Abstract
Nano-sized polymersomes functionalized with peptides or proteins are being increasingly studied for targeted delivery of diagnostic and therapeutic molecules. Earlier computational studies have suggested that ellipsoidal nanoparticles, compared to spherical ones, display enhanced binding efficiency with target cells, but this has not yet been experimentally validated. We hypothesize that hydrophilic polymer chains coupled to vesicle-forming polymers would result in ellipsoidal polymersomes. In addition, ellipsoidal polymersomes modified with cell adhesion peptides bind with target cells more actively than spherical ones. We examine this hypothesis by substituting polyaspartamide with octadecyl chains and varying numbers of poly(ethylene glycol) (PEG) chains. Increasing the degree of substitution of PEG from 0.5 to 1.0 mol% drives the polymer to self-assemble into an ellipsoidal polymersome with an aspect ratio of 2.1. Further modification of these ellipsoidal polymersomes with peptides containing an Arg-Gly-Asp sequence (RGD peptides) lead to a significant increase in the rate of association and decrease in the rate of dissociation with a substrate coated with αvβ3 integrins. In addition, in a circulation-mimicking flow, the ellipsoidal polymersomes linked with RGD peptides adhere to target tissues more favorably than their spherical equivalents do. Overall, the results of this study will greatly serve to improve the efficiency of targeted delivery of a wide array of polymersomes loaded with various biomedical modalities.
Keywords: Biomedical Applications, Bionanotechnology, Polymeric Materials, Self-Assembly, Surface Plasmon Resonance
1. Introduction
The targeted delivery of biomedical modalities, including imaging contrast agents and drugs, has been extensively studied to improve the quality of diagnoses and treatments of various chronic and malignant diseases.[1] Specifically, nano-sized polymeric vesicles, often called polymersomes, have been increasingly studied for use in the targeted delivery of hydrophilic biomedical molecules, including protein drugs.[2] It is common to tune the targeting capability of polymersomes by varying the number of peptides or antibodies conjugated to the particles’ surfaces.[3] However, this approach often produces only limited improvement in the targeting capability of the polymersomes because of steric interference among molecules on the particle surface. In addition, a large number of targeting peptides or proteins on the particle surface may instead induce non-specific interactions between particles and non-target cells.[4]
Therefore, it would be advantageous to develop a strategy to improve the targeting efficiency of polymersomes with a set number of peptides or proteins. Earlier studies have demonstrated that the shape of a microparticle may be a significant factor in modulating the particle’s targeting capability.[5] For example, elliptical disks injected into a blood vessel-mimicking microchannel were reported to accumulate on target sites more favorably than spherical microparticles.[5a] Numerical analysis also suggests that ellipsoidal particles placed in a microchannel drift from the center to the wall more readily than spherical particles do.[5b] It is conceivable that the important role of microparticle shape extends to nano-sized polymersomes as well. However, this hypothesis has not been examined to date, because of our limited control over the shape of polymersomes.
We hypothesized that the chemical conjugation of hydrophilic polymers to a vesicle-forming polymer would result in polymersomes with an ellipsoidal shape due to changes in the curvature of polymeric bilayers, and furthermore, that ellipsoidal polymersomes modified with cell adhesion peptides would bind with target cells more favorably than spherical polymersomes. We examined these hypotheses by conjugating varying numbers of poly(ethylene glycol) (PEG) chains to a vesicle-forming poly(2-hydroxyethyl aspartamide) substituted with octadecyl chains (PHEA-g-C18), then monitoring the shape change of the resulting polymersome. The polyaspartamide polymersomes were further modified with oligopeptides containing an Arg-Gly-Asp (RGD) sequence, which binds with cells that overexpress integrins. The binding affinities of these PHEA-g-C18 polymersomes with controlled shapes and cell adhesion molecules were evaluated by quantifying the association and dissociation rates of the polymersomes onto target substrates using surface plasmon resonance (SPR) spectroscopy. Finally, the enhanced targeting capabilities of ellipsoidal polymersomes were assessed by quantifying the number of polymersomes adhered to a target cell layer which had been stimulated to overexpress integrins.
