Summary
The decision to initiate DNA replication is a critical step in the cell cycle of all organisms. Cells often delay replication in the face of stressful conditions, but the underlying mechanisms remain incompletely defined. Here, we demonstrate in Caulobacter crescentus that proteotoxic stress induces a cell cycle arrest by triggering the degradation of DnaA, the conserved replication initiator. A depletion of available Hsp70 chaperone, DnaK, either through genetic manipulation or heat shock, induces synthesis of the Lon protease, which can directly degrade DnaA. Unexpectedly, we find that unfolded proteins, which accumulate following a loss of DnaK, also allosterically activate Lon to degrade DnaA, thereby ensuring a cell cycle arrest. Our work reveals a new mechanism for regulating DNA replication under adverse growth conditions. Additionally, our data indicate that unfolded proteins can actively and directly alter substrate recognition by cellular proteases.
Introduction
Whether, and when, to initiate DNA replication are important decisions facing nearly all cells. In particular, cells must be able to control the onset of DNA replication in response to changing environmental conditions. Under favorable conditions, cells often strive to grow and proliferate, and hence will promote DNA replication initiation. Conversely, in stressful conditions, cells must prevent the initiation of replication and instead shift their priorities to cytoprotective functions. The molecular mechanisms used to transduce information about prevailing environmental conditions to the replication machinery remain incompletely understood, particularly in bacteria.
In all organisms, replication initiation depends on the formation of highly organized nucleoprotein complexes at chromosomal origins (Kawakami and Katayama, 2010). In eukaryotes, the multi-subunit origin recognition complex (ORC) binds to origins and drives assembly of pre-initiation replication complexes (Bell, 2002). In bacteria, DNA replication requires the AAA+ ATPase DnaA (Leonard and Grimwade, 2011). When bound to ATP, DnaA forms an oligomeric structure along the origin, unwinds the DNA duplex, and recruits other components of the replication machinery, thereby initiating DNA replication. Given its central role, DnaA represents a major target for regulatory mechanisms that dictate the timing of DNA replication. Although major progress has been made in elucidating the mechanisms controlling DnaA during exponential growth (Ishida et al., 2004; Kato and Katayama, 2001; Merrikh and Grossman, 2011; Murray and Errington, 2008; Noirot-Gros et al., 2002; Zawilak-Pawlik et al., 2007), it remains largely unexplored how DnaA and cell cycle progression are regulated in response to nutrient limitation or stressful environmental conditions.
The α-proteobacterium Caulobacter crescentus is a model for dissecting the regulation of the bacterial cell cycle. Caulobacter cells divide asymmetrically, producing daughter cells that differ with respect to morphology and replicative fate (Tsokos and Laub, 2012). The daughter stalked cell immediately enters S-phase after cell division whereas the daughter swarmer cell enters a G1 phase and cannot initiate DNA replication until after differentiating into a stalked cell. Caulobacter cells initiate replication once, and only once, per cell cycle (Marczynski, 1999). As in most bacteria, DNA replication in Caulobacter requires DnaA (Gorbatyuk and Marczynski, 2001) and the availability of active DnaA dictates the timing of replication initiation (Jonas et al., 2011). In rich media, the abundance of Caulobacter DnaA does not change significantly during the cell cycle (Gorbatyuk and Marczynski, 2005; Jonas et al., 2011; Taylor et al., 2011), indicating that DnaA is controlled mainly at the level of activity in these conditions. Like in E. coli, Caulobacter DnaA can switch between an active, ATP-bound form and an inactive, ADP-bound form (Fernandez-Fernandez et al., 2011; Jonas et al., 2011). The protein HdaA, a homolog of E. coli Hda, stimulates ATP hydrolysis by DnaA in Caulobacter (Collier and Shapiro, 2009; Jonas et al., 2011). Additional factors regulating DnaA activity likely exist, but remain to be identified.
Growth of Caulobacter cells in certain stressful conditions, including nutrient starvation and stationary phase, results in a substantial reduction in DnaA levels and a cessation of DNA replication (Gorbatyuk and Marczynski, 2005; Lesley and Shapiro, 2008). The decrease in DnaA likely constitutes an important adaptation to stress and starvation by preventing cell cycle progression when cells need to devote their energy and resources to survival strategies rather than proliferation. The drop in DnaA levels results in part from the proteolysis of DnaA (Gorbatyuk and Marczynski, 2005; Lesley and Shapiro, 2008), but the protease(s) involved and the mechanisms through which stress ultimately triggers proteolysis remain elusive.
