Abstract
Coxsackieviruses (CV) A1, CV-A19 and CV-A22 have historically comprised a distinct phylogenetic clade within Enterovirus (EV) C. Several novel serotypes that are genetically similar to these three viruses have been recently discovered and characterized. Here, we report the coding sequence analysis of two genotypes of a previously uncharacterized serotype EV-C113 from Bangladesh and demonstrate that it is most similar to CV-A22 and EV-C116 within the capsid region. We sequenced novel genotypes of CV-A1, CV-A19 and CV-A22 from Bangladesh and observed a high rate of recombination within this group. We also report genomic analysis of the rarely reported EV-C104 circulating in the Gambia in 2009. All available EV-C104 sequences displayed a high degree of similarity within the structural genes but formed two clusters within the non-structural genes. One cluster included the recently reported EV-C117, suggesting an ancestral recombination between these two serotypes. Phylogenetic analysis of all available complete genome sequences indicated the existence of two subgroups within this distinct Enterovirus C clade: one has been exclusively recovered from gastrointestinal samples, while the other cluster has been implicated in respiratory disease.
Introduction
The genus Enterovirus consists of 12 species, designated Enterovirus (EV) A–J, and Rhinovirus (HRV) A–C (http://www.picornaviridae.com). Of these, only EV-A, -B, -C, -D and HRV-A, -B, -C are associated with human infection and disease. Typically, an EV genome consists of a 7.3–7.5 kb-long, positive-sense, single-stranded RNA molecule with a single open reading frame (ORF). The ORF encodes a polyprotein that is organized into three regions, designated P1, P2 and P3. The P1 encodes VP4, VP2, VP3 and VP1 proteins that comprise the viral capsid. The P2 and P3 encode non-structural proteins involved in enzymic activity and processing (2A, 2B, 2C and 3A, 3B, 3C, 3D). Historically, serology was used to differentiate EV serotypes; more recently molecular methods are used with novel serotypes possessing <75 % nucleotide (nt) and <85 % amino acid (aa) identity within the VP1 relative to known EVs (Oberste et al., 1999a, b; Brown et al., 2009; Knowles et al., 2012). Over 120 EV serotypes have been reported, of which 23 are classified within the EV-C (http://www.picornaviridae.com). Phylogenetic analysis of early EV-C prototype strains revealed that it consists of two distinct subgroups; one includes Polio 1–3, coxsackieviruses (CV) A11, A13, A17, A20, A21 and A24, and EV-C95, 96, 99 and 102; the other includes CV-A1, CV-A19 and CV-22 (Brown et al., 2003). The prototype strains of CV-A1, A19 and A22 are distinct from the former subgroup: they are monophyletic in P1–P3, and show little evidence for recombination with other EV-Cs. In addition, unlike most EVs that can be cultured in a wide variety of standard cell lines, the prototype strains of CV-A1, A19 and A22 have not been grown in cell culture (Schmidt et al., 1975; Lipson et al., 1988).
Although the prototype strains of CV-A1 (strain Tomkins, isolated in 1947), CV-A19 (strain 8663, isolated in 1952) and CV-A22 (strain Chulman, isolated in 1955) were reported in the 1950s (Brown et al., 2003; Knowles et al., 2012), these serotypes are only rarely identified during EV surveillance, which has classically depended on growing the virus in cell culture (Khetsuriani et al., 2006). Although they have been implicated in enteric and neurological disease, little data exists on the genetic diversity, prevalence and spectrum of disease associated with infection (Begier et al., 2008; Kapusinszky et al., 2010; Chitambar et al., 2012; Tapparel et al., 2013). With more widespread implementation of molecular diagnostics, novel strains of these viruses have been discovered (Witsø et al., 2006; Benschop et al., 2010; Oberste et al., 2013). Nonetheless, phylogenetic and genetic information remains limited. Full-length genome sequences are available for only four CV-A22, three CV-A1 and the prototype CV-A19 virus.
Recently, several novel serotypes genetically similar to CV-A1, CV-A19 and CV-A22 have been discovered using molecular methods. The first was EV-C104, discovered in Switzerland in patients with respiratory disease; thereafter, EV-C109 was identified in respiratory disease samples from Nicaragua (Tapparel et al., 2009; Yozwiak et al., 2010). In 2012, four additional serotypes (EV-C105, EV-C116, EV-C117, EV-C118) were reported and characterized; none were grown in cell culture (Daleno et al., 2012, 2013; Lukashev et al., 2012; Tokarz et al., 2013). Analysis of phylogenetic and replication properties, including these novel serotypes, indicated three distinct groups recognized within EV-C (Lukashev et al., 2012). Each group makes up a distinct phylogenetic cluster based on the P1 sequence. These groups also differ in 5′ untranslated region (UTR), growth in cell culture, and maintain reproductive isolation indicated by rare recombination across other groups. The novel serotypes EV-C104, C105, C109, C117 and C118 represent group III, with an unconventional 5′ UTR. Group II consists of CV-A1, CV-A19 and CV-A22, as well as one novel serotype EV-C116 and are non-cytopathogenic in RD cells. Group I is composed of the remaining serotypes, with a conventional 5′ UTR and cytopathogenicity in RD cell culture.
In surveys of samples from human subjects with respiratory and enteric disease, we detected and identified EV-C serotypes from groups II and III. In an effort to shed light on genetic relationships of these viruses, we sequenced genomes of several representatives of these groups. In this report, we analyse genotypes of a new member of group II, EV-C113, and novel genotypes of CV-A1, CV-A19 and CV-A22; all were detected in stool samples from Bangladesh. We also describe strains of the rarely reported EV-C104 circulating in the Gambia in 2009 and analyse the potential for recombination and reproductive isolation within both viral groups.
