Abstract
The inhibitor of apoptosis (IAP) family of anti-apoptotic proteins regulate programmed cell death and/or apoptosis. One such protein, X-linked IAP (XIAP), inhibits the activity of the cell death proteases, caspase-3, -7, and -9. In this study, using constitutively active mutants of caspase-3, we found that XIAP promotes the degradation of active-form caspase-3, but not procaspase-3, in living cells. The XIAP mutants, which cannot interact with caspase-3, had little or no activity of promoting the degradation of caspase-3. RING finger mutants of XIAP also could not promote the degradation of caspase-3. A proteasome inhibitor suppressed the degradation of caspase-3 by XIAP, suggesting the involvement of a ubiquitin-proteasome pathway in the degradation. An in vitro ubiquitination assay revealed that XIAP acts as a ubiquitin-protein ligase for caspase-3. Caspase-3 was ubiquitinated in the presence of XIAP in living cells. Both the association of XIAP with caspase-3 and the RING finger domain of XIAP were essential for ubiquitination. Finally, the RING finger mutants of XIAP were less effective than wild-type XIAP at preventing apoptosis induced by overexpression of either active-form caspase-3 or Fas. These results demonstrate that the ubiquitin-protein ligase activity of XIAP promotes the degradation of caspase-3, which enhances its anti-apoptotic effect.
Apoptosis is a physiological cell suicide program critical to the development and homeostasis of all animals (1). Abnormal inhibition of apoptosis is a hallmark of cancer and autoimmune disease, whereas excessive cell death has been implicated in neurodegenerative disorders (2). The caspases, a family of intracellular cysteine proteases, are the central executioners of apoptosis (3). Effector caspases, such as caspase-3 and -7, are activated by initiator caspases, such as caspase-9, through proteolytic cleavage (3). Once activated, the effector caspases are responsible for the proteolytic cleavage of a diverse array of structural and regulatory proteins, resulting in an apoptotic phenotype (4).
Inhibitor of apoptosis proteins (IAPs), originally found in baculoviruses, have been conserved in a number of species, ranging from insects to humans throughout evolution and play a role in regulating apoptosis (5, 6). Several members of the human IAP family proteins, including X-linked IAP (XIAP), c-IAP1 and c-IAP2, have been shown to be potent direct inhibitors of caspase-3, -7, and -9 (7–9). Among the above IAPs, XIAP is the most potent inhibitor of caspases and apoptosis (6). The structure of XIAP is characterized by three tandem repeats of the baculovirus IAP repeat (BIR) domain at its NH2 terminus and a RING finger domain near its COOH terminus. Deletional analysis indicates that the second BIR domain (BIR2) of XIAP is sufficient to inhibit caspase-3 and -7 (10), whereas a XIAP fragment encompassing the third BIR domain (BIR3) and the RING finger domain specifically inhibits caspase-9 (11). A recent report indicates that only the BIR3 domain of XIAP is required to inhibit caspase-9 and that the RING finger domain is unnecessary for potent caspase-9 inhibition (12). Collectively, these results suggest that the BIR domains of IAP are essential in inhibiting caspases.
Recent results implicate the RING finger domain in specific ubiquitination events (13–15). Protein ubiquitination begins with the formation of a thiol-ester linkage between the COOH terminus of ubiquitin and the active site cysteine of the ubiquitin-activating enzyme (E1). Ubiquitin then is transferred to a ubiquitin-conjugating enzyme (Ubc or E2), again through a thiol-ester linkage. Ubiquitin-protein ligases (E3s), which are primarily responsible for providing specificity to ubiquitin conjugation, interact with E2 and the substrate, facilitating the formation of isopeptide bonds between the COOH terminus of ubiquitin and lysines either on a target protein or on the last ubiquitin of a protein-bound multiubiquitin chain. Multiubiquitin chains are potent targeting signals for protein degradation in proteasomes (16). E3s that are highly homologous to IAP with respect to their RING finger domains, such as Mdm2 and Siah, promote degradation of both themselves and specific substrates, such as p53, and the deleted in colorectal cancer gene product (DCC), respectively (17–20). Recent reports indicate that XIAP and c-IAP1 promote self-degradation in response to apoptotic stimuli (21), and that c-IAP2 promotes in vitro monoubiquitination of caspase-3 and -7 (22). In this study, we investigated whether XIAP promotes degradation of caspase-3 through its E3 activity and whether its E3 activity affects its anti-apoptotic effect.
Materials and Methods
Plasmid Constructions.