To better understand how DnaA is regulated in Caulobacter, we isolated mutations that restore the viability of a strain overproducing DnaA; an oversupply of DnaA drives the overinitiation of replication and is lethal (Jonas et al., 2011). Strikingly, all of the mutations mapped to dnaK, dnaJ, and grpE, which encode for the Hsp70 molecular chaperone system (Genevaux et al., 2007; Hartl and Hayer-Hartl, 2009). We find that this chaperone is required for DnaA accumulation and the initiation of DNA replication. Cells lacking active DnaK target DnaA for degradation by the protease Lon in two mutually reinforcing ways. (1) Loss of DnaK activity triggers the synthesis of Lon. (2) Unfolded proteins that accumulate following a loss of DnaK activity, and that are themselves degraded by Lon, allosterically activate the protease to degrade DnaA. Collectively, our findings indicate that the Caulobacter cell cycle coordinates DNA replication with the status of intracellular protein folding through the highly conserved Hsp70 chaperone and a AAA+ protease. Our work further suggests that unfolded proteins are not simply passive substrates of Lon, but can play an active role in determining substrate recognition following heat shock and other stresses, a phenomenon that may be broadly conserved.
Results
Mutations in dnaK, dnaJ and grpE suppress the lethality of dnaA overexpression
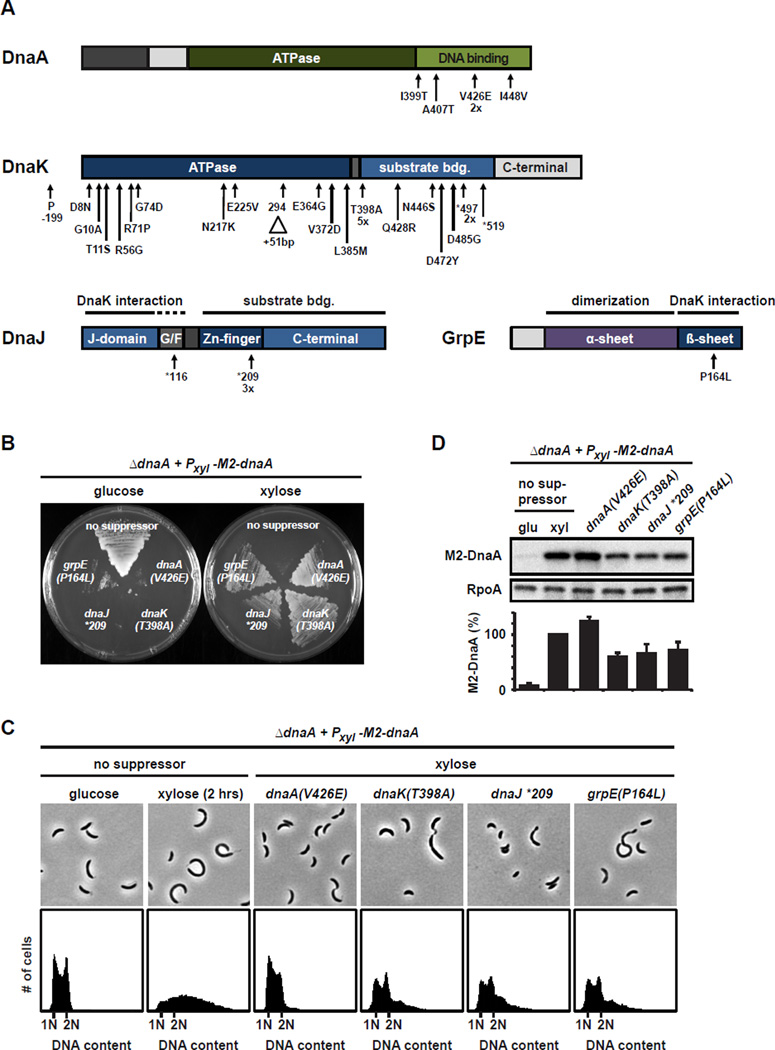
Overexpression of Caulobacter dnaA results in the overinitiation of DNA replication and is lethal (Jonas et al., 2011). We hypothesized that mutations bypassing the lethality of dnaA overexpression would provide insight into the regulation of DnaA and DNA replication. To isolate such mutations we grew a culture of cells lacking the chromosomal copy of dnaA and carrying a plasmid with M2-dnaA driven by a xylose-inducible promoter (Pxyl). On plates containing xylose, most cells died, as expected; however, prolonged incubation resulted in a few slow growing colonies, harboring putative suppressor mutations. Because the chromosomal copy of dnaA had been deleted and because cells lacking a functional copy of dnaA cannot survive (Gorbatyuk and Marczynski, 2001), we ruled out the possibility that colony growth was due to plasmid loss or mutations that abolish DnaA activity. To eliminate clones with plasmid-based mutations that simply reduced Pxyl-M2-dnaA expression, we assessed the lethality of the plasmid by re-transforming it into wild type cells and sequencing Pxyl-M2-dnaA. Most clones contained mutations in Pxyl and were therefore excluded from further analysis. In five clones we identified mutations in the plasmid-borne dnaA leading to the amino acid substitutions I399T, A407T, V426E (discovered twice independently) and I448V (Fig. 1A). All four residues are highly conserved and reside in the DNA binding domain of DnaA, suggesting that these mutations reduce DNA binding.