2. Results and Discussion
2.1. Preparation and characterization of the ellipsoidal polymersomes
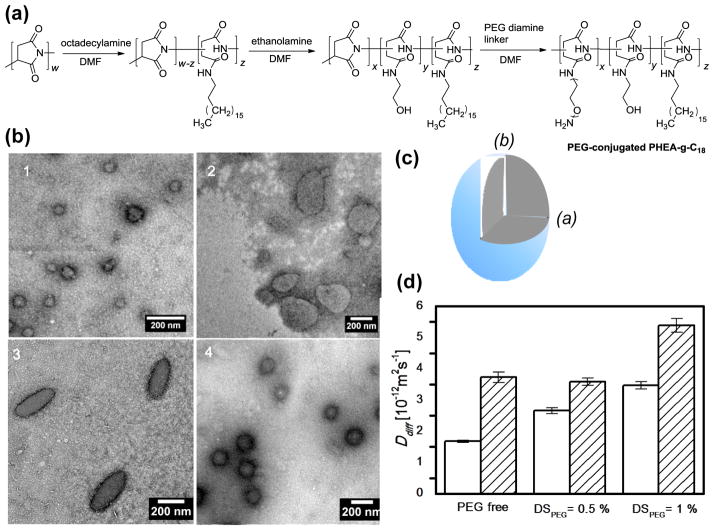
First, poly(2-hydroxyethyl aspartamide) substituted with octadecyl chains, termed PHEA-g-C18, was synthesized by modifying poly(succinimide) (PSI) in a top-down manner. PSI prepared via the acid-catalyzed polycondensation of L-aspartic acid reacted with designated amounts of octadecylamine and ethanolamine to prepare PHEA-g-C18 through nucleophilic substitution to PSI (Fig. 1a).[6] Next, the PHEA-g-C18 was chemically linked with PEG chains via additional nucleophilic substitution with PEG bis(amine) (Fig. 1a). The degree of substitution of PEG (DSPEG) to PHEA-g-C18 was varied from 0 to 0.5 to 1.0 mol% by altering the molar ratio between PEG bis(amine) and PHEA-g-C18. The chemical structure of the resulting polymers were confirmed by 1H NMR spectra (Fig. S1). The degree of substitution of octadecyl chains (DSC18) to the PHEA-g-C18 backbone, defined as a mole percentage of succinimide units substituted with octadecyl chains, was in the range of 24 to 28 mol%, according to the integrals of the characteristic NMR peaks at 0.85 to 0.95 ppm and 4.3 to 4.7 ppm (Table 1). The peak at 0.85 to 0.95 ppm represents protons of methyl groups at the ends of the substituted octadecyl chains, and the peak at 4.3 to 4.7 ppm represents protons on the polyaspartamide backbone (Fig. S1). A minimal fraction of succinimide rings in the PHEA-g-C18, as confirmed by the disappearance of the characteristic peak of succinimide rings at 5.3 ppm (Fig. S1).[7] The resulting DSPEG was quantified with chemical assays to count the number of free amine groups of PEG diamines (Table 1). Increasing DSPEG from 0.5 to 1.0 mol% doubled the critical aggregation concentration (CAC) of PEG-conjugated PHEA-g-C18, which was marked by the significant increase in the emission intensity ratio (I3/I1) of pyrene between 385 (I3) and 373 (I1) nm (Table 1 and Fig. S2). There was a minimal difference in CACs between PHEA-g-C18 dissolved in DI water and that in phosphate buffered saline (PBS) (Fig. S3).
Figure 1.
((Syntheses and characterizations of PHEA-g-C18 polymersomes. (a) The overall reaction scheme of PHEA substituted with octadecyl chains (DSC18) and varying degrees of substitution of PEG-NH2 chains (DSPEG). The DSC18, denoted as ‘z’ in the scheme, was kept constant at 24 to 28 mol%. The DSPEG, denoted as ‘x’ in the scheme, was varied from 0 to 1.0 mol%. (b) TEM micrographs of PHEA-g-C18 polymersomes with varying DSPEG of 0 (b-1), 0.5 (b-2), and 1.0 mol% (b-3). Micrographs (b-4) represents polymersomes formed from mixture of PHEA-g-C18 and free PEG chains. (c) The scheme of ellipsoidal polymersome shown in (b-3) with the polar radius (b) and the equatorial radius (a). (d) Diffusion coefficients (Ddiff) of PHEA-g-C18 polymersomes substituted with varying DSPEG in PBS (opened bar) and plasma solution (filled bar).))
Table 1.
((Molecular characterizations of PHEA-g-C18 conjugated with PEG.))
| Stoichiometric DSPEG [mol%] | DSPEG [mol%][a] | DSC18 [mol%][b] | CACs [mg/mL][c] |
|---|---|---|---|
| 0 | - | 24.0 | 3.57×10−3 |
| 0.5 | 0.54 | 28.5 | 3.73×10−3 |
| 1.0 | 1.05 | 27.3 | 6.84×10−3 |
((Determined based on TNBS assay.))
((Determined based on 1H NMR spectra of the polymer.))
((Determined based on fluorescent emission spectra of pyrene.))
Incorporation of the resulting PEG-conjugated PHEA-g-C18 with DSC18 of 24 to 28 mol% into aqueous media resulted in intermolecular self-assembly into polymersomes (Fig 1b). In contrast, the PEG-conjugated PHEA-g-C18 with DSC18 of 15 mol% formed micelles (Fig. S4).[8] Additionally, the shape of these polymersomes was found to be mediated by the DSPEG of PHEA-g-C18. The polymers with DSPEG of 0 and 0.5 mol% formed spherical polymersomes with an average radius of 57 (± 28) and 117 (± 35) nm, respectively, as imaged by transmission electron microscope (TEM) (Fig. 1b-1 & 1b-2). In contrast, the PHEA-g-C18 substituted with DSPEG of 1.0 mol% self-assembled to form ellipsoidal polymersomes with an average polar radius (b) of 109 (± 36) nm, average equatorial radius (a) of 52 (± 16) nm (Fig. 1c), and average aspect ratio of 2.1 (Fig. 1b-3). The average surface area of the ellipsoidal polymersomes was between PEG-free PHEA-g-C18 and PHEA-g-C18 with DSPEG of 0.5 mol%. In contrast, simply mixing PEG with PHEA-g-C18 only minimally altered the spherical shape of PHEA-g-C18 polymersomes (Fig. 1b-4). Polymersomes formed in PBS also exhibited the ellipsoidal shape at DSPEG of 1.0 mol% (Fig. S5). These results confirm that the number of PEG chains chemically linked to PHEA-g-C18 is a key factor in driving the sphere-to-ellipsoid transition of PHEA-g-C18 polymersomes.