Results
Characterization of EV-C113 as a member of group II of EV-C
Of 149 stool samples from Bangladesh, 52 were positive for at least one EV; four samples represented mixed infections. An EV-C virus was identified in 19 samples, of which 13 belonged to group II and none to group III. Viruses identified in three samples exhibited low nucleotide (nt) homology (<80 %) to CV-A1, CV-A19 and CV-A22 within the VP4/2 region, suggesting they may represent a unique EV serotype. Amplification and sequence analysis of the VP1 gene indicated that these viruses were approximately 90 % identical to 324 nt VP1 sequences obtained from stool samples from Bolivia (GenBank accession number JX219564) that included a virus designated EV-C113 (http://www.picornaviridae.com). The viruses closest to EV-C113 were EV-C116 and CV-A22, and placed this uncharacterized serotype along with CV-A1 and CV-A19 within group II of EV-C. To compare the complete phylogenetic relationship of EV-C113 to other viruses within this clade, we obtained the full-length coding sequence of EV-C113 viruses from two samples, designated BBD48 and BBD83 (GenBank accession numbers KC344833 and KC344834) (Table 1). The third EV-C113-positive sample contained the same genotype as BBD48 (99 % nt identity within the VP4/2) and was not analysed further. BBD48 and BBD83, obtained from samples in 2006 and 2009, respectively, represented two genotypes of EV-C113; the 6618 nt ORFs of the two viruses were 91.7 % identical to each other (99.1 % aa identity). Within the P1 region, the two viruses had 93.3 % nt identity (99.6 aa identity). The complete 888 nt of VP1 sequence of EV-C113 BBD83 was 64 % similar on the nt level and 81 % similar on the aa level relative to the nearest serotype, CV-A22 ban99-10427, also from Bangladesh (DQ995647).
Table 1. Group II EV-C viruses analysed in this study.
| Sample | Serotype | Origin | Date of collection | Sample type |
| BBD48 | EV-C113 | Bangladesh | July 2006 | Stool |
| BBD83 | EV-C113 | Bangladesh | November 2009 | Stool |
| BBD01 | CV-A22 | Bangladesh | January 2009 | Stool |
| BBD58 | CV-A22 | Bangladesh | June 2009 | Stool |
| BBD34 | CV-A1 | Bangladesh | April 2009 | Stool |