Fig. 1. Mutations in dnaK, dnaJ, grpE and dnaA suppress the lethality of dnaA overexpression.
(A) The location of suppressor mutations indicated on domain structures of DnaA, DnaK, DnaJ and GrpE. Amino acid substitutions resulting from missense mutations are listed below each gene. Nonsense mutations are marked with asterisks. A 51-bp duplication within dnaK is marked with a triangle. (B) Viability of dnaA overexpressing cells without or with the suppressor mutations indicated in the presence of glucose or xylose. Strains were streaked on plates and incubated for four days at 22°C. (C) Flow cytometry profiles and phase contrast microscopy images of dnaA overexpressing cells with the suppressor mutations indicated. Strains were grown in the presence of xylose to induce dnaA overexpression. The parent dnaA overexpression strain is shown for comparison when grown in PYE with glucose or with xylose for 2 h to induce dnaA. (D) Western blots showing M2-DnaA levels in the parent dnaA overexpression strain grown in glucose or in xylose for 2 h, and in the suppressor strains grown in xylose. RpoA was a loading control. Band intensities were quantified (bottom); error bars represent standard deviations (n=3).
In 30 clones, there were no mutations in Pxyl, dnaA, or elsewhere on the plasmid, suggesting that each harbored a chromosomal suppressor. We sequenced the genomes of 10 of these clones. Strikingly, in all 10, the mutations mapped to dnaK, which encodes the major protein chaperone Hsp70 (Fig. 1A). Together with the co-chaperone DnaJ and the nucleotide exchange factor GrpE, this highly conserved chaperone promotes the folding of a wide range of proteins (Genevaux et al., 2007; Hartl and Hayer-Hartl, 2009). We then sequenced dnaK, dnaJ and grpE in our remaining 20 clones finding that each had a mutation in one of these three genes. In total, we identified 25 mutations in dnaK, 4 in dnaJ, and 1 in grpE (Fig. 1A, Table S1). Most mutations in dnaK resulted in substitutions of conserved residues in the ATPase and substrate binding domains. We also identified three nonsense mutations producing truncations and one 51 bp duplication within the dnaK coding region. All of the mutations identified in dnaJ were nonsense mutations that would retain the N-terminal J-domain, which interacts with DnaK (Genevaux et al., 2007).
In contrast to the parent dnaA overexpression strain, each suppressor strain tolerated growth in xylose when Pxyl-M2-dnaA was induced, but not in glucose when dnaA expression was only driven by the leaky expression of the Pxyl promoter (Fig 1B), indicating that the suppressor mutations caused a reduction in active DnaA. For each suppressor strain, flow cytometry and phase microscopy indicated that the replication overinitiation phenotype of the parent strain in xylose had been greatly reduced. In the parent strain, growth for two hours in xylose led to a significant accumulation of extra chromosomes per cell and an inhibition of cell division (Jonas et al., 2011) (Fig. 1C). In contrast, the suppressor strains harboring mutations in dnaA, dnaK, dnaJ, or grpE showed only a mild accumulation of chromosomes and were able to divide, although cultures of strains with mutations in dnaK, dnaJ, or grpE grew slowly and were often heterogeneous, with some filamentous and small cells. Quantitative immunoblotting revealed that the suppressor strains with mutations in dnaK, dnaJ and grpE consistently contained lower levels of M2-DnaA when grown in xylose (Fig. 1D), suggesting that mutations in the chaperone system alter DnaA steady-state levels.