This shape transition of the polymersomes driven by PEG chains is attributed to changes in the bending modulus and spontaneous curvature of the polymeric bilayer. Earlier studies have reported that hydrophilic polymer tails linked with lipid molecules increase the bending modulus of the micro-sized liposome which subsequently shifts the bilayer curvature of the liposome into an ellipsoidal shape.[9] The increased bending modulus has an even greater effect when the number of hydrophilic chains exceeds the critical value at which the hydrophilic tails contact each other. We therefore suggest that PEG chains linked to PHEA-g-C18 contact each other at a DSPEG between 0.5 and 1.0 mol% and drive the PHEA-g-C18 to self-assemble into an ellipsoidal polymersome, an effect similar to that of PEG chains on the liposome. We also propose that the critical concentration range of DSPEG for the sphere-to-ellipsoid transition will decrease with increasing molecular weights of PEG tails, which needs further investigation in future studies.
The resulting ellipsoidal PHEA polymersomes substituted with DSPEG of 1.0 mol% exhibited higher diffusivity than either spherical vesicles prepared with unmodified PHEA-g-C18 or PHEA-g-C18 substituted with DSPEG of 0.5 mol%, as characterized with the dynamic light scattering unit (Fig. 1d). The diffusion coefficients (Ddiff) of polymersomes suspended in PBS was proportional to DSPEG. In contrast, Ddiff of polymersomes suspended in a blood-mimicking plasma solution disproportionately increased as DSPEG was increased from 0.5 to 1.0 mol%. These results demonstrate that the ellipsoidal PHEA polymersomes have a higher than spherical ones in various media, likely due to their ellipsoidal shape. We suggest that the higher Ddiff of ellipsoidal polymersomes should be advantageous in their transportation through capillaries, where the effects of particle diffusion on velocity becomes significant.[10]
2.2. Modification of the ellipsoidal PHEA-g-C18 polymersomes with cell adhesion peptides
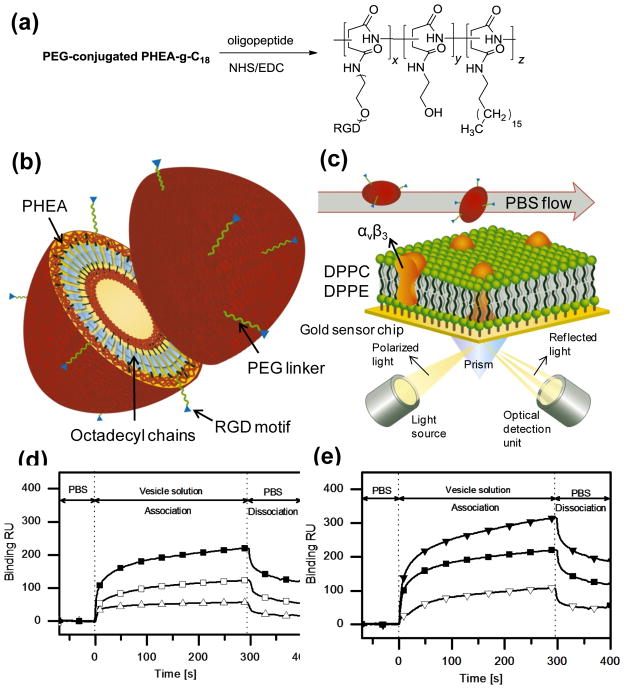
Both spherical and ellipsoidal PHEA-g-C18 polymersomes were further modified with varying numbers of oligopeptides containing an Arg-Gly-Asp sequence, termed RGD peptides, so that the modified polymersomes would adhere to a target tissue that over-expressed integrins. It is well known that several pathologic tissues, including tumor and inflammatory tissues, contain more cells that overexpress integrins, such as αvβ3, than normal tissue.[11] In addition, RGD peptides have often been used for targeted drug delivery; they are coupled to drug molecules or drug-encapsulating microparticles and injected into the circulation.[12] In this study, the RGD peptides were linked exclusively to the ends of PEG tails on the polymersome surface via a chemical reaction between the carboxyl ends of the peptides and the amine groups of the PEG tails (Fig. 2a & 2b).
Figure 2.
((Characterizations of spherical/ellipsoidal polymersomes modified with RGD peptides. (a) The reaction scheme to chemically link RGD peptides to PEG of PHEA-g-C18 while varying the degrees of substitution of RGD peptides (DSRGD). (b) Schematic description of the structure of the resulting PHEA-g-C18 polymersomes modified with PEG chains and RGD peptides. (c) Schematic description of the experimental set-up for SPR analysis to evaluate binding kinetics of polymersomes. The bilayer of DPPC (dipalmitoyl phosphatidylcholine), between which the integrins αvβ3 were inserted, was built on the gold SPR chip, and the polymersomes modified with PEG and RGD peptides were added to the flow at a rate of 0.3 ml/hr. (d) Effects of the shape of polymersomes controlled with DSPEG on the binding kinetics of polymersomes to αvβ3 integrins.
 represents PHEA-g-C18 with DSPEG of 1 mol% and DSRGD of 0.5 mol%,
represents PHEA-g-C18 with DSPEG of 1 mol% and DSRGD of 0.5 mol%,
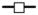 represents PHEA-g-C18 with DSPEG of 0.5 mol% and DSRGD of 0.5 mol%, and
represents PHEA-g-C18 with DSPEG of 0.5 mol% and DSRGD of 0.5 mol%, and
 represents PHEA-g-C18 with DSPEG of 0.5 mol% and DSRGD of 0 mol%. (e) Effects of DSRGD of polymersomes on the binding kinetics of polymersomes to αvβ3 integrins. DSPEG was kept constant at 1 mol% to maintain the ellipsoidal morphology of polymersomes.
represents PHEA-g-C18 with DSPEG of 0.5 mol% and DSRGD of 0 mol%. (e) Effects of DSRGD of polymersomes on the binding kinetics of polymersomes to αvβ3 integrins. DSPEG was kept constant at 1 mol% to maintain the ellipsoidal morphology of polymersomes.
 represents PHEA-g-C18 with DSRGD of 1.0 mol%,
represents PHEA-g-C18 with DSRGD of 1.0 mol%,
 represents PHEA-g-C18 with DSRGD of 0.5 mol%, and
represents PHEA-g-C18 with DSRGD of 0.5 mol%, and
 represents PHEA-g-C18 with DSRGD of 0 mol%.))
represents PHEA-g-C18 with DSRGD of 0 mol%.))