| BBD26 | CV-A19 | Bangladesh | March 2009 | Stool |
| BBD66 | CV-A19 | Bangladesh | October 2006 | Stool |
The stool sample with EV-C113 BBD66 also contained a CV-A1 virus. We detected CV-A1 in three other samples: one of these contained an EV-B virus. CV-A22 was present in two samples: one of them also contained an EV-B virus. Two distinct genotypes of CV-A19 were detected amongst four samples. We sequenced the complete coding sequence of both genotypes, designated strains BBD26 and BBD66. Complete coding sequence was also obtained for CV-A1 (one sample: BBD34) and CV-A22 viruses (two samples: BBD1 and BBD58) (Table 1).
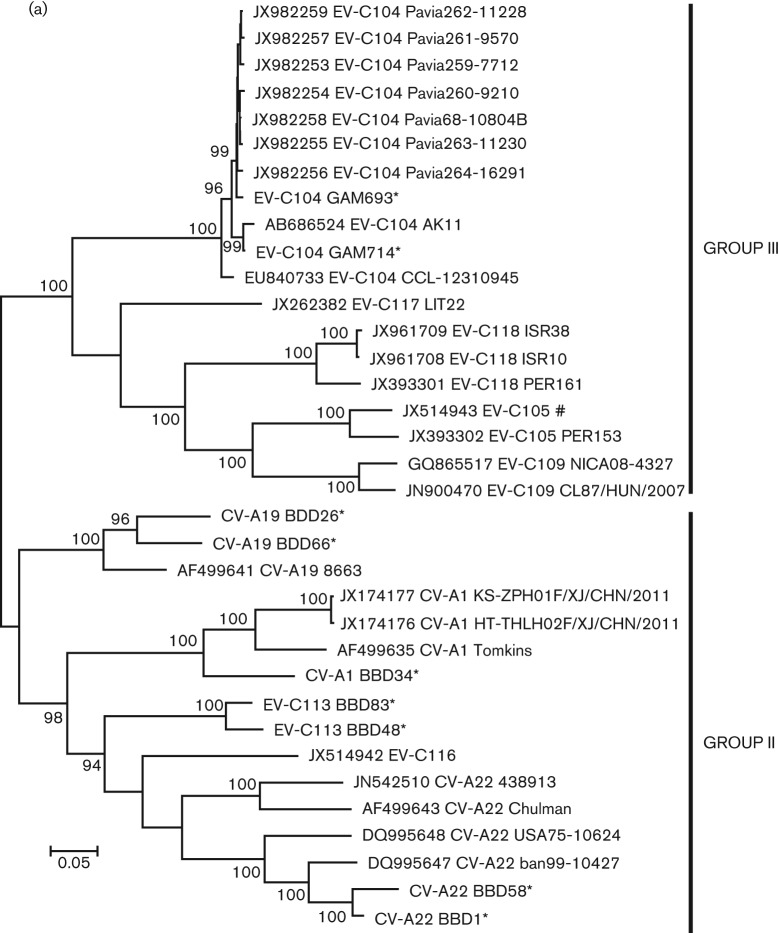
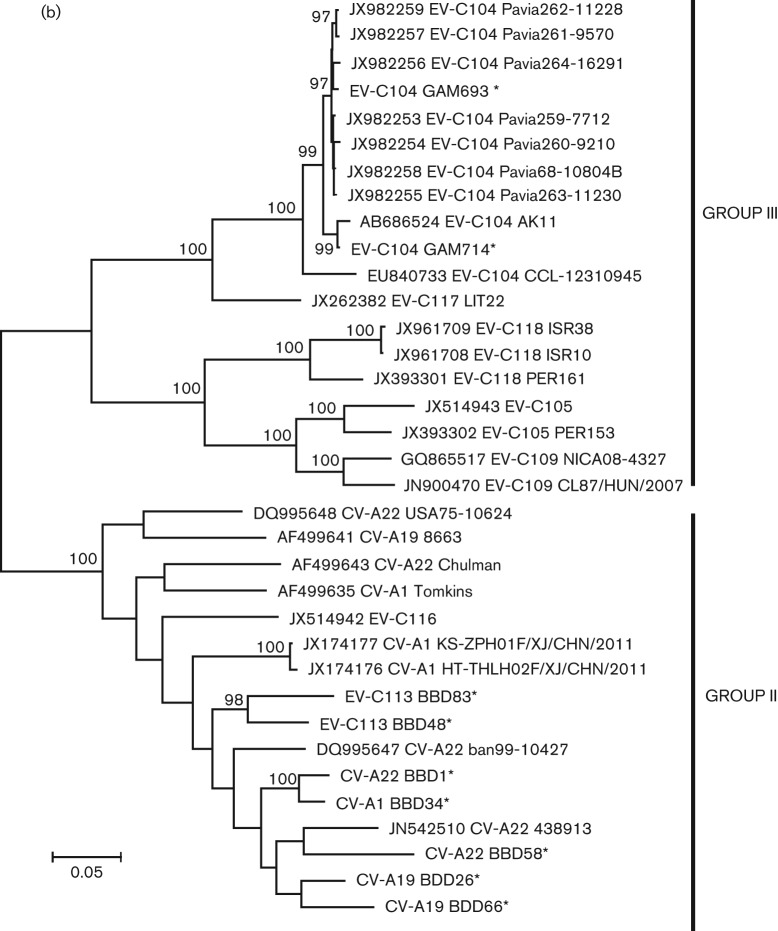
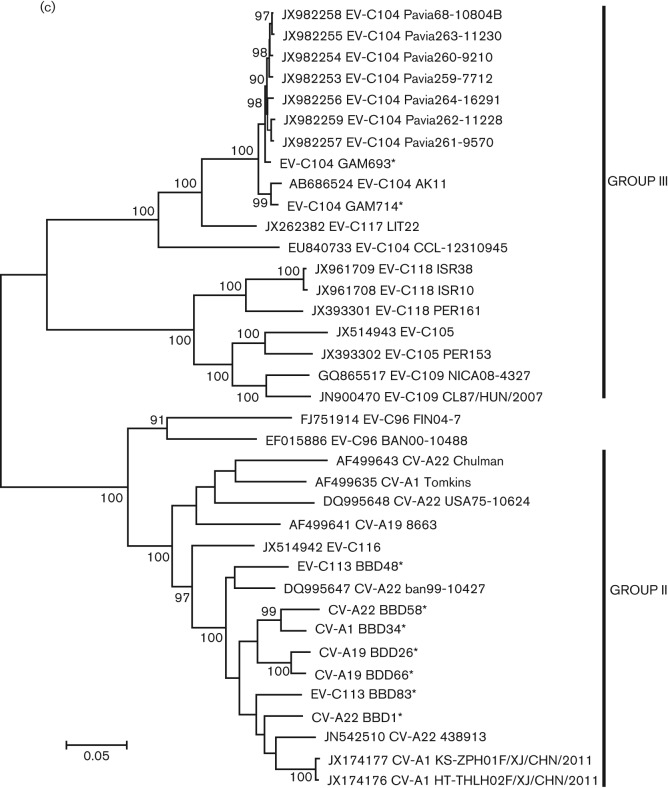
Within the P1 region, CV-A19 viruses formed a distinct phylogenetic branch from the other four serotypes of group II and exhibited the highest degree of aa conservation in relation to their prototype strain (Fig. 1a). Throughout the P1 region, the prototype strain had 82.4 % nt identity (98.1 % aa identity) to BBD26 and 83.4 % nt identity (99.0 % aa) to BBD66; BBD26 and BBD66 had 85.1 % nt identity (98.3 % aa identity) to each other. CV-A1 BBD34 had <85 % nt identity (<93 % aa identity) to the three other CV-A1 genomes. CV-A22 BBD01 and CV-A22 BBD58 were both obtained from samples collected in 2009 and had 94.1 % nt identity (96.5 % aa identity) to each other. Both were most similar to the CV-A22 detected in 1999 in Bangladesh. BBD01 was nearly identical on the aa level (99.1 %); BBD58 had 96.2 % aa identity. Overall, among the six full genomes of CV-A22, the P1 aa identity among strains ranged from 99.1 % to 86.2 %.
Fig. 1.
Maximum-likelihood phylogenetic tree constructed from all available complete P1 (a) P2 (b) and P3 (c) sequences of group II (non-respiratory tropic) and group III (respiratory tropic) viruses. For each virus, the accession number, serotype and strain name are provided. * indicates sequences identified for this study; # indicates the lone group III sequence obtained from a stool sample. Part (c) contains P3 sequences from recombinant EV-C96, which is not part of either group.