DnaK is required for DNA replication and DnaA stability
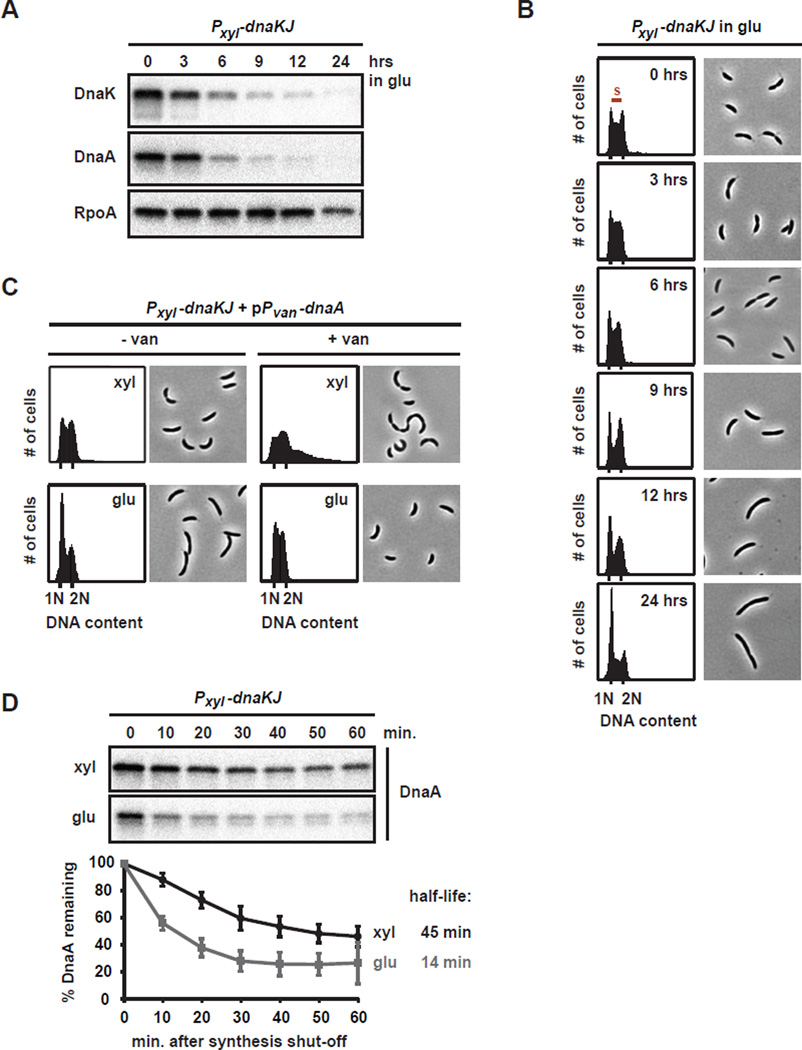
DnaK is essential for viability in C. crescentus (da Silva et al., 2003). To investigate the role of DnaK in regulating DNA replication and DnaA, we used a depletion strain, in which the co-operonic dnaK and dnaJ genes are under the control of Pxyl at the native dnaKJ locus. Shifting cells from xylose to glucose resulted in the depletion of DnaK within 9 hours (Fig. 2A). Flow cytometry revealed that over the course of DnaK/J depletion fewer cells entered S-phase while cells with a single chromosome accumulated, indicating that loss of DnaK/J resulted in an inability to initiate DNA replication (Fig. 2B). Consistent with previous data (Susin et al., 2006), prolonged depletion of DnaK/J led to filamentous cells without mid-cell division septa, a phenotype typical for G1 arrested Caulobacter cells.
Fig. 2. DnaK/J depletion results in the rapid degradation of DnaA and a G1 arrest.
(A) Western blots of DnaK, DnaA and RpoA in the DnaK/J depletion strain after growth in PYE with glucose for the time indicated. (B) Flow cytometry profiles and phase contrast microscopy images of the DnaK/J depletion strain grown in PYE with glucose for the times indicated. Cells with a DNA content between 1N and 2N are in S-phase. (C) Flow cytometry profiles and phase contrast microscopy images of the DnaK/J depletion strain overexpressing dnaA from a vanillate-inducible promoter on a plasmid. Cells were grown in PYE with glucose for 5 hrs, or in PYE with xylose, before vanillate was added to induce expression of dnaA for 4 hrs. (D) In vivo degradation assays showing DnaA stability in the DnaK/J depletion strain. Cells were grown in PYE with xylose (black) or PYE with glucose for 5.5 hrs (grey) to deplete DnaK/J, before chloramphenicol was added to shut-off protein synthesis. DnaA was monitored by Western Blotting (top). Band intensities were quantified (bottom); error bars represent standard deviations (n=3).
Overexpressing dnaA from a plasmid concomitant with the depletion of DnaK/J prevented cells from arresting in G1 (Fig. 2C). In contrast to cells that possess DnaK/J, overproducing DnaA in DnaK/J-depleted cells did not cause replication overinitiation (Fig. 2C), consistent with the finding that point mutations in dnaK, dnaJ or grpE also suppressed replication overinitiation (Fig. 1). Together, our data demonstrate that the DnaK (Hsp70) chaperone is required for DNA replication and that overproducing DnaA can bypass the need for DnaK/J in replication initiation.