The binding affinities of the resulting RGD peptide-conjugated polymersomes to a target cell were evaluated by measuring association and dissociation rates of the polymersomes with a model target cell membrane using surface plasmon resonance (SPR) spectroscopy. The target cell membrane was built by assembling a monolayer consisting of lipid molecules and αvβ3 integrins on the SPR chip (Fig. 2c). The subsequent flow of the PHEA-g-C18 polymersomes modified with PEG chains and RGD peptides increased the ka of the polymersomes with the model cell membrane, compared to the PHEA-g-C18 modified only with PEG chains (Fig. 2d & 2e, Table 2). This increase in ka became much larger as DSPEG was increased from 0.5 to 1.0 mol%, over which the polymersome shape changed from a sphere to an ellipsoid. No significant change in kd was found with increasing DSPEG from 0.5 to 1.0 mol%.
Table 2.
((SPR analysis of the association rate (ka) and the dissociation rate (kd) of PHEA-g-C18 modified with varying DSPEG and DSRGD over the model cell membrane assembled with lipid molecules and αvβ3 integrins.))
| DSPEG [mol%] | DSRGD [mol%] | Association rate ka [M−1s−1] | Dissociation rate kd [s−1] | Binding RU |
|---|---|---|---|---|
| 0.5 | 0 | 446 | 3.38×10−3 | 16 |
| 0.5 | 528 | 2.87×10−3 | 58 | |
| 1 | 0 | 205 | 5.03×10−3 | 48 |
| 0.5 | 810 | 1.19×10−3 | 123 | |
| 1 | 751 | 1.01×10−5 | 193 |
Subsequently, the binding resonance unit (RU) value was doubled by increasing DSPEG from 0.5 to 1.0 mol% at a DSRGD of 0.5 mol%. Note that change in the binding RU value corresponds to change in the mass density of the molecules on the SPR chip.[13] Therefore, this result indicates that increasing DSPEG led to an increase of the number of polymersomes adhered to the artificial cell membrane, due to the increase in ka. In addition, at a DSPEG of 1.0 mol%, an increase in DSRGD from 0.5 to 1.0 mol% resulted in a decrease in kd by two orders of magnitude, which contributed to the increase in the binding RU value.
For control experiments, the model cell membrane assembled with only lipid molecules was also exposed to polymersomes modified with both PEG and RGD peptides. This experiment displayed a decrease in ka and binding RU values as DSRGD was increased from 0 to 0.5 mol% (Table S1). These results suggest that adhesion of RGD peptide-modified polymersomes to the integrin-coated substrate results from specific binding between peptides and integrins. However, the increase in the binding RU values resulting from the increase of DSRGD from 0.5 to 1.0 mol% suggests that a large population of RGD peptides on the polymersome surface may preferentially induce the non-specific adhesion of the polymersome. These results therefore emphasize the importance in developing a strategy to fine-tune the adhesion of nanoparticles to the target site with a given number of peptides or proteins.
2.3. In vitro evaluation of the targeting capability of PHEA-g-C18 polymersomes
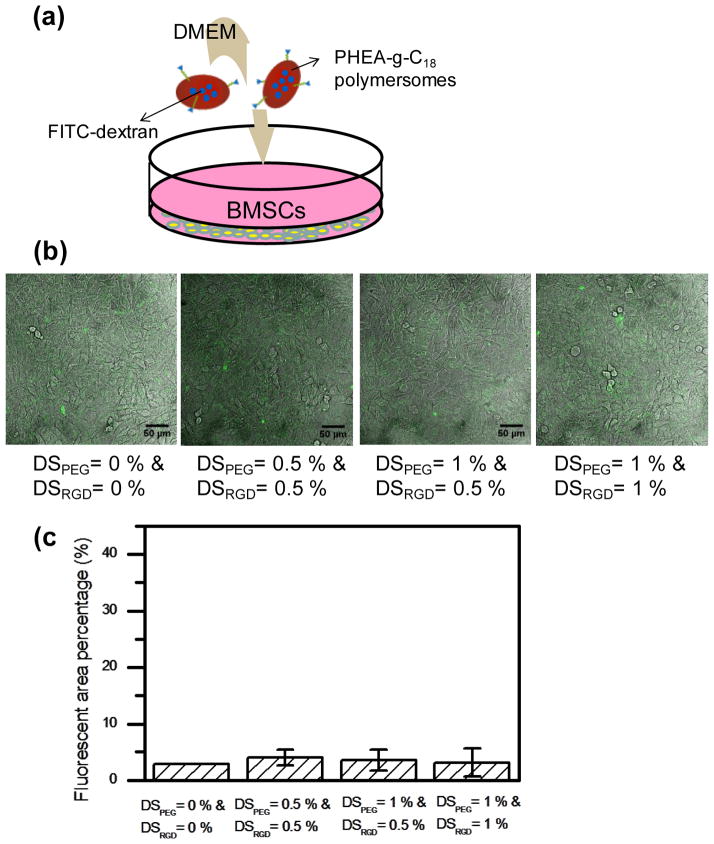
First, the targeting capability of PHEA-g-C18 polymersomes modified with both PEG and RGD peptides was evaluated using a model tissue under static conditions. The model tissue was prepared by plating bone marrow stromal cells (BMSCs) on the surface of a cell culture well at confluency (Fig. 3a). These cells presented minimal integrin expression on their exposed surfaces, which was characterized by immunostaining for αvβ3 integrins.[14] Furthermore, to identify the polymersome adherent to BMSC surfaces, fluorescein-conjugated dextrans (FITC-dextran) was loaded into PHEA polymersomes via in situ encapsulation during polymersome formation. The polymersomes minimally adhered to the model tissue, as confirmed by minimal fluorescence from the cells (Fig. 3b). The number of polymersomes adherent to the model tissue was not dependent on either DSPEG or DSRGD.