While all the new viruses analysed here clustered with their prototype strains within the P1, none did so in P2 or P3, suggesting these viruses have undergone a number of recombination events (Fig. 1b, c). Both EV-C113 strains cluster together within the P1 and P2, but BBD83 from 2009 clusters with CV-A22 BBD01 from 2009 while BBD48 from 2006 clusters with CV-A22 from 1999. In both P2 and P3, CV-A1 BBD34 clusters with CV-A22 BBD58; both CV-A1 BBD34 and CV-A22 BBD58 came from samples collected in 2009. Interestingly, unlike the other serotypes, the two new CV-A19 genotypes clustered together within both P2 and P3 regions, which suggests at least some selective reproductive isolation for this serotype. Overall, despite the apparent recombination, the P3 aa sequence of all five serotypes was highly conserved, with all viruses being within 97.9 % identical to each other.
Analysis of EV-C104 from the Gambia
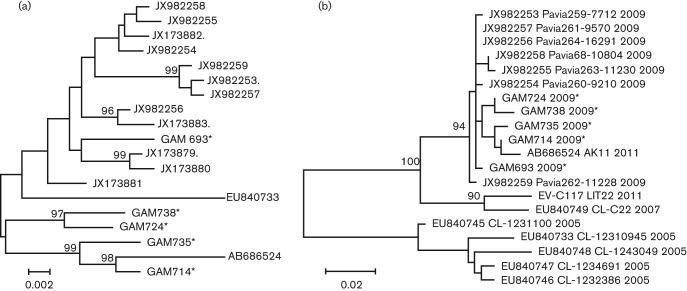
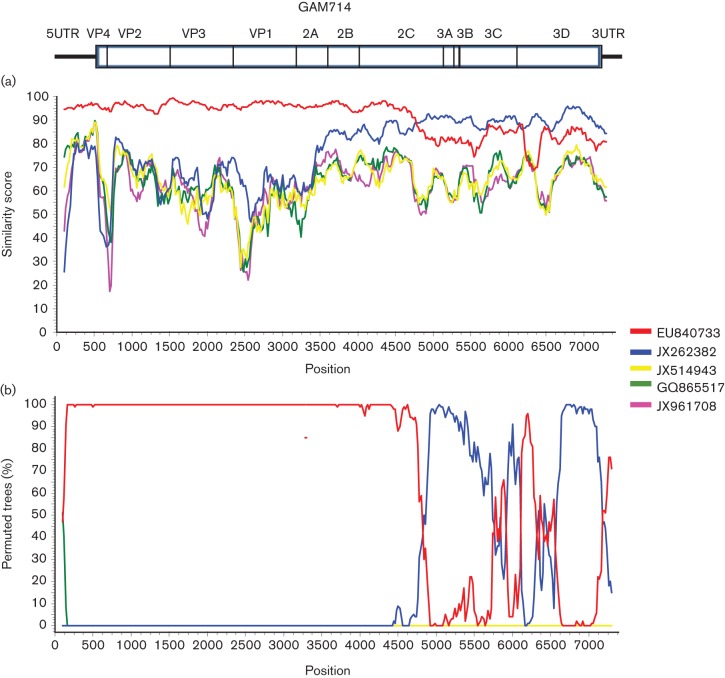
EV-C104 is a rare EV serotype; thus far, it was only reported from cases of respiratory disease in Switzerland, Italy and Japan (Tapparel et al., 2009; Kaida et al., 2012; Piralla et al., 2012). During a case-control study of paediatric respiratory disease in the Gambia, we detected EV-C104 in 5 out of 819 samples. The five samples, designated GAM693, GAM714, GAM724, GAM735 and GAM738 were obtained between 26 March and 2 June 2009, all from individuals residing within the Greater Banjul area (Table 2). Three samples came from children with pneumonia, while two EV-C104-positive samples were identified among control children recruited from the community. To more closely characterize the strains circulating in the Gambia, we sequenced the complete coding sequence of GAM693 and GAM714, as well as partial genome fragments from the remaining three samples. The complete VP1 sequences of the GAM strains had a nucleotide divergence of 3.8 % to 1.1 % (99–100 % aa identity). Analysis of all VP1 sequences currently available revealed that GAM693 clustered with strains reported from Europe between 2005 and 2009, while the remaining four viral sequences from the Gambia clustered with the strain recently described from Japan in 2011 (Fig. 2a). Analysis of the P1–P3 regions from all available EV-C104 genomes revealed high similarity within the P1, with >5 % nt divergence among all identified strains (Fig. 1a). Within the P2 and P3 regions, all the GAM strains clustered with the strains from Japan and Italy, but were significantly divergent from the prototype strain described in Switzerland (nt divergence: 6 % within the P2, 17 % within the P3) (Fig. 1b, c). Only a single complete genome is available from EV-C104 identified in Switzerland, but several 3Dpol partial sequences exist and were used for phylogenetic analysis. The analysis revealed two clusters; all EV-C104 strains from Switzerland detected in samples originating in 2005 clustered together, while a second cluster consisted of all strains recovered in 2007 and beyond (single strain from Switzerland from 2007; all Italian and Gambian strains from 2009; a single 2011 Japanese strain) (Fig. 2b). Additionally, the phylogenetic tree of P3 from all EV-C serotypes from groups II and III indicated that strains from the Gambia, Italy and Japan cluster with the recently described EV-C117 serotype rather than the prototype EV-C104 from Switzerland. This suggests that a possible recombination may have occurred between ancestral EV-C104 strains and an EV-C117-like virus. Bootscan analysis did reveal a probable recombination event between EV-C104 and EV-C117 around position 4750 within the P2 region, which mapped to an area within the 2C gene (Fig. 3).