cDNA encoding reversed-caspase-3 (revCasp3), a constitutively active mutant of caspase-3, was generated by PCR according to Srinivasula et al. (23). Using the resultant plasmid as a template, PCR was performed with the following primers: revCasp3-forward, GGAATTCAGTGGTGTTGATGATGACAT; revCasp3-reverse, ATTCTCGAGTCAGTCTGTCTCAATGCCAC. The PCR product was double-digested with EcoRI–XhoI and cloned into the EcoRI–XhoI sites of pcDNA3-Myc-N. To generate pcDNA3-FLAG-XIAP, the EcoRI–XhoI fragment of the full-length XIAP (residues 1–497) of pcDNA3-Myc-XIAP was cloned into pcDNA3-FLAG-N. The RING finger domain-deleted form of XIAP (1–437; XIAPΔRING) was generated by PCR using the following primers: forward, CGGAATTCATGACTTTTAACAGTTTTGAAGG; reverse, CTCCTCGAGTTAAGTACTAATCTCTTTCTG. All of the point mutants were generated by using a QuickChange site-directed mutagenesis kit (Stratagene). Full-length XIAP (residues 1–497) was cloned into the EcoRI–XhoI sites of pGEX4T1 (Amersham Pharmacia). pGEX4T1-XIAP has been described (7). UbcH5B and UbcH7 were cloned by reverse transcription–PCR. Amino-terminal His6-tagged UbcH5B and UbcH7 constructs were generated by ligating PCR fragments into the EcoRI–XhoI sites of pET28a (Novagen). pCGN-hemagglutinin (HA)-ubiquitin was a gift from Shigetsugu Hatakeyama, Kyusyu University, Fukuoka, Japan. Proper construction of all of the plasmids was confirmed by DNA sequencing.
Transfection, Immunoblot, and Immunoprecipitation Analysis.
293T and 293 cells were transfected by using either Lipofectamine Plus (GIBCO/BRL) or FuGENE6 (Roche Diagnostics) according to the manufacturer's instructions. A total of 4–5 × 105 cells were plated in 6-well plates, washed the next day, and then transfected by using a total of 2.5 μg of plasmid DNA. After 3-h incubation, the medium was replaced with fresh complete medium. At 40–48 h after transfection, the cells were washed with PBS and recovered by scraping. Precipitated cells were lysed in 50 μl of lysis buffer [20 mM Hepes, pH 7.4/120 mM NaCl/5 mM EDTA/1% Triton X-100/10% glycerol/protease inhibitor (Complete, Roche Diagnostics)], with or without 20 μM of the proteasome inhibitor MG132 (Peptide Institute, Osaka), at 4°C. Cellular debris was removed by centrifugation at 15,000 × g for 30 min, then 5–10 μl of samples were separated by SDS/PAGE using 12% or 16% polyacrylamide gel and transferred to poly(vinylidene difluoride) membranes (Immobilon, Millipore). For coimmunoprecipitation, 20–40 μl of lysates in 100 μl of lysis buffer was incubated with 5 μl of Anti-FLAG affinity gels (Sigma) or agarose-conjugated 9E10 anti-Myc antibody (Santa Cruz Biotechnology), at 4°C for 4 h. Immunoprecipitates were washed five times with 500 μl of lysis buffer without protease inhibitor at 4°C and analyzed by immunoblot analysis with the appropriate antibodies. For immunoprecipitation of endogenous caspase-3, 293 cells were lysed in RIPA buffer [20 mM Hepes, pH 7.4/150 mM NaCl/2 mM EDTA/1% Nonidet P-40/1% sodium deoxycholate/0.1% SDS/protease inhibitor (Complete, Roche Diagnostics)] with 20 μM of the proteasome inhibitor, MG132 (Peptide Institute) and 0.5 mM of the isopeptidase inhibitor N-ethylmaleimide. Cellular debris was removed by centrifugation at 15,000 × g for 30 min, then the supernatant (2 mg protein) was precleared with 5 μg of control IgG for 5 h at 4°C. The precleared lysates were incubated with 3 μg of anti-caspase-3 mAb (Transduction Laboratories, Lexington, KY) coupled with 5 μl of protein G Sepharose (Amersham Pharmacia) for overnight at 4°C. Immunoprecipitates were washed five times with 1 ml of RIPA buffer without protease inhibitor at 4°C and analyzed by immunoblot analysis with the appropriate antibodies. Antibody detection was performed by using an enhanced chemiluminescence detection kit (Amersham Pharmacia).
Expression and Purification of Proteins.
The glutathione S-transferase (GST) fusion proteins were prepared as described (7), with minor modifications. The GST fusion form of full-length XIAP was induced with 0.01 mM isopropyl-1-thio-β-d-galactpyranoside (IPTG) at 25°C for 1 h. NH2-terminal His6-tagged UbcH5B and UbcH7 were induced with 1 mM IPTG at 30°C for 3 h, then prepared by using Ni-NTA Agarose (Qiagen, Chatsworth, CA) according to the manufacturer's instructions. Concentrations of the purified proteins were determined by the Coomassie Plus Protein Assay Reagent Kit (Pierce).
In Vitro Translation of Caspases.
Wild-type and mutant caspases were translated in vitro in the presence of [35S]methionine in rabbit reticulocyte lysate with a TNT T7 Quick Coupled Transcription/Translation System (Promega).
In Vitro Ubiquitination Assay.
35S-methionine-labeled, in vitro-translated revCasp3 was added to GST-XIAP (250 ng) immobilized on glutathione-Sepharose beads. The mixture was incubated on ice for 1 h to form a GST-XIAP/revCasp3 complex. After extensive washing, the GST-XIAP/revCasp3 complex was used to perform the ubiquitination assay, which was carried out in ubiquitination buffer (50 mM Tris⋅HCl, pH 7.5/2 mM ATP/5 mM MgCl2/2 mM DTT) containing yeast E1 (200 ng), E2 (150 ng), and His6-ubiquitin (10 μg), in a total volume of 30 μl. The mixture was incubated at 30°C for 90 min with agitation to keep the beads in suspension. After the reaction, the samples were resolved on SDS/PAGE using a 5–15% linear gradient gel and analyzed by autoradiography.