To test if the abundance of DnaA was affected by the loss of DnaK/J, we measured DnaA levels by immunoblotting. Strikingly, as DnaK/J were depleted, the level of DnaA also strongly declined. After 6 hours of DnaK/J depletion, DnaA was reduced by ~80% and after 9 hours it was barely detectable (Fig. 2A). In contrast to DnaA, levels of the loading control protein RpoA remained high. The decrease in DnaA coincided with the cessation of replication that follows depletion of DnaK/J (Fig. 2B) indicating that an absence of DnaA is a likely cause of the replication arrest. To assess whether the reduction in DnaA results from increased proteolysis, we measured DnaA degradation rates in vivo using chloramphenicol shut-off assays. In the presence of DnaK/J, DnaA had a half-life of ~45 min (Fig. 2D), slightly faster than previously reported (Gorbatyuk and Marczynski, 2005). In contrast, in cells depleted of DnaK/J for 5.5 hours, the half-life of DnaA was only 14 min, demonstrating that loss of DnaK/J results in more rapid degradation of DnaA in vivo.
Prolonged depletion of DnaK/J also reduced growth rate and led to lower rates of DnaA synthesis (Fig. S1, data not shown). However, DnaA synthesis did not decrease more than the synthesis of other proteins such as RpoA (Fig. S1), suggesting that active proteolysis is the primary mechanism driving a drop in DnaA levels following depletion of DnaK/J.
DnaA degradation depends on the protease Lon
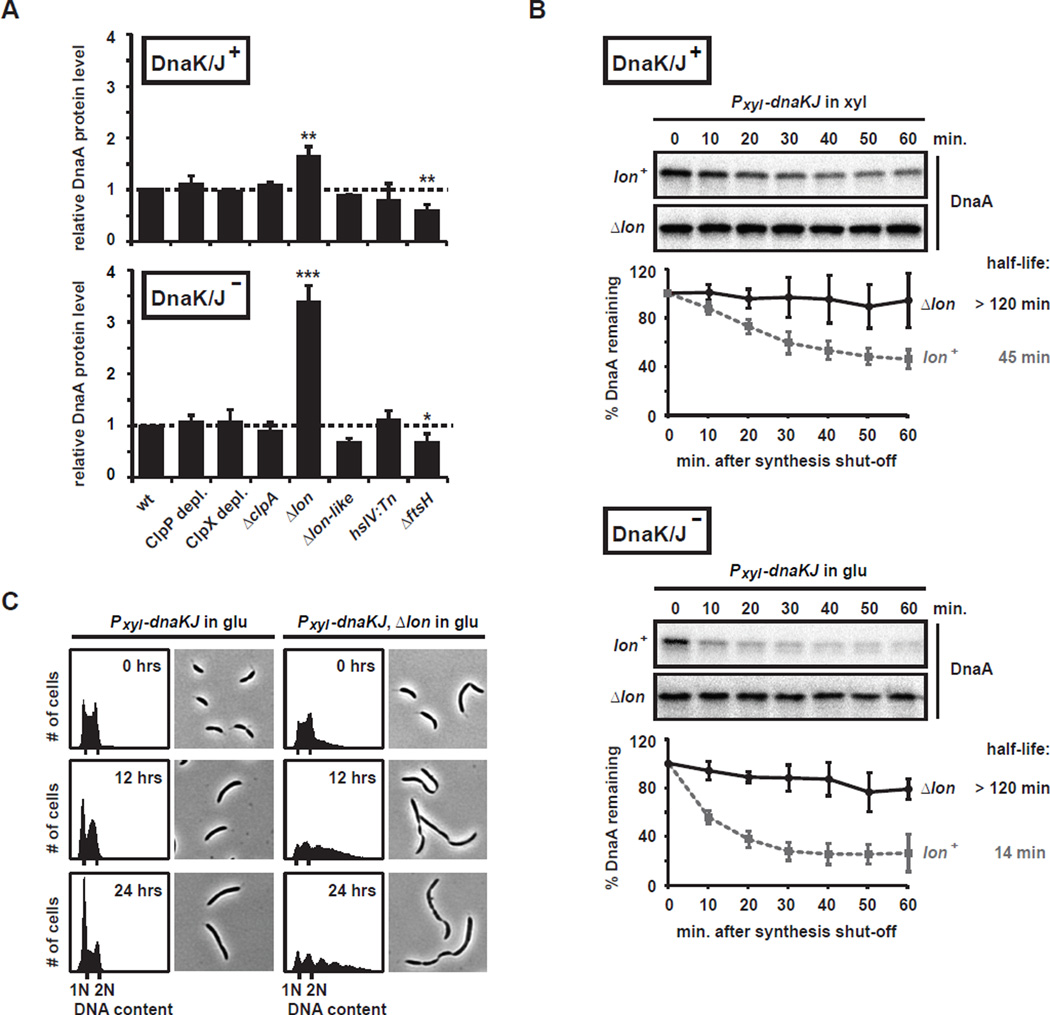
To determine which protease is responsible for DnaA degradation, we individually eliminated each of the five major ATP-dependent proteases in a DnaK/J depletion strain and measured the effect on DnaA steady-state levels by immunoblotting. We did not detect a significant change in steady-state DnaA levels after depleting cells of the essential protease subunits ClpP or ClpX for 8 hours, or in strains lacking functional ClpA, HslV, or a Lon-like protein (CCNA_00108), regardless of whether DnaK/J was depleted (Fig. 3A, Fig. S2). The only protease mutant that substantially increased DnaA levels was Δlon (Fig. 3A, Fig. S2). In the presence of DnaK/J, DnaA protein was slightly, but significantly, more abundant (~1.7-fold) in Δlon cells compared to cells harboring wild-type lon. After depleting DnaK/J for 6 to 9 hours, steady-state DnaA levels were 3–4 times higher in the Δlon mutant compared to cells expressing wild-type lon. Because DnaA synthesis rates were not affected by the Δlon mutation (data not shown), the observed changes in DnaA steady state levels are likely caused by stabilization of the protein. A deletion of ftsH resulted in a modest (30–40%) reduction in DnaA levels although this decrease may be indirect as Lon levels are increased ~50% in an ftsH mutant (data not shown).