Figure 3.
((In vitro analysis of the binding affinity of polymersomes to the model target tissue in static condition. (a) Schematic description of the in vitro static experimental set-up. The bone marrow stromal cells (BMSCs) were plated at confluency on the petri-dish, and PHEA-g-C18 polymersomes with varying DSPEG and DSRGD were added into the DMEM. The FITC-dextran was encapsulated into polymersomes. (b) Confocal microphotographs of BMSCs incubated for 10 minutes with DMEM containing PHEA-g-C18 polymersomes. (c) Effects of the DSPEG and DSRGD on the number of polymersomes bound to target BMSCs, in the absence of flow. Each value and error bar in the plot represents the mean and standard deviation from three independent experiments.))
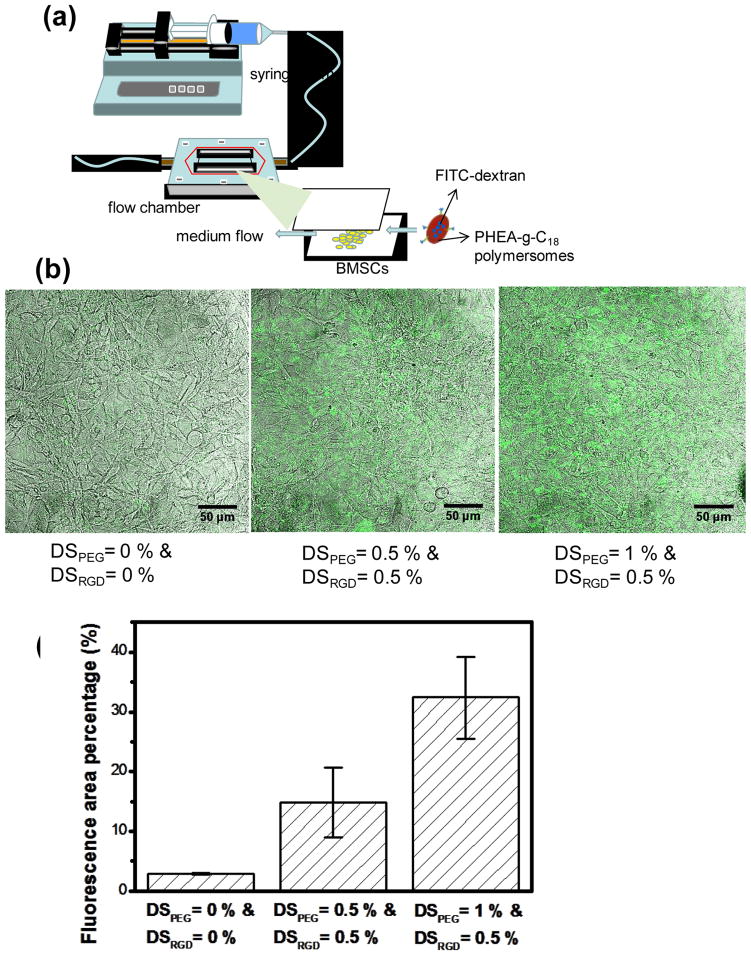
Next, the model tissue was exposed to shear flow in a flow chamber at a rate of 200 ml/hr in order to elevate cellular αvβ3 integrin expression on the cell surface exposed to the flow (Fig. 4a).[15] PHEA-g-C18 polymersomes encapsulated with FITC-dextran were injected into the flow chamber to evaluate the difference in adhesions among polymersomes with various densities of PEG and RGD. The polymersomes modified with PEG and RGD peptides, each at a DS of 0.5 mol%, adhered to the tissue more actively than the polymersomes modified only with PEG chains, as characterized by an increase in the fluorescence from the cells (Fig. 4b & 4c). In addition, at a given DSRGD of 0.5 mol%, an increase in DSPEG from 0.5 to 1.0 mol% resulted in a two-fold increase in the fluorescence from the cells (Fig. 4c).
Figure 4.
((In vitro analysis of the binding affinity of polymersomes to the model target tissue in a flow chamber. (a) The experimental set-up of a flow chamber designed to evaluate binding affinity of PHEA-g-C18 polymersomes to BMSCs. The polymersomes encapsulated with FITC-dextran were added to DMEM that flowed at a rate of 200 ml/hr. (b) Confocal microphotographs of BMSCs exposed in the flow chamber for 10 minutes to fluorescent PHEA polymersomes modified with varying DSPEG and DSRGD. (c) Quantitative analysis of the effects of DSPEG and DSRGD on the number of PHEA-g-C18 polymersomes adhered to BMSCs. The differences of the fluoresce area percentages between any two conditions were statistically significant (p* < 0.05; p** < 0.05). Each value and error bar in the plot represents the mean and standard deviation from three independent experiments.))