DEVDase Activity.
Caspase activities were assayed as described (7) with some modification. In brief, caspase activity was measured at 30°C in 100 μl of caspase buffer (20 mM Pipes/100 mM NaCl/1 mM EDTA/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/10% sucrose/10 mM DTT, pH 7.2) containing 20 μg of cell lysate and 100 μM of the fluorogenic peptide substrate, acetyl-DEVD-MCA (Peptide Institute). Activity was measured continuously over the indicated time by the release of 7-amino-4-methylcoumarin from acetyl-DEVD-MCA as emission at 460 nm on excitation at 355 nm by using a fluorometer, Fluoroskan Ascent FL (Labsystems, Chicago) in the kinetic mode.
Apoptosis Assay.
A total of 2 × 105 293 cells were plated in 6-well plates. After 2 days, the cells were cotransfected by using Lipofectamine Plus (GIBCO/BRL), as described by the manufacturer, with the following DNA concentrations: 0.2 μg of pDsRed1-N1 marker plasmid (CLONTECH), 15 ng of pcDNA3-revCasp3 plasmid, or 0.2 μg of pCMV-Fas plasmid with various pcDNA3-XIAP constructs or the control plasmid pcDNA3 at 0.75–0.8 μg. At 24–36 h after transfection, all cells were collected and fixed in 80% ethanol. Apoptosis of transiently transfected 293 cells was determined with an In Situ Cell Death Kit (Roche Diagnostics) based on TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling). TUNEL-positive cells were viewed at excitation 488 nm/emission 512 nm by fluorescence microscopy. Transfection efficiency was determined by counting DsRed-positive cells. The percentage of TUNEL-positive cells among transfected cells was determined under each condition.
Results
XIAP Specifically Regulates the Level of Active-Form Caspase-3 Protein Expression.
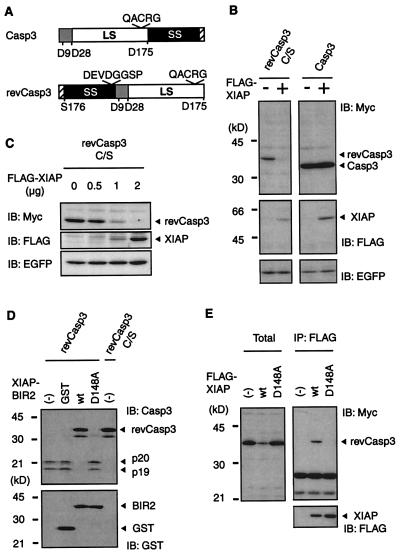
To investigate whether XIAP decreases the steady-state level of caspases, we constructed constitutively active form of caspase-3 (revCasp3) (23) by switching the order of its two subunits (Fig. 1A). RevCasp3 is capable of autocatalytic processing during an in vitro translation reaction and rapidly induces apoptosis in MCF-7 cells (23). Because undetectable expression levels of revCasp3 were found to strongly induce apoptosis, we made the construct, revCasp3 C/S, in which its active sites cysteine residue was mutated to serine to abrogate its enzymatic activity. When 293T cells were cotransfected, a specific decrease in revCasp3 C/S was observed because of the expression of XIAP, whereas the protein expression levels of procaspase-3 did not significantly decrease (Fig. 1B). To confirm the above observation, we examined the reduction in revCasp3 in the presence of increasing amounts of XIAP in a cotransfection assay of 293 cells. The expression of XIAP led to a dose-dependent decrease in the level of revCasp3 C/S protein (Fig. 1C). Because XIAP binds to the active form of caspase-3, but not to procaspase-3 (7) (data not shown), the complex formation of XIAP with caspase-3 seems to be important for the active form-specific reduction in caspase-3 because of XIAP. To examine this idea, we have constructed a point mutant of full-length XIAP (D148A), which should not bind to the active from of caspase-3, based on the fact that the XIAP-BIR2 fragment with D148A mutation (BIR2-D148A) lost the inhibitory activity to caspase-3 (24). An in vitro-translated revCasp3 is able to undergo autocatalytic processing (23) (Fig. 1D). BIR2 inhibited the autoprocessing of in vitro-translated revCasp3, whereas BIR2-D148A mutant did not, indicating that BIR2-D148A mutant does not bind to and inhibit active caspase-3 (Fig. 1D). Moreover, this result confirms the previous finding that revCasp3 is enzymatically active without the separation of the two subunits (23), providing evidence that revCasp3 C/S as well as active revCasp3 is in the correct conformation of active-form enzyme. As expected, cotransfection with XIAP-D148A mutant, unlike wild-type XIAP, did not decrease the expression level of revCasp3 C/S (Fig. 1E). Although we also constructed revCasp7 and revCasp9 and examined whether XIAP decreases the amount of these caspases, no obvious decline in these caspases was detected, probably because the low affinity of these caspases to XIAP in cells (data not shown). These results indicate that XIAP specifically decreases the level of active-form caspase-3 protein expression through complex formation with this protease.