Fig. 3. DnaA degradation in DnaK/J depleted cells depends on Lon.
(A) DnaA steady-state levels in various protease mutants expressing DnaK/J (top) or depleted for 6 hrs (bottom) as determined by Western blotting (Fig. S2). For the double depletion strains Plac-clpP; Pxyl-dnaKJ and Plac-clpX; Pxyl-dnaKJ, the protease subunits ClpP or ClpX, respectively, were pre-depleted for 3 hrs before depletion of DnaK/J was initiated. Bar graphs show relative DnaA levels normalized to the parent DnaK/J depletion strain. Error bars represent standard deviations (n ≥ 3, except for the lon-like mutant where n=2). Statistical significance was assessed by t-test (***, p<0.001; **, p<0.01; *, p<0.05). (B) In vivo degradation assay showing the effect of a Δlon mutation on DnaA stability in the DnaK/J depletion strain. DnaA stability in the DnaK/J depletion strain with wild type lon (lon+) is shown for comparison (Fig. 2D). Cells were grown in PYE with xylose (top), or in PYE with glucose for 5.5 hrs to deplete DnaK/J (bottom), before adding chloramphenicol to shut-off protein synthesis. DnaA levels were monitored by Western Blotting. Band intensities were quantified; error bars represent standard deviations (n=3). (C) Flow cytometry profiles and phase contrast images showing the phenotype of the DnaK/J depletion strain containing a lon deletion after 0, 6 and 12 hrs of DnaK/J depletion. For comparison, data for the DnaK/J depletion strain expressing wild type lon are shown (Fig. 2B).
To examine the effect of Lon on DnaA stability we performed in vivo degradation assays. As noted, the depletion of DnaK/J results in the rapid degradation of DnaA, with a protein half-life of ~14 minutes (Fig. 2D, 3B). Consistent with the observed increase in DnaA steady-state levels (Fig. 3A, Fig. S2), we found that a deletion of lon caused a dramatic stabilization of DnaA, both in the presence and absence of DnaK/J (Fig. 3B). In contrast to DnaA, CtrA (a ClpXP substrate) was not stabilized by a Δlon mutation (Fig. S3A). We also measured DnaA stability in cells depleted of ClpP, but did not see an effect after 8 hours of ClpP depletion in rich media (Fig. S3B–C). A previous study suggested that ClpP, but not ClpX or ClpA, affects DnaA stability (Gorbatyuk and Marczynski, 2005), although that work examined cells in minimal media depleted of ClpP for 12 hours which leads to a significant loss in cell viability (Jenal and Fuchs, 1998) and used a different antibody. We conclude that Lon is the primary protease for DnaA, but do not rule out a minor role for ClpP or a role in alternative conditions.
To characterize the phenotypic consequences of a Δlon mutation in a DnaK/J depletion strain, we analyzed DNA replication, and cell morphology over the course of DnaK/J depletion. Strikingly, the stabilization of DnaA following DnaK/J depletion in a Δlon strain was sufficient to rescue the replication initiation defect of a DnaK/J depletion alone (Fig. 3C). Instead of arresting with a single chromosome, cells harbored multiple chromosomes indicating that the stabilized DnaA protein was active for replication initiation. These cells also became filamentous with multiple division septa suggesting that a loss of DnaK/J and Lon somehow affects cell division. In contrast to Δlon, none of the other protease mutations had a replication phenotype that differed substantially from the DnaK/J depletion alone (data not shown).
Taken together, our data indicate that (i) Lon is a primary protease for DnaA and (ii) depletion of the DnaK/J chaperone promotes DnaA proteolysis by Lon.