Overall, this study is the first to demonstrate that ellipsoidal polymersomes can be prepared by chemically linking hydrophilic PEG chains to self-associating PHEA-g-C18. This result is highly distinctive from previous studies that largely focused on controlling micelle formation using PHEA substituted with hydrophobic alkyl chains.[16] Additionally, SPR and in vitro results clearly demonstrate that the shape of nano-sized polymersomes is one of the key factors in controlling their binding affinity to target cells. We propose that the enhanced binding affinity of ellipsoidal polymersomes should be attributed to increases in the lateral drifting rate and adhesion probability when compared with spherical vesicles. Several computational studies have previously suggested that the lateral drifting velocity of non-spherical particles is proportional to their aspect ratio.[17] In addition, it has been suggested that ellipsoidal particles have a higher probability of adhesion than spherical particles, because the longer axis of the particles can align with the substrate and subsequently form a greater number of receptor-ligand bonds.[18]
Recently, a few studies using many model particles, including rod-shaped gold nanoparticles and polystyrene microparticles, have experimentally demonstrated the importance of a particle’s morphology in tuning its binding affinity to target substrates.[19] However, our study is the first to demonstrate that the ellipsoidal particle shape is also important in improving the targeting capability of the polymersome, which can be used as a carrier of hydrophilic biomedical molecules in several clinical applications. The enhanced targeting capability of cell-adherent ellipsoidal polymersomes needs to be further examined in vivo; we suggest that the polymersomes developed in this study have a strong potential to improve the quality of targeted delivery of various imaging contrast agents and therapeutic molecules.
3. Conclusions
This study demonstrates a new method to fabricate ellipsoidal polymersomes and subsequently enhance the targeting capability of surface-functionalized polymersomes. Chemically conjugating hydrophilic PEG chains to PHEA-g-C18 polymersomes can drive the sphere-to-ellipsoid transition of the polymersomes, likely due to increases in the bending modulus and spontaneous curvature of the polymeric bilayer. Furthermore, compared to spherical polymersomes, ellipsoidal polymersomes functionalized with RGD peptides demonstrated enhanced adhesion to a model target tissue in a circulation-mimicking flow. This enhanced targeting capability is attributed to the larger association rate with a target tissue, according to analysis conducted with SPR. We propose that the resulting ellipsoidal cell-adherent polymersomes will be broadly useful in improving the efficiency of targeted delivery of a wide variety of imaging contrast agents and therapeutic molecules, and ultimately in the quality of clinical diagnoses and treatments. In addition, we suggest that this strategy to synthesize ellipsoidal PHEA-g-C18 polymersomes can be readily used to control the morphologies of various nanoparticles formed from self-assembly between lipids, block copolymers, and graft polymers.[5b]
4. Experimental
Synthesis of PHEA-g-C18 conjugated with varying number of PEG chains (Fig. 1(a))
Poly(succinimide) (PSI, Mw 19,000 g/mol, PDI 1.5) was synthesized and purified as described in the previous literature [7]. PHEA derivates, including PHEA-g-C18 at DSPEG at 0, 0.5, and 1.0 mol%, were prepared and purified by adopting the reported procedure [20]. In short, PHEA polymers were prepared by aminolysis of PSI with designated amounts of octadecylamine, ethanolamine. and poly(ethylene glycol) bis(amine) (Mw 2000, Aldrich), sequentially. Details on the synthesis are described in supporting information.
Modification of PHEA-g-C18 polymersomes with RGD peptides
PHEA-g-C18 conjugated with varying numbers of PEG chains was dissolved in dimethyl sulfoxide (DMSO) (Fisher Scientific), and the polymer solutions were dropped in deionized water to drive intermolecular self-assembly. The resulting polymersomes were dialyzed for one day to remove the DMSO and then lyophilized. Afterward, the solid samples were dissolved in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Aldrich). Then, 1-hydroxybenzotriazole (Fluka), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (Thermo Scienti c), and peptides with a sequence of Arg-Gly-Asp-Ser (Sigma) were added to the polymer solution. After reacting for one day, the mixture was dialyzed against deionized water for one day and lyophilized. The final products were stored in the form of powder at −20 °C until characterization.
Characterization of PHEA-g-C18 structure
1H NMR spectra of PHEA-g-C18 were collected using a Varian Unity 500 MHz spectrometer. The polymers were dissolved in dimethyl sulfoxide-d6. The integrals of characteristic peaks were used to quantify the DS of octadecyl chains in each sample using Eq. (1).
| (1) |
Quantification of DSPEG in PHEA-g-C18
The number of free amine groups at the end of PEG chains conjugated to PHEA-g-C18 was counted via reactions with 2,4,6-trinitrobenzene sulfonic acid (TNBS, 5 % solution, Aldrich). In brief, PHEA-g-C18 dissolved in sodium bicarbonate buffer (0.01 M, pH 8.5) was mixed with TNBS for 15 minutes. Then the absorbance of the mixture was measured at 335 nm using a microplate reader (Synergy HT Multi-Mode Microplate Reader, Biotek Instruments). The measured absorbance was back-calculated to the number of unreacted amines using a calibration curve prepared with standard solutions of varying cysteine (Aldrich) concentrations.