Figure 1.
Regulation of caspase-3 protein expression by XIAP. (A) Schematic representation of the constitutively active caspases used in this study. The procaspase-3 and the constitutively active caspases-3 (revCasp3) are indicated. Locations of the large subunit (LS), small subunit (SS), and prodomain, of each caspase, are illustrated by the white box, black box, and gray box, respectively. Numbers indicate their amino acid position. The cysteine residue of their active site is shown by QACRG. We also constructed inactive mutant of the revCasp3 by substituting its cysteine residue with serine. The hatched boxes represent a Myc tag. (B) Active-form caspase-3 protein expression is specifically decreased in the presence of wild-type XIAP. 293T cells were cotransfected with a pcDNA3 expression vector containing cDNAs (0.5 μg Myc-revCasp3 C/S; 0.5 μg Procaspase-3-Myc; 2 μg, FLAG-XIAP) as indicated, and pEGFP (0.25 μg). The total amount of plasmid was adjusted to 2.75 μg with pcDNA3. Forty-eight hours after transfection, cell lysates were subjected to immunoblot (IB) analysis with anti-Myc, anti-FLAG, or anti-EGFP antibody as indicated. (Bottom) Probes with anti-EGFP antibody verify equivalent transfection efficiencies. (C) 293 cells were cotransfected with 0.25 μg of Myc-revCasp3 C/S, 0.25 μg of pEGFP, and increasing amounts of the XIAP expression plasmids, as indicated. The total amount of plasmid was adjusted to 2.5 μg with pcDNA3. (Bottom) Equivalent transfection efficiencies. (D) The inhibitory effect of wild-type or mutant XIAP on the autoprocessing of revCasp3 in an in vitro translation reaction. RevCasp3 or active site Cys to Ala mutant in pcDNA3-myc constructs were in vitro-translated in the absence or presence of 100 nM of GST-tagged recombinant XIAP-BIR2 proteins. The translation products were analyzed by immunoblot with anti-caspase-3 antibody (Upper). (Lower) The equivalent amounts of GST-proteins were used. (E) The association of XIAP with caspase-3 is essential for the reduction in caspase-3 protein expression. Cell lysates from 293T cells cotransfected with the indicated expression vectors were subjected to immunoblot analysis with anti-Myc (Left). The conditions of transfection were similar to those indicated in B. The same lysates were immunoprecipitated with anti-FLAG antibody followed by probing with anti-Myc (Right) and anti-FLAG antibody (Lower).
Ubiquitin-Proteasome Pathway in the Regulation of Caspase-3 and XIAP Proteins.
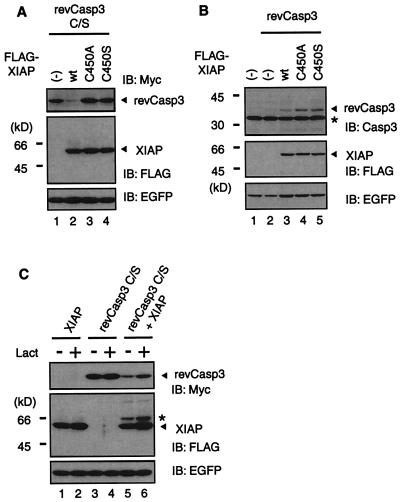
Previous studies have shown that the RING finger domains of other proteins, such as Siah, c-Cbl, and Mdm2, which are very similar to that of XIAP, are essential for the proteasome-dependent degradation of their target proteins (18, 20, 25–27). Therefore, we next examined whether the intact RING finger domain of XIAP is required to decrease caspase-3 and the possibility that the ubiquitin-proteasome pathway is involved in the process. 293T cells were cotransfected with plasmids encoding wild-type or RING finger mutants of XIAP together with Myc-tagged revCasp3 C/S mutant and the expression level of each protein was examined by immunoblot analysis. As expected, a decrease in caspase-3 was not observed in the presence of the RING-finger mutants of XIAP (Fig. 2A). Essentially the same data were obtained when we used enzymatically active revCasp3 (Fig. 2B). The relatively low level of revCasp3 expression in the absence of XIAP, compared with that of revCasp3 C/S mutant, was caused by cytotoxicity of this active enzyme. Supporting this idea, the expression of enhanced green fluorescent protein (EGFP) in the presence of revCasp3 alone was lower than that of the others (Fig. 2B Bottom). These results indicate that the RING finger domain of XIAP is essential for the reduction in caspase-3. Treatment with a proteasome inhibitor, lactacystin, partially blocked the decrease in caspase-3 observed in the presence of wild-type XIAP (Fig. 2C Top). The expression level of XIAP itself also was affected by lactacystin treatment in the presence of revCasp3, whereas the level of EGFP was not, suggesting the involvement of ubiquitin-proteasome pathway-mediated degradation of XIAP itself as described (21) (Fig. 2C Middle). This result suggests that the degradation of caspase-3 is proteasome-dependent.
Figure 2.