Table 2. EV-C104-positive samples from the Gambia.
NPA, Nasopharyngeal aspirate; na, not available.
| Strain | Origin | Date of collection | Sample source | Clinical outcome | Temp. (°C) | Viral load* | Age (months) | Sex |
| GAM693 | Gambia | 26 March 2009 | NPA | Pneumonia | 36.5 | 1.7×105 | 25 | M |
| GAM714 | Gambia | 28 April 2009 | NPA | Control | na | 4.3×105 | 25 | F |
| GAM724 | Gambia | 12 May 2009 | NPA | Control | na | 3.6×103 | 7 | F |
| GAM735 | Gambia | 1 June 2009 | NPA | Pneumonia | 38.8 | 8.3×103 | 19 | M |
| GAM738‡ | Gambia | 2 June 2009 | NPA | Pneumonia | 40.0 | na | 15 | F |
Represents copy number/100 μl of sample.
Sample also contained rhinovirus C.
Fig. 2.
Analysis of EV-C104 from the Gambia. (a) Maximum-likelihood phylogenetic tree constructed on the complete VP1 sequence. (b) Maximum-likelihood phylogenetic tree constructed on a 398 bp fragment of all available sequences within the 3Dpol gene. For the 3Dpol tree, the year of collection is provided next to the accession numbers, serotype and strain designation.
Fig. 3.
Similarity plot (a) and bootscan plot (b) of EV-C104 GAM714 compared to other group III viruses.
Analysis of complete genomes indicated that viruses in group III form two distinct clusters within both P2 and P3 regions (Fig. 1b, c). EV-C104 and EV-C117 form one cluster, while EV-C105, EV-C109, EV-C118 form the second, more divergent cluster. In P3, the viruses in each cluster diverge by >30 % nt and >13 % aa from viruses in the other cluster.
Incidence and tropism of viruses in groups II and III of EV-C
Analysis of the literature and available sequence data reveals an overall low incidence of group III viruses. EV-C104 was the lone group III virus detected in the samples from the Gambia. We also did not detect any group III viruses in 1003 nasopharyngeal aspirate (NPA) samples from South Africa obtained from 2000–2001 or from 940 NPA samples from New York City in 2009–2010 (Tokarz et al., 2011, 2012). Only single strains of EV-C105 and EV-C118 were detected within 163 respiratory samples from Peru (Tokarz et al., 2013). We did not detect group II viruses in any of these sample sets. In addition, group II viruses have not been identified in any prior surveys of respiratory disease where molecular EV typing was performed and have not been linked with respiratory disease (Piralla et al., 2012; Xiang et al., 2012; Tapparel et al., 2013). Analysis of sample origin for all VP1 sequences from group II and III of EV-C (deposited in GenBank as of 15 January 2013), indicates that every sequence for a group II virus was obtained from stool samples and none from the respiratory tract (Table S1, available in JGV Online). Clinical status varied widely. Some subjects were asymptomatic whereas others had GI disease or flaccid paralysis. Conversely, viruses from group III have predominantly been detected in nasal swabs or NPAs from subjects who were asymptomatic or had respiratory disease. The lone exception is one EV-C105 strain detected in stools of a patient with flaccid paralysis from the Democratic Republic of Congo (Lukashev et al., 2012).
Discussion
CV-A1, CV-A19 and CV-A22 have historically comprised a distinct clade within EV-C based on their phylogeny, lack of evidence for recombination, and inability to grow in tissue culture (Brown et al., 2003). With the characterization of EV-C113, ten serotypes are now recognized within this group. Initially, Lukashev et al. (2012) divided these viruses into two subgroups based on cytopathic effect (or lack thereof), reproductive isolation, and the presence of a 5′ UTR sequence distinct with respect to other EV-C viruses. Our results confirm this analysis and indicate that the two subgroups also differ in tropism and clinical outcome. Group II viruses, which typically replicate in the gastrointestinal tract, are most frequently implicated with gastroenteritis and herpangina; CV-A19 and CV-A22 have also been linked to aseptic meningitis (Tapparel et al., 2013). In contrast, group III viruses are more commonly associated with respiratory disease and all but a single virus have been detected in nasal or nasopharyngeal samples. Both groups have rarely been reported due to the fact that EV surveillance historically relied on viruses being isolated in culture. Ideally, surveillance and molecular typing of viruses in both respiratory and stool samples from the same geographical area would determine the prevalence and confirm the tropism of these viruses.
Differential tropism among viruses within the same EV species has been documented. Within EV-D, enterovirus 68 (EV-D68) has been associated with respiratory disease, while enterovirus 70 is primarily linked to acute haemorrhagic conjunctivitis (Schieble et al., 1967; Mirkovic et al., 1973). A number of factors can contribute to differential tropisms amongst EVs. One of the main determinants is the receptor or co-receptor used for cellular entry. Several EV-C serotypes, such as CV-A21, use intercellular adhesion molecule 1 (ICAM-1) as a cellular receptor and are mainly associated with mild respiratory disease (Xiao et al., 2001). ICAM-1 is also utilized by major group rhinoviruses, which are predominantly found in the respiratory tract and associated with respiratory disease (Greve et al., 1989). Utilization of different receptors may also have a role in tropism of group II and III viruses.