Imaging of polymersomes with transmission electron microscopy (TEM)
The morphology of self-assembled PHEA-g-C18 polymersomes was observed using TEM (JEOL 2100 with LaB6 emitter). A drop of PHEA polymersome suspension containing 0.1% phosphotungstic acid (Acros) was placed on a formvar-coated grid (SPI Supplies, West Chester, PA). After drying in air, the samples were imaged at 120 kV. The geometry of polymersomes was quantified using ImageJ software. Thirty polymersomes were analyzed for each condition.
Measurement of the diffusion coefficients of polymersomes
Diffusion coefficients (Ddiff) of polymersomes were evaluated using a dynamic light scattering–based 90Plus particle size analyzer (Brookhaven Instruments Co., Holtsville, NY) equipped with a 35 mW solid-state laser. The scattered light of the sample was measured at a 90° angle to the incident beam, and the distribution of decay rates of scattered light was quantified using Eq. (2),
| (2) |
where τ is a given delay time, g(2)(τ) is the intensity of the scattered light during the interval of τ, β is a factor that depends on the experimental geometry, Γ̄ is the average decay rate, and κ represents the constants. The diffusion coefficient of polymersomes was then calculated from Γ̄ using Eq. (3),
| (3) |
where q is the magnitude of the scattering wave vector [21].
Measurement of the critical aggregation concentration (CAC) of polymersomes
Pyrenes (Acros) were suspended in the PHEA-g-C18 solution at a concentration of 10−4 mg/ml. The fluorescent spectra of the suspensions with varying PHEA-g-C18 concentrations were collected using a FluoroMax®-4 spectrometer (HORIBA Jobin Yvon). Excitation wavelength was set at 330 nm and the slit widths for excitation and emission were both fixed at 2 nm. The resulting emission between 350 and 450 nm was collected. The CAC was determined by the polymer concentration at the point where the emission intensity ratio (I3/I1) between the third vibronic peak at 385 nm (I3) and the first vibronic peak at 373 nm (I1) was significantly increased [22].
Measurements of the association/dissociation rates of the polymersomes using surface plasmon resonance (SPR)
A gold sensor chip (GE Healthcare, USA) was modified to present integrin αvβ3 in the DPPC-DPPE modeled bilayers, and the overall experimental setup on the gold sensor chip is shown in Fig. 2c. Details on surface modification of the chip are described in supporting information. The PHEA-g-C18 polymersomes suspended in PBS at a concentration of 0.5 mg/ml were injected into the flow cell to examine the association and dissociation rates of the polymersomes with the gold sensor chip modified with DPPC and integrins. The media flow rate was kept constant at 5.0 μL/min. The kinetic data from SPR sensorgrams were obtained with the assistance of BIAevaluation version 4.1, where a 1:1 Langmuir binding model was applied to quantify the association and dissociation rates [23].
Analysis of polymersome adhesion onto a cell layer under static conditions
Mouse bone marrow stromal cells (BMSCs, ATTC) with a passage number between 22 and 30 were seeded on glass bottom dishes (MatTek, Ashland, MA) and incubated in Dulbecco’s Modified Eagle Medium (DMEM) supplemented by 10% fetal bovine serum and 1% penicillin/streptomycin (all from GIBCO) at 37 °C until reaching confluency. Spherical (DSPEG at 0 or 0.5 mol%) or ellipsoidal (DSPEG at 1 mol%) PHEA-g-C18 polymersomes encapsulated with fluorescein-conjuated dextrans (FITC-dextran, Mw 40000, Sigma) were added into serum-free DMEM. Next, the cells and polymersomes were incubated for 10 minutes at 37 °C. Afterward, the mixture was removed, and cells were washed with PBS three times. Then the cells were fixed with 10 % neutral buffered formalin (NBF) and fluorescence from the polymersomes bound with cells was captured using a laser-scanning confocal microscope (Leica SP2). Finally, the fluorescence yield per image was quantified by counting the number of pixels showing more green fluorescence than specific threshold values using the image processing software ImageJ. Five different areas of the cell layer were analyzed with at least three different samples per condition. The statistical significance between each two data populations was evaluated using an unpaired, two-tailed Student’s t-test in Microsoft Excel. Differences were considered statistically significant for p < 0.05.
Analysis of polymersome adhesion onto a cell layer under flow
Alternatively, the BMSCs were plated onto a glass substrate fixed in a custom-built flow chamber in order to examine the adhesion of polymersomes to a cell layer under shear flow. The glass substrates were sterilized and coated with collagen (Advanced BioMatrix) before cell seeding. After the formation of a cell layer, the substrate was placed into the chamber and exposed to shear flow at a rate of 200 ml/hr. The suspension of PHEA-g-C18 polymersomes encapsulated with FITC-dextran was injected into the chamber via syringe, and the polymersomes were circulated for 10 minutes. Then, the cells on the glass substrate were fixed with 10 % NBF and adhesion of polymersomes to the cell layer was examined by measuring cell fluorescence under the confocal microscope. Finally, the fluorescence yield was quantified using ImageJ software. Five different areas of the cell layer were analyzed with at least three different samples per condition.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (1R01 HL109192 and 1R21HL097314 to H.K. and 1R25 CA154015A to R.D) and the American Heart Association (Scientist Development Grant to H.K).
Contributor Information
Mei-Hsiu Lai, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana IL, 61801, USA.
Dr. Jae Hyun Jeong, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana IL, 61801, USA
Ross J. DeVolder, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana IL, 61801, USA
Christopher Brockman, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana IL, 61801, USA.