The RING mutations of XIAP abolish its ability to decrease caspase-3 protein expression. (A) RING finger-dependent degradation of revCasp3 C/S by XIAP. 293 cells were cotransfected with a pcDNA3 expression vector containing cDNAs (0.2 μg Myc-revCasp3 C/S; 1.6 μg, FLAG-XIAP) as indicated, and pEGFP (0.2 μg). The total amount of plasmid was adjusted to 2 μg with pcDNA3. Forty-eight hours after transfection, cell lysates were subjected to immunoblot (IB) analysis with anti-Myc, anti-FLAG, or anti-EGFP antibody as indicated. (Bottom) Probe with anti-EGFP antibody verifies equivalent transfection efficiencies. (B) RING finger-dependent degradation of constitutively active caspase-3 (revCasp3) by XIAP. 293 cells were cotransfected with a pcDNA3 expression vector containing cDNAs (15 ng Myc-revCasp3; 0.8 μg, FLAG-XIAP) as indicated, and pEGFP (0.2 μg). Sixteen hours after transfection, cell lysates were subjected to immunoblot analysis with anti-caspase-3, anti-FLAG, or anti-EGFP antibody as indicated. * indicates endogenous caspase-3. (C) Proteasome-dependent degradation of revCasp3 C/S by XIAP. 293 cells were cotransfected with a pcDNA3 expression vector containing cDNAs (0.4 μg Myc-revCasp3 C/S; 1.6 μg, FLAG-XIAP) as indicated, and pEGFP (0.2 μg). The total amount of plasmid was adjusted to 2 μg with pcDNA3. Twenty-four hours after transfection, cells were treated with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) 30 μM lactacystin (Lact), a specific inhibitor of proteasome. After an additional 10 h of incubation, the expression levels of Myc-revCasp3 C/S (Top) and FLAG-tagged XIAP (Middle) were assessed by immunoblot analysis. (Bottom) Equivalent transfection efficiencies. * indicates a nonspecific band.
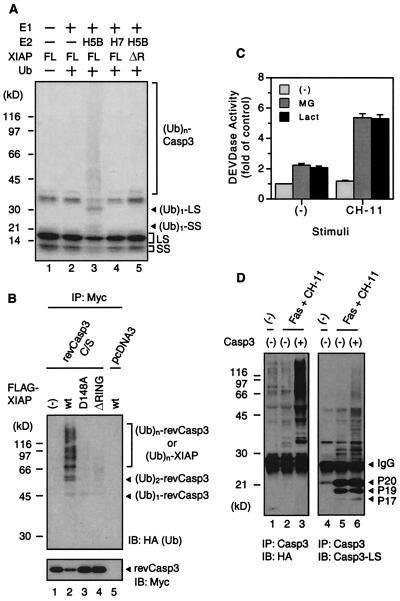
Ubiquitination marks proteins for degradation by the proteasome (16). Previous studies have shown that IAP family proteins act as ubiquitin-protein ligases (E3) for themselves and caspase-3 (21, 22). To examine whether caspase-3 is ubiquitinated by XIAP, and XIAP has E3 activity capable of targeting caspase-3, we performed in vitro ubiquitination assays. 35S-labeled, in vitro-translated revCasp3 and GST-XIAP, immobilized on glutathione-Sepharose beads, were mixed to form a revCasp3/GST-XIAP complex and washed extensively. Then, the complex was incubated in the presence or absence of yeast ubiquitin-activating enzyme (E1), recombinant human ubiquitin-conjugating enzyme (E2; UbcH5B or UbcH7), and His6-ubiquitin. The higher molecular weight species of caspase-3 was detected only in the presence of all of the components, including UbcH5B as an E2 (Fig. 3A, lane 3), but not in the absence of E2 and presence of UbcH7 (Fig. 3A, lanes 2 and 4). The reduction in nonubiquitinated form and the presence of possible monoubiquitinated species of large or small subunits of caspase-3 (about 30 kDa or 23 kDa, respectively) suggest that both large and small subunits of this enzyme are ubiquitinated (Fig. 3A, lane 3). Moreover, the higher molecular weight species of caspase-3 was not detected when the reaction was performed in the presence of GST-XIAPΔRING (Fig. 3A, lane 5). These results demonstrate that XIAP has E3 activity for caspase-3.
Figure 3.