Another explanation for the paucity of group III viruses in gastrointestinal samples might be acid lability as is the case for rhinoviruses and EV-D68. Although acid sensitivity would restrict group III viruses to the respiratory tract, a single representative of this group has been detected in a stool sample (Lukashev et al., 2012). This is not unusual, however, as rhinoviruses can be detected in stool samples (Harvala et al., 2012; Oberste et al., 2013). In the stool samples from Bangladesh, we identified seven samples containing rhinoviruses. We cannot experimentally test the acid lability hypothesis because group II and III viruses have historically not been propagated inculture. The recent discovery of CV-A1 strains capable of growth in RD cell lines, marks a potential breakthrough in studying group II and III viruses, which would be an important first step for elucidating components of the life cycle of these viruses (Sun et al., 2012).
Beginning with EV-C104 in 2009, five novel serotypes of group III have been reported at the time of writing. While clearly displaying an association with respiratory disease, in most cases these viruses were identified not in large clusters but in rare individual cases within a large cohort. Rarity of detection, in combination with difficulties in propagating these viruses in culture, limits our ability to investigate their biology and genetic diversity. From a phylogenetic and clinical perspective, EV-C104 is the best characterized virus in group III. In addition to the three cases described in this report, only 16 other probable cases have been documented. EV-C104 was found in eight patients in Switzerland with otitis media or pneumonia and in seven patients in Italy with upper respiratory tract (four cases, two immumocompromised), or with lower respiratory tract (three cases, two immunocompromised) disease (Tapparel et al., 2009; Piralla et al., 2012). A single case of upper respiratory tract infection was reported from Japan (Kaida et al., 2012). Based on available clinical data, EV-C104 infection ranges from asymptomatic to pneumonia, although the majority of cases appear to result in mild respiratory disease. All three symptomatic cases from the Gambia presented with pneumonia; however, we cannot rule out that EV-C104 may have represented a secondary infection, as we did not observe a difference in viral load between clinical cases and controls. No additional viral agents were found in two of the three cases; nonetheless, a primary or secondary bacterial infection cannot be excluded.
Genetic diversity in enteroviruses is generated by genetic drift and recombination. Both inter- and intratypic recombination occurs frequently (Santti et al., 1999; Brown et al., 2003; Oberste et al. 2004a, b; Simmonds & Welch, 2006; Bessaud et al., 2011; Combelas et al., 2011). The majority of recombination occurs within P2 and P3 regions, with little or no recombination within the P1. Although molecular typing using sequence divergence within VP1 is commonly used for enterovirus identification, such assays limit the phylogenetic and biological information generated by analysing complete genomes (Lukashev, 2005). Recombination can alter a virus by conferring properties of another virus that may offer selective advantages and increase fitness. One of the criteria for demarcation of the three EV-C groups was reproductive isolation, due to very rare recombination among viruses in different groups. Thus far, only a single serotype, EV-C96, had shown signs of recombination between groups I and II (Smura et al., 2007; Brown et al., 2009). It is apparent that viruses in group II undergo frequent recombination similar to viruses in group I. Recently, a CV-A22 from Hong Kong and CV-A1 from China both were shown to have undergone recombination with other group II viruses (Yip et al., 2011; Sun et al., 2012). Our results show that amongst the group II viruses from Bangladesh there were many rearrangements within the P2 and P3 indicative of recombination although it was also restricted among group II viruses with no evidence of recombination with other groups. Within viruses of group III, recombination was previously proposed but has not been conclusively shown (Lukashev et al., 2012; Tokarz et al., 2013). Analysis of the P3 region of EV-C104 indicates that this virus has undergone a rearrangement within the last decade, likely due to recombination with a EV-C117-like virus. While this is the first example of recombination among viruses in group III, we expect with the availability of more full-length genomes more frequent evidence of recombination will be observed. Such studies will allow for a more thorough phylogenetic analysis of both groups of EV-C viruses.
Methods
Stool samples (n = 149) obtained from children <5 years old with gastrointestinal disease in Bangladesh were screened for EVs using primers Fwd 5′-TCCTCCGGCCCCTGAATGCGGCTAATCC-3′ and Rev 5′-GAAACACGGWCACCAAAGTASTCG-3′. EV-positive samples were typed using PCR assays targeting the VP4/2, and 5′ UTR of EVs (Tokarz et al., 2012, 2013). The complete coding sequences of selected viruses were obtained by consensus PCR. Full genomes of group II and III viruses were aligned, and multiple consensus primers were designed (Table S2). Overlapping PCR products were generated and used to resolve full-length sequences. In cases where multiple viruses were present in the same sample, PCR products were cloned and multiple fragments sequenced to obtain specific virus sequence.
Samples with EV-C104 were obtained during the course of a case-control study of paediatric respiratory disease in the Gambia. All samples were collected between 2007 and 2009 and screened for respiratory pathogens using MassTag PCR (Briese et al., 2005). EV-positive samples were typed as above; EV-C104 sequence was amplified by consensus PCR using EV-C104 genomes AK11 and CL-1 as reference (AB686524 and EU840733).
Alignments, phylogenetic trees, and distance matrices were obtained with Mega5 software (Tamura et al., 2007). Phylogenetic trees were generated using the maximum-likelihood method with 1000 bootstrap replicates. Similarity plots and recombination analysis were performed using SimPlot 3.5 with manual bootscaning using the Kimura distance model with a 200 nt-window and a step size of 20nt.
Acknowledgements
This work was supported by grants from the National Institutes of Health AI057158 (North-east Biodefence Center-Lipkin) and the Defense Threat Reduction Agency.
Footnotes
Two supplementary tables are available with the online version of this paper.