Prof. Charles Schroeder, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana IL, 61801, USA
Prof. Hyunjoon Kong, Department of Chemical and Biomolecular Engineering, Institute for Genomic Biology, University of Illinois at Urbana-Champaign, 600 South Matthews Avenue, Urbana IL, 61801, USA
References
- 1.a) Patra CR, Bhattacharya R, Mukhopadhyay D, Mukherjee P. Adv Drug Delivery Rev. 2010;62:346. doi: 10.1016/j.addr.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vladimir TP. Adv Drug Delivery Rev. 2002;54:235. doi: 10.1016/s0169-409x(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Ahmed F. Annu Rev Biomed Eng. 2006;8:323. doi: 10.1146/annurev.bioeng.8.061505.095838. [DOI] [PubMed] [Google Scholar]
- 3.Egli S, Nussbaumer MG, Balasubramanian V, Chami M, Bruns N, Palivan C, Meier W. J Am Chem Soc. 2011;133:4476. doi: 10.1021/ja110275f. [DOI] [PubMed] [Google Scholar]
- 4.Irvine DJ. Nat Mater. 2011;10:342. doi: 10.1038/nmat3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Muro S, Garnacho C, Champion JA, Leferovich J, Gajewski C, Schuchman EH, Mitragotri S, Muzykantov VR. Mol Ther. 2008;16:1450. doi: 10.1038/mt.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MMC, Liu X, Ferrari M, Decuzzi P. J Biomech. 2008;41:2312. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]; c) Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, Ferrari M. J Control Release. 2010;141:320. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Kang HS, Yang SR, Kim JD, Han SH, Chang IS. Langmuir. 2001;17:7501. [Google Scholar]
- 7.Tomida M, Nakato T, Matsunami S, Kakuchi T. Polymer. 1997;38:4733. [Google Scholar]
- 8.a) Lee HJ, Yang SR, An EJ, Kim JD. Macromolecules. 2006;39:4938. [Google Scholar]; b) Zhang L, Lin J, Lin S. J Phys Chem B. 2007;111:9209. doi: 10.1021/jp068429l. [DOI] [PubMed] [Google Scholar]; c) Jeong JH, Cha C, Kaczmarowski A, Haan J, Oh S, Kong H. Soft Matter. 2012;8:2237. doi: 10.1039/C2SM06763C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Marsh D. Biophys J. 2001;81:2154. doi: 10.1016/S0006-3495(01)75863-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huang C. Appl Phys Lett. 2011;98:043702. doi: 10.1063/1.3554381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan A, David T, Homer-Vanniasinkam S, Graham A, Walker P. Biorheology. 2004;41:641. [PubMed] [Google Scholar]
- 11.Brooks P, Clark R, Cheresh D. Science. 1994;264:569. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 12.a) Arap W, Pasqualini R, Ruoslahti E. Science. 1998;279:377. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]; b) Zhang Y-f, Wang J-c, Bian D-y, Zhang X, Zhang Q. Eur J Pharm Biopharm. 2010;74:467. doi: 10.1016/j.ejpb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Stenberg E, Persson B, Roos H, Urbaniczky C. J Colloid Interface Sci. 1991;143:513. [Google Scholar]
- 14.Gerber DJ, Pereira P, Huang SY, Pelletier C, Tonegawa S. Proc Natl Acad Sci USA. 1996;93:14698. doi: 10.1073/pnas.93.25.14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Yuan W, Wang J. Biomech Model Mechanobiol. 2010;9:659. doi: 10.1007/s10237-010-0206-x. [DOI] [PubMed] [Google Scholar]
- 16.a) Craparo EF, Ognibene MC, Casaletto MP, Pitarresi G, Teresi G, Giammona G. Nanotechnology. 2008;19:485603. doi: 10.1088/0957-4484/19/48/485603. [DOI] [PubMed] [Google Scholar]; b) Lee HJ, Jang KS, Jang S, Kim JW, Yang HM, Jeong YY, Kim JD. Chem Commun. 2010:46. doi: 10.1039/b925106e. [DOI] [PubMed] [Google Scholar]
- 17.a) Gavze E, Shapiro M. J Fluid Mech. 1998;371:59. [Google Scholar]; b) Kojić M, Filipović N, Stojanović B, Kojić N. John Wiley & Sons; 2008. [Google Scholar]
- 18.a) Shah S, Liu Y, Hu W, Gao J. J Nanosci Nanotechnol. 2011;11:919. doi: 10.1166/jnn.2011.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Champion JA, Mitragotri S. Proc Natl Acad Sci U S A. 2006;103:4930. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Doshi N, Prabhakarpandian B, Rea-Ramsey A, Pant K, Sundaram S, Mitragotri S. J Control Release. 2010;146:196. doi: 10.1016/j.jconrel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Toy R, Hayden E, Shoup C, Baskaran H, Karathanasis E. Nanotechnology. 2011;22:115101. doi: 10.1088/0957-4484/22/11/115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caliceti P, Quarta SM, Veronese FM, Cavallaro G, Pedone E, Giammona G. Biochem Biophys Acta. 2001;1528:177. doi: 10.1016/s0304-4165(01)00191-x. [DOI] [PubMed] [Google Scholar]
- 21.Frisken BJ. Appl Opt. 2001;40:4087. doi: 10.1364/ao.40.004087. [DOI] [PubMed] [Google Scholar]
- 22.Kalyanasundaram K, Thomas JK. J Am Chem Soc. 1977;99:2039. [Google Scholar]
- 23.Karlsson R, Michaelsson A, Mattsson L. J Immunol Methods. 1991;145:229. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.