The ubiquitination of caspase-3 in vitro and in cells. (A) In vitro ubiquitination of caspase-3. In vitro-translated, 35S-labeled revCasp3 was pulled down by bacterially expressed GST-tagged full-length XIAP (FL) and XIAPΔRING (ΔR). After extensive washing, the complex was incubated in the presence or absence of E1, E2, and His6-ubiquitin at 30°C for 90 min. After the reaction, the samples were resolved on SDS/PAGE using a 5–15% linear gradient gel and analyzed by autoradiography. Details are described in Materials and Methods. The position of the molecular standard marker (kDa) is indicated on the left. (B) The association of XIAP and RING finger-dependent ubiquitination of caspase-3 in living cells. 293T cells were cotransfected with the indicated expression plasmids, pEGFP and pCGN-HA-ubiquitin (0.25 μg). The conditions of transfection, including the amount of plasmid, were similar to those indicated in Fig. 1B except for the HA-ubiquitin expression plasmid. Twenty-four hours after transfection, cells were treated with 50 μM lactacystin. After an additional 18 h of incubation, cell lysates were immunoprecipitated with anti-Myc antibody followed by probing with anti-HA (Upper) or anti-Myc (Lower) antibody. The position of the molecular standard marker (kDa) is indicated on the left. (C) Proteasome inhibitors potentiate caspase-3-like protease activity induced by Fas. 293 cells were transfected with pCMV-Fas expression plasmid. Twenty-four hours after transfection, cells were treated with or without 10 ng/ml CH-11 anti-Fas antibody, proteasome inhibitor MG132 (3 μM, MG), or lactacystin (30 μM, Lact) as indicated. After an additional 12 h of incubation, cell lysates were prepared, and then DEVDase activity of each lysates were measured as described in Materials and Methods and the data (mean ± SE; n = 3) presented here as a fold of nontreated cells (control). (D) The ubiquitination of endogenous caspase-3. 293 cells were transfected with XIAP plasmid and with or without pCMV-Fas expression plasmid or active-site mutant of caspase-3 as indicated. Twenty-four hours after transfection, cells were treated with 3 μM MG132 alone or with 100 ng/ml CH-11 anti-Fas antibody and 3 μM MG132. After an additional 18 h of incubation, cell lysates were immunoprecipitated with anti-caspase-3 antibody followed by probing with anti-HA (Left) or anti-cleaved caspase-3 (Casp3-LS; Right) antibody. IB, immunoblot; IP, immunoprecipitation.
To test whether caspase-3 is ubiquitinated by XIAP in cells, revCasp3 was coexpressed in 293T cells along with a construct encoding FLAG-tagged wild-type or mutant XIAP and HA epitope-tagged ubiquitin in the presence of a proteasome inhibitor. Lysates were prepared from the cells, and then revCasp3 protein was collected by immunoprecipitation with anti-Myc antibody. When probed with anti-HA antibody, which recognizes exogenously expressed HA-tagged ubiquitin, the signal corresponding to higher molecular weight species of revCasp3 was detected (Fig. 3B Upper) only in the presence of wild-type XIAP. Because HA-ubiquitin is a 9.5-kDa protein, the ubiquitinated revCasp3 proteins appear as 45- or 56-kDa bands (Fig. 3B Upper, lane 2). The 67- and 78-kDa bands correspond to the mixture of ubiquitinated caspase-3 and XIAP proteins. These results suggest that caspase-3 is ubiquitinated in living cells. Therefore, we further examined whether the ubiquitination of endogenous caspase-3 occurs in 293 cells, which express endogenous XIAP. When 293 cells were transfected with pCMV-Fas expression plasmid and then stimulated with or without of either MG132 or lactacystin, the proteasome inhibitors, these proteasome inhibitors potentiated Fas-induced DEVDase activity (Fig. 3C). The effect of proteasome inhibitors was prominent when stimulated with CH-11 anti-Fas agonistic antibody (Fig. 3C). Treatment of 293 cells, which were not transfected with pCMV-Fas, with above proteasome inhibitors alone did not affect the DEVDase activity (data not shown). These data strongly suggest that activated caspase-3 might be degraded through a ubiquitin-proteasome pathway at least in a setting of Fas-induced apoptosis. So we tried to detect the ubiquitination of endogenous caspase-3 under conditions similar to those in Fig. 3C. The XIAP and HA-ubiquitin expression plasmids were cotransfected in 293 cells with or without pCMV-Fas, and then stimulated with MG132 alone or MG132 and CH-11. Lysates were prepared from the cells, and then caspase-3 protein was collected by immunoprecipitation with anti-caspase-3 antibody. When probed with anti-HA antibody, which recognizes exogenously expressed HA-tagged ubiquitin, or with anti-cleaved caspase-3 antibody, which recognizes the large subunit of activated caspase-3, the signal corresponding to higher molecular weight species of caspase-3 was detected (Fig. 3D, lanes 2 and 5). When cotransfected with active site-mutated caspase-3, the ubiquitination of caspase-3 was enhanced probably because the large and small subunits of catalytically inactive caspase-3 were increased in amount and contributed to the increase of ubiquitinated species (Fig. 3D, lanes 3 and 6). The ubiquitination of caspase-3 in its small subunit may explain the difference of position of signals probed with anti-HA and anti-cleaved caspase-3 antibodies. Taken together, these results demonstrated that caspase-3 is ubiquitinated by XIAP in vitro and endogenous caspase-3 is indeed ubiquitinated in apoptotic cells.
E3 Activity of XIAP and Its Anti-Apoptotic Effect.