References
- Begier E. M., Oberste M. S., Landry M. L., Brennan T., Mlynarski D., Mshar P. A., Frenette K., Rabatsky-Ehr T., Purviance K. & other authors (2008). An outbreak of concurrent echovirus 30 and coxsackievirus A1 infections associated with sea swimming among a group of travelers to Mexico. Clin Infect Dis 47, 616–623 10.1086/590562 [DOI] [PubMed] [Google Scholar]
- Benschop K., Minnaar R., Koen G., van Eijk H., Dijkman K., Westerhuis B., Molenkamp R., Wolthers K. (2010). Detection of human enterovirus and human parechovirus (HPeV) genotypes from clinical stool samples: polymerase chain reaction and direct molecular typing, culture characteristics, and serotyping. Diagn Microbiol Infect Dis 68, 166–173 10.1016/j.diagmicrobio.2010.05.016 [DOI] [PubMed] [Google Scholar]
- Bessaud M., Joffret M. L., Holmblat B., Razafindratsimandresy R., Delpeyroux F. (2011). Genetic relationship between cocirculating Human enteroviruses species C. PLoS ONE 6, e24823 10.1371/journal.pone.0024823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Palacios G., Kokoris M., Jabado O., Liu Z., Renwick N., Kapoor V., Casas I., Pozo F. & other authors (2005). Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis 11, 310–313 10.3201/eid1102.040492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B., Oberste M. S., Maher K., Pallansch M. A. (2003). Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J Virol 77, 8973–8984 10.1128/JVI.77.16.8973-8984.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. A., Maher K., Flemister M. R., Naraghi-Arani P., Uddin M., Oberste M. S., Pallansch M. A. (2009). Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J Gen Virol 90, 1713–1723 10.1099/vir.0.008540-0 [DOI] [PubMed] [Google Scholar]
- Chitambar S., Gopalkrishna V., Chhabra P., Patil P., Verma H., Lahon A., Arora R., Tatte V., Ranshing S. & other authors (2012). Diversity in the enteric viruses detected in outbreaks of gastroenteritis from Mumbai, Western India. Int J Environ Res Public Health 9, 895–915 10.3390/ijerph9030895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combelas N., Holmblat B., Joffret M. L., Colbère-Garapin F., Delpeyroux F. (2011). Recombination between poliovirus and coxsackie A viruses of species C: a model of viral genetic plasticity and emergence. Viruses 3, 1460–1484 10.3390/v3081460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleno C., Piralla A., Scala A., Baldanti F., Usonis V., Principi N., Esposito S. (2012). Complete genome sequence of a novel human enterovirus C (HEV-C117) identified in a child with community-acquired pneumonia. J Virol 86, 10888–10889 10.1128/JVI.01721-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleno C., Greenberg D., Piralla A., Scala A., Baldanti F., Principi N., Esposito S. (2013). A novel human enterovirus C (EV-C118) identified in two children hospitalised because of acute otitis media and community-acquired pneumonia in Israel. J Clin Virol 56, 159–162 10.1016/j.jcv.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve J. M., Davis G., Meyer A. M., Forte C. P., Yost S. C., Marlor C. W., Kamarck M. E., McClelland A. (1989). The major human rhinovirus receptor is ICAM-1. Cell 56, 839–847 10.1016/0092-8674(89)90688-0 [DOI] [PubMed] [Google Scholar]
- Harvala H., McIntyre C. L., McLeish N. J., Kondracka J., Palmer J., Molyneaux P., Gunson R., Bennett S., Templeton K., Simmonds P. (2012). High detection frequency and viral loads of human rhinovirus species A to C in fecal samples; diagnostic and clinical implications. J Med Virol 84, 536–542 http://www.picornaviridae.com 10.1002/jmv.23203 [DOI] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Sekiguchi J., Hase A., Iritani N. (2012). Enterovirus 104 infection in adult, Japan, 2011. Emerg Infect Dis 18, 882–883 10.3201/eid1805.111890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusinszky B., Szomor K. N., Farkas A., Takács M., Berencsi G. (2010). Detection of non-polio enteroviruses in Hungary 2000-2008 and molecular epidemiology of enterovirus 71, coxsackievirus A16, and echovirus 30. Virus Genes 40, 163–173 10.1007/s11262-009-0440-4 [DOI] [PubMed] [Google Scholar]
- Khetsuriani N., Lamonte-Fowlkes A., Oberst S., Pallansch M. A., Centers for Disease Control and Prevention (2006). Enterovirus surveillance – United States, 1970-2005. MMWR Surveill Summ 55, 1–20 [PubMed] [Google Scholar]
- Knowles N. J., Hovi T., Hyypia T., King A. M. Q., Lindberg A. M., Pallansch M. A., Palmenberg A. C., Simmonds P., et al. (2012). Picornaviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses . Edited by King A. M. J., et al. San Diego, CA: Elsevier Academic Press [Google Scholar]
- Lipson S. M., Walderman R., Costello P., Szabo K. (1988). Sensitivity of rhabdomyosarcoma and guinea pig embryo cell cultures to field isolates of difficult-to-cultivate group A coxsackieviruses. J Clin Microbiol 26, 1298–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev A. N. (2005). Role of recombination in evolution of enteroviruses. Rev Med Virol 15, 157–167 10.1002/rmv.457 [DOI] [PubMed] [Google Scholar]
- Lukashev A. N., Drexler J. F., Kotova V. O., Amjaga E. N., Reznik V. I., Gmyl A. P., Grard G., Taty Taty R., Trotsenko O. E. & other authors (2012). Novel serotypes 105 and 116 are members of distinct subgroups of human enterovirus C. J Gen Virol 93, 2357–2362 10.1099/vir.0.043216-0 [DOI] [PubMed] [Google Scholar]
