson D. E., Chippendale G. R., Lockatell C. V., Jones B. D., Warren J. W. (1990). Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58, 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. A., Lopez-Boado Y. S., Wilson C. L., Roth R., Parks W. C., Heuser J., Hultgren S. J. (1998). Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282, 1494–1497. 10.1126/science.282.5393.1494 [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. (1984). Standard reference strains of Escherichia coli from natural populations. J Bacteriol 157, 690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørskov F., Ørskov I. (1992). Escherichia coli serotyping and disease in man and animals. Can J Microbiol 38, 699–704. 10.1139/m92-115 [DOI] [PubMed] [Google Scholar]
- Palaszynski S., Pinkner J., Leath S., Barren P., Auguste C. G., Burlein J., Hultgren S., Langermann S. (1998). Systemic immunization with conserved pilus-associated adhesins protects against mucosal infections. Dev Biol Stand 92, 117–122. [PubMed] [Google Scholar]
- Picard B., Garcia J. S., Gouriou S., Duriez P., Brahimi N., Bingen E., Elion J., Denamur E. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67, 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L. W., Remis R. S., Helgerson S. D., McGee H. B., Wells J. G., Davis B. R., Hebert R. J., Olcott E. S., Johnson L. M. & other authors (1983). Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308, 681–685. 10.1056/NEJM198303243081203 [DOI] [PubMed] [Google Scholar]
- Rushton H. G. (1997). Urinary tract infections in children. Epidemiology, evaluation, and management. Pediatr Clin North Am 44, 1133–1169. 10.1016/S0031-3955(05)70551-4 [DOI] [PubMed] [Google Scholar]
- Russo T. A., Johnson J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5, 449–456. 10.1016/S1286-4579(03)00049-2 [DOI] [PubMed] [Google Scholar]
- Stapleton A., Moseley S., Stamm W. E. (1991). Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J Infect Dis 163, 773–779. 10.1093/infdis/163.4.773 [DOI] [PubMed] [Google Scholar]
- Svanborg Edén C., Hull R., Falkow S., Leffler H. (1983). Target cell specificity of wild-type E. coli and mutants and clones with genetically defined adhesins. Prog Food Nutr Sci 7, 75–89. [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullus K., Hörlin K., Svenson S. B., Källenius G. (1984). Epidemic outbreaks of acute pyelonephritis caused by nosocomial spread of P fimbriated Escherichia coli in children. J Infect Dis 150, 728–736. 10.1093/infdis/150.5.728 [DOI] [PubMed] [Google Scholar]
- Wright K. J., Seed P. C., Hultgren S. J. (2007). Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol 9, 2230–2241. 10.1111/j.1462-5822.2007.00952.x [DOI] [PubMed] [Google Scholar]
- Xie Y., Yao Y., Kolisnychenko V., Teng C. H., Kim K. S. (2006). HbiF regulates type 1 fimbriation independently of FimB and FimE. Infect Immun 74, 4039–4047. 10.1128/IAI.02058-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Terai A., Yuri K., Kurazono H., Takeda Y., Yoshida O. (1995). Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12, 85–90. 10.1111/j.1574-695X.1995.tb00179.x [DOI] [PubMed] [Google Scholar]