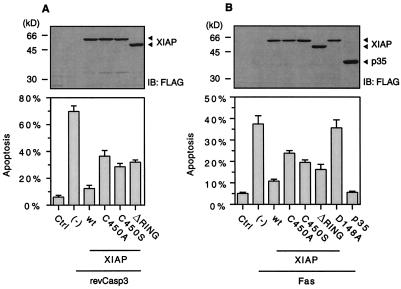
The finding that the E3 activity of XIAP promotes the proteasomal degradation of both caspase-3 and itself brings the question of whether its E3 activity could enhance or attenuate its anti-apoptotic effect. Therefore, we examined the effect of wild-type or RING-mutated XIAP on revCasp3- or Fas-induced apoptosis by means of the TUNEL technique (Fig. 4 A and B Lower). We have chosen Fas overexpression as an apoptosis stimulus in addition to revCasp3 for the following reasons. In the Fas system, caspase-3 is directly cleaved and activated by caspase-8, which is not inhibitable by XIAP. In this setting, XIAP can inhibit cell death at the level of caspase-3, but not caspase-9, enabling us to examine the effect of XIAP-E3 on caspase-3 (11). Moreover, because Fas stimulation is more natural death induction than revCasp3 overexpression, it is a more appropriate system to show the biological significance of XIAP-E3 activity. About 70% or 40% of 293 cells transfected with revCasp3 alone or Fas alone, respectively, were apoptotic (TUNEL-positive). When cotransfected with the combination of revCasp3 and wild-type, a C450A mutant, or a ΔRING mutant XIAP, 12.3%, 28.5%, or 31.9% of all of the transfected cells, respectively, were apoptotic (Fig. 4A). The RING-mutated XIAPs, which lack E3 activity for caspase-3, inhibited apoptosis significantly more weakly than wild-type XIAP did. The similar result was obtained when overexpression of Fas, instead of revCasp3, was used as a death-inducing stimulus (Fig. 4B). It is noteworthy that the XIAP D148A mutant, which does not associate with caspase-3, did not suppress Fas-induced apoptosis (Fig. 4B), suggesting that XIAP inhibits Fas-induced apoptosis at the point of caspase-3 in 293 cells. p35, a baculovirus caspase inhibitor, inhibited Fas-induced apoptosis more strongly than wild-type XIAP, probably because p35 inhibited caspase-3 more strongly than XIAP and p35 inhibited caspase-8, whereas XIAP does not. To check the expression level of XIAP protein, cell lysates were prepared and analyzed by immunoblot with anti-FLAG antibody. The RING mutants were expressed in the similar level to wild-type XIAP (Fig. 4 A and B Upper), which is consistent with the data obtained by using an inactive mutant of revCasp3 (Fig. 2A). These results indicate that the E3 activity of XIAP enhances its anti-apoptotic effect.
Figure 4.
The RING finger domain of XIAP enhances its anti-apoptotic effect through degradation of caspase-3. DNA fragmentation analysis of 293 cells cotransfected with constitutively active caspase-3 (revCasp3) (A) or Fas (B) along with wild-type or RING-mutated forms of XIAP, and DsRed plasmids, as indicated. Twenty four to 36 h after transfection, a TUNEL assay was performed. The percentage of apoptotic DsRed-positive cells (mean ± SEM) is indicated. The protein expression of XIAP variants were assessed by immunoblot (IB) analysis using anti-FLAG antibody (Upper).
Discussion
Recent studies have revealed that IAP family proteins act as potent inhibitors of the cell death proteases known as caspases (6). E3s, such as Siah and Mdm2, which are highly related to XIAP at their RING finger domains, promote the degradation of both themselves and their substrates (18, 20, 28). XIAP also promotes self-degradation in response to apoptotic stimuli through its E3 activity (21), but whether it has substrate(s) other than itself remains to be determined. In this study, we demonstrated that XIAP, a most potent inhibitor of mammalian caspase-3, also acts as an E3 for caspase-3 and thus, promotes the proteasomal degradation of this protease. Moreover, its E3 activity for caspase-3 enhances its anti-apoptotic effect on Fas- as well as revCasp3-induced apoptosis.
Very recently, the crystal structure of the BIR2 domain of XIAP in complex with caspase-3 revealed the structural basis for inhibition (29). The data show that BIR2 is a noncovalent and fully reversible inhibitor of caspase-3 (29). In contrast, the crystal structure of p35, a broad spectrum caspase inhibitor, in complex with caspase-8 revealed that the caspase is inhibited in the active site through a covalent thioester linkage to p35 (30). Because XIAP is a reversible inhibitor of caspase-3, free active enzymes might be released from XIAP/caspase-3 complex if caspase-3 is not degraded. The E3 activity of XIAP for caspase-3 may be necessary to ensure the complete inhibition of caspase-3.
XIAP also binds to and inhibits caspase-7 and -9 (7, 9). Although we could not detect the ubiquitination of caspase-7 or -9 in cells up to now, the ubiquitination of these caspases were detected by an in vitro ubiquitination assays under the similar condition described in Fig. 3A (data not shown). Whether these caspases are ubiquitinated by XIAP in vivo under certain conditions requires further experiments.
IAPs exert their anti-apoptotic activities, at least in part, by preventing the activation of procaspases, and by inhibiting the enzymatic activity of mature caspases (7–9, 31, 32). Such anti-caspase activities of IAPs reside in NH2-terminal BIR domains (8, 10, 33, 34). Although the role that E3 activity plays in the anti-apoptotic effect of IAPs has not been determined, the COOH-terminal RING finger domain either positively (14, 34–36) or negatively (21, 33) affects the anti-apoptotic activity of IAPs. For XIAP, the RING-less form is degraded to a lesser extent and confers better protection from dexamethasone-induced apoptosis in T hybridoma cells, than the wild-type protein (21). In contrast, the RING deletion mutant protects 293 cells from ICE-induced death less efficiently than wild-type XIAP (36), which is consistent with our results showing that wild-type XIAP protects cells from constitutively active caspase-3 and Fas-induced cell death better than RING mutant forms. These differences seem to depend on the cell type and the apoptotic stimuli. In Drosophila, the RING-less form of DIAP1 blocks HID-dependent death in the eye much more efficiently than wild-type DIAP1 (33). The point mutation of DIAP1's RING domain at the conserved cysteine residues also suppresses HID-induced death in the eye more efficiently, whereas the same mutation enhances both REAPER and GRIM-induced death (37).