- Mirkovic R. R., Kono R., Yin-Murphy M., Sohier R., Schmidt N. J., Melnick J. L. (1973). Enterovirus type 70: the etiologic agent of pandemic acute haemorrhagic conjunctivitis. Bull World Health Organ 49, 341–346 [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R., Pallansch M. A. (1999a). Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol 73, 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Kilpatrick D. R., Flemister M. R., Brown B. A., Pallansch M. A. (1999b). Typing of human enteroviruses by partial sequencing of VP1. J Clin Microbiol 37, 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Maher K., Pallansch M. A. (2004a). Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J Virol 78, 855–867 10.1128/JVI.78.2.855-867.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Peñaranda S., Maher K., Pallansch M. A. (2004b). Complete genome sequences of all members of the species Human enterovirus A. J Gen Virol 85, 1597–1607 10.1099/vir.0.79789-0 [DOI] [PubMed] [Google Scholar]
- Oberste M. S., Feeroz M. M., Maher K., Nix W. A., Engel G. A., Hasan K. M., Begum S., Oh G., Chowdhury A. H. & other authors (2013). Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J Virol 87, 558–571 10.1128/JVI.00837-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piralla A., Lilleri D., Sarasini A., Marchi A., Zecca M., Stronati M., Baldanti F., Gerna G. (2012). Human rhinovirus and human respiratory enterovirus (EV68 and EV104) infections in hospitalized patients in Italy, 2008-2009. Diagn Microbiol Infect Dis 73, 162–167 10.1016/j.diagmicrobio.2012.02.019 [DOI] [PubMed] [Google Scholar]
- Santti J., Hyypiä T., Kinnunen L., Salminen M. (1999). Evidence of recombination among enteroviruses. J Virol 73, 8741–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieble J. H., Fox V. L., Lennette E. H. (1967). A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85, 297–310 [DOI] [PubMed] [Google Scholar]
- Schmidt N. J., Ho H. H., Lennette E. H. (1975). Propagation and isolation of group A coxsackieviruses in RD cells. J Clin Microbiol 2, 183–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Welch J. (2006). Frequency and dynamics of recombination within different species of human enteroviruses. J Virol 80, 483–493 10.1128/JVI.80.1.483-493.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smura T., Blomqvist S., Paananen A., Vuorinen T., Sobotová Z., Bubovica V., Ivanova O., Hovi T., Roivainen M. (2007). Enterovirus surveillance reveals proposed new serotypes and provides new insight into enterovirus 5′-untranslated region evolution. J Gen Virol 88, 2520–2526 10.1099/vir.0.82866-0 [DOI] [PubMed] [Google Scholar]
- Sun Q., Zhang Y., Zhu S., Cui H., Tian H., Yan D., Huang G., Zhu Z., Wang D. & other authors (2012). Complete genome sequence of two coxsackievirus A1 strains that were cytotoxic to human rhabdomyosarcoma cells. J Virol 86, 10228–10229 10.1128/JVI.01567-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tapparel C., Junier T., Gerlach D., Van-Belle S., Turin L., Cordey S., Mühlemann K., Regamey N., Aubert J. D. & other authors (2009). New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis 15, 719–726 10.3201/eid1505.081286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C., Siegrist F., Petty T. J., Kaiser L. (2013). Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol 14, 282–293 10.1016/j.meegid.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Tokarz R., Kapoor V., Wu W., Lurio J., Jain K., Mostashari F., Briese T., Lipkin W. I. (2011). Longitudinal molecular microbial analysis of influenza-like illness in New York City, May 2009 through May 2010. Virol J 8, 288 10.1186/1743-422X-8-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R., Firth C., Madhi S. A., Howie S. R., Wu W. Y., Sall A. A., Haq S., Briese T., Lipkin W. I. (2012). Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol 93, 1952–1958 10.1099/vir.0.043935-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R., Hirschberg D. L., Sameroff S., Haq S., Luna G., Bennett A. J., Silva M., Leguia M., Kasper M. & other authors (2013). Genomic analysis of two novel human enterovirus C genotypes found in respiratory samples from Peru. J Gen Virol 94, 120–127 10.1099/vir.0.046250-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witsø E., Palacios G., Cinek O., Stene L. C., Grinde B., Janowitz D., Lipkin W. I., Rønningen K. S. (2006). High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J Clin Microbiol 44, 4095–4100 10.1128/JCM.00653-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., Gonzalez R., Wang Z., Ren L., Xiao Y., Li J., Li Y., Vernet G., Paranhos-Baccalà G. & other authors (2012). Coxsackievirus A21, enterovirus 68, and acute respiratory tract infection, China. Emerg Infect Dis 18, 821–824 10.3201/eid1805.111376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Bator C. M., Bowman V. D., Rieder E., He Y., Hébert B., Bella J., Baker T. S., Wimmer E. & other authors (2001). Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J Virol 75, 2444–2451 10.1128/JVI.75.5.2444-2451.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. C., Lau S. K., Woo P. C., Chan K. H., Yuen K. Y. (2011). Complete genome sequence of a coxsackievirus A22 strain in Hong Kong reveals a natural intratypic recombination event. J Virol 85, 12098–12099 10.1128/JVI.05944-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yozwiak N. L., Skewes-Cox P., Gordon A., Saborio S., Kuan G., Balmaseda A., Ganem D., Harris E., DeRisi J. L. (2010). Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J Virol 84, 9047–9058 10.1128/JVI.00698-10 [DOI] [PMC free article] [PubMed] [Google Scholar]