Loss of DnaK/J promotes Lon synthesis through σ32
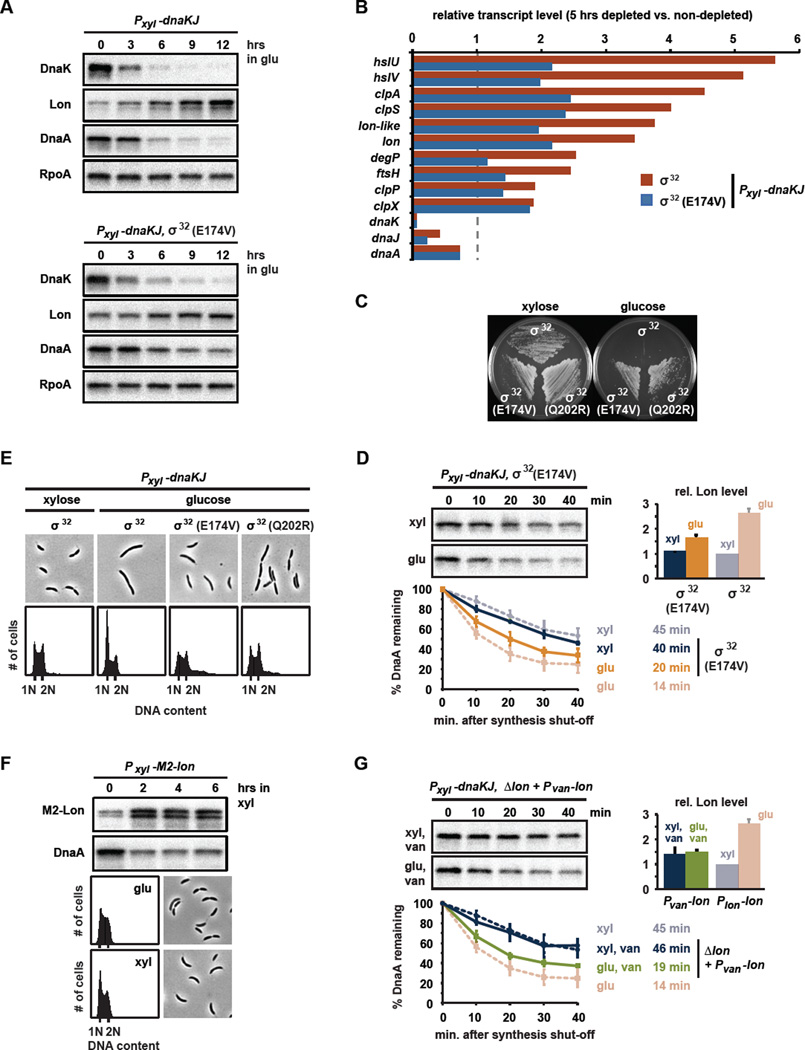
How does the absence or disruption of DnaK/J trigger increased degradation of DnaA by Lon? We initially hypothesized that the loss of DnaK/J might affect the abundance of Lon. Indeed, Lon levels steadily increased over the course of DnaK/J depletion (Fig. 4A). We suspected that the depletion of DnaK/J may induce Lon as part of the heat shock regulon that responds to proteotoxic stress. In E. coli, DnaK/J normally binds and destabilizes the sigma factor σ32, which can otherwise activate the heat shock regulon (Arsene et al., 2000), including lon (Phillips et al., 1984). In Caulobacter, DnaK likely operates similarly to prevent the accumulation of active σ32 under non-stress conditions (da Silva et al., 2003); hence, the depletion of DnaK should induce a heat shock response. Consistent with this hypothesis, we found, using DNA microarrays that the depletion of DnaK/J for 5 hours resulted in an upregulation of genes encoding many known heat-shock proteins (Fig. 4B, Table S4). Among the induced genes were all of the ATP-dependent proteases (Fig. 3A), including lon, which was 3.7-fold upregulated (Fig. 4B).
Fig. 4. DnaK/J prevents σ32-mediated upregulation of Lon synthesis.
(A) Western blots showing levels of DnaK, Lon, DnaA and RpoA while depleting DnaK/J in strains containing wild type σ32 (upper blots) or σ32(E174V) (lower blots), respectively. (B) Transcript levels in cells depleted of DnaK/J for 5 hrs, harboring wild type σ32 (red) or σ32(E174V) (blue) as determined by microarray analysis. Levels are relative to non-depleted cells expressing wild type σ32. (C) Viability of the DnaK/J depletion strain with or without the suppressor mutations σ32(E174V) and σ32(Q202R) in the presence of xylose or glucose. Plates were incubated for three days at 22°C. (D) DnaA stability in DnaK/J depleted cells containing the mutation σ32(E174V). DnaA degradation was monitored for cells grown in xylose (blue) or in glucose for 5.5 hrs to deplete DnaK/J (orange). For comparison, data for the DnaK/J depletion strain with wild type σ32 are shown in faded color. Bar graph shows Lon levels in each condition. Means and standard deviations (n≥2) are shown. (E) Flow cytometry and phase contrast images of the DnaK/J depletion strain containing suppressor mutations that produce σ32(E174V) or σ32(Q202R). Strains were grown in PYE with glucose. For comparison, data for the DnaK/J depletion strain with wild type σ32 before and after depletion for 24 hrs are shown (Fig. 2A). (F) DnaA and Lon levels were monitored by Western blotting after inducing lon from Pxyl on a medium copy vector for the times indicated. Flow cytometry and phase contrast images show the effects on DNA replication and cell morphology, respectively. (G) DnaK/J depletion promotes DnaA degradation despite unchanged levels of lon transcription. Pvan-lon was induced with vanillate from a low copy plasmid in the DnaK/J depletion strain containing a chromosomal deletion of lon. DnaA degradation was monitored in cells grown in xylose (blue) or in glucose for 5.5 hrs (green) to deplete DnaK/J. For comparison, data for the DnaK/J depletion strain expressing lon from its native promoter (Plon) are shown in faded color. The bar graph shows Lon levels in each condition. Means and standard deviations (n≥2) are shown.
Importantly, the transcriptional changes induced by the depletion of DnaK/J were attenuated in a strain producing the mutant σ32(E174V), indicating that DnaK/J affects the heat shock regulon at least partly through σ32 (Fig. 4B). We had identified σ32(E174V), along with σ32(Q202R), as suppressors of the lethality of a DnaK/J depletion strain (Fig. 4C). In Caulobacter, σ32 is essential for viability so these mutations are likely partial loss-of-function alleles. These residues (E174 and Q202) are located in the conserved region 3 of σ32, which is required for binding to RNA polymerase. In E. coli this region also directly interacts with DnaK (Rodriguez et al., 2008).
Consistent with the microarray data, we found that Lon protein increased only modestly over the course of DnaK/J depletion in a strain harboring σ32(E174V) (Fig. 4A). DnaA stability was slightly increased in the σ32(E174V) strain compared to DnaK/J depleted cells with wild-type σ32 (Fig. 4D). DnaA steady-state levels also remained slightly higher during DnaK/J depletion (Fig. 4A) in the σ32(E174V) strain and these cells were able to initiate DNA replication despite the loss of DnaK/J (Fig. 4E). These results are consistent with a model in which the depletion of DnaK/J normally induces the σ32-dependent synthesis of Lon, helping to drive increased degradation of DnaA. Importantly, however, depleting DnaK/J in a strain expressing σ32(E174V) still led to increased degradation of DnaA and a drop in DnaA steady-state levels (Fig. 4A,C), suggesting that an increase in Lon may not be the only mechanism affecting DnaA stability.
To directly test if the upregulation of Lon was sufficient to cause increased DnaA proteolysis and a G1-arrest, we overexpressed lon from an inducible promoter on a medium-copy vector. Two hours after induction Lon levels were 3- to 4-fold elevated, similar to the upregulation of Lon after 6 hours of DnaK/J depletion (Fig. 4A,F). We found that the higher levels of Lon indeed caused a drop in DnaA levels to ~35% of initial steady-state levels (Fig. 4F). However, unlike DnaK/J depleted cells, cells overproducing Lon were still able to grow and initiate replication. Thus, the transcriptional induction of Lon alone enhances DnaA degradation, but does not fully eliminate DnaA or induce a G1-arrest as seen in cells depleted of DnaK/J, suggesting that another mechanism may contribute to DnaA degradation. To further test this possibility, we assessed DnaA degradation following the depletion of DnaK/J in a strain that constitutively expresses lon. Indeed, for cells engineered to constitutively express lon, DnaA was degraded more rapidly in DnaK/J-depleted cells than in cells expressing DnaK/J even though Lon levels were constant and similar in the two conditions (Fig. 4G). Collectively, these data suggested that another mechanism, in addition to increased Lon synthesis, helps destabilize DnaA following the depletion of DnaK/J and onset of proteotoxic stress.
Unfolded proteins can allosterically activate Lon to degrade DnaA
To better understand the mechanisms that influence Lon-mediated degradation of DnaA, we attempted to reconstitute the reaction in vitro. Purified Lon alone did not rapidly degrade DnaA in conditions that did support efficient degradation of another Lon substrate, SciP (Fig. 5A, Fig. S4C) (Gora et al., 2013), suggesting that additional factors are required that promote the degradation of DnaA by Lon.