Recent reports indicate that Mdm2 is conjugated with SUMO-1 at Lys-446, which plays a critical role in Mdm2 self-ubiquitination. Whereas mutant Mdm2 (K446R) is stabilized, it elicits an increased degradation of p53 and a concomitant inhibition of p53-mediated apoptosis (38). This result suggests the possibility that the ubiquitination of E3 itself is not essential for its ability to ubiquitinate its substrates. Because IAPs attenuate their anti-apoptotic effect, in part, by promoting self-ubiquitination, which is followed by a reduced expression of their own protein, it is possible that sumoylation of IAPs or blockage of self-ubiquitination by mutagenesis enhances their activity. Inversely, a SUMO-1 modification of p53 activates its transcriptional activity by inhibiting its degradative pathway (39, 40). Our results demonstrate that caspase-3 is ubiquitinated and degraded by XIAP. Whether IAPs or caspases can be modified by ubiquitin-like proteins remains to be determined.
Acknowledgments
We thank J. C. Reed for providing the expression construct of Fas, P. Friesen for p35 cDNA, G. S. Salvesen for cDNA of caspases, S. Yonehara for the 293 and 293T cells, S. Hatakeyama for helpful advice and providing the cDNA of ubiquitin, R. Araya for technical assistance, and Y. Imai for technical advice during experiments. We also thank all of our laboratory members for helpful discussion. This work is supported, in part, by grants from Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and from the Ministry of Health and Welfare of Japan (to R.T.).
Abbreviations
- IAP
inhibitor of apoptosis protein
- XIAP
X-linked IAP
- BIR
baculovirus IAP repeat
- EGFP
enhanced green fluorescent protein
- GST
glutathione S-transferase
- revCasp3
reversed-caspase-3
- HA
hemagglutinin
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jacobson M D, Weil M, Raff M C. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 4.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 5.Miller L K. Trends Cell Biol. 1999;9:323–328. doi: 10.1016/s0962-8924(99)01609-8. [DOI] [PubMed] [Google Scholar]
- 6.Deveraux Q L, Reed J C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 7.Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 8.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deveraux Q L, Roy N, Stennicke H R, Van Arsdale T, Zhou Q, Srinivasula S M, Alnemri E S, Salvesen G S, Reed J C. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen G S, Reed J C. J Biol Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 11.Deveraux Q L, Leo E, Stennicke H R, Welsh K, Salvesen G S, Reed J C. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun C, Cai M, Meadows R P, Xu N, Gunasekera A H, Herrmann J, Wu J C, Fesik S W. J Biol Chem. 2000;275:33777–33781. doi: 10.1074/jbc.M006226200. [DOI] [PubMed] [Google Scholar]
- 13.Borden K L. J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 14.Freemont P S. Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 15.Joazeiro C A, Weissman A M. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 16.Ciechanover A. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 18.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 19.Hu G, Zhang S, Vidal M, Baer J L, Xu T, Fearon E R. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu G, Fearon E R. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Fang S, Jensen J P, Weissman A M, Ashwell J D. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 22.Huang H, Joazeiro C A, Bonfoco E, Kamada S, Leverson J D, Hunter T. J Biol Chem. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasula S M, Ahmad M, MacFarlane M, Luo Z, Huang Z, Fernandes-Alnemri T, Alnemri E S. J Biol Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 24.Sun C, Cai M, Gunasekera A H, Meadows R P, Wang H, Chen J, Zhang H, Wu W, Xu N, Ng S C, Fesik S W. Nature (London) 1999;401:818–822. doi: 10.1038/44617. [DOI] [PubMed] [Google Scholar]
- 25.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterman H, Levkowitz G, Alroy I, Yarden Y. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 27.Levkowitz G, Waterman H, Ettenberg S A, Katz M, Tsygankov A Y, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Guenther M G, Carthew R W, Lazar M A. Genes Dev. 1998;12:1775–1780. doi: 10.1101/gad.12.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riedl S J, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik S W, Liddington R C, Salvesen G S. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 30.Xu G, Cirilli M, Huang Y, Rich R L, Myszka D G, Wu H. Nature (London) 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- 31.Wang S L, Hawkins C J, Yoo S J, Muller H A, Hay B A. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 32.Meier P, Silke J, Leevers S J, Evan G I. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hay B A, Wassarman D A, Rubin G M. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 34.Vucic D, Kaiser W J, Miller L K. J Biol Chem. 1998;273:33915–33921. doi: 10.1074/jbc.273.51.33915. [DOI] [PubMed] [Google Scholar]
- 35.Clem R J, Miller L K. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanna M G, Duckett C S, Richter B W, Thompson C B, Ulevitch R J. Proc Natl Acad Sci USA. 1998;95:6015–6020. doi: 10.1073/pnas.95.11.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lisi S, Mazzon I, White K. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 39.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz S E, Scheffner M, Del Sal G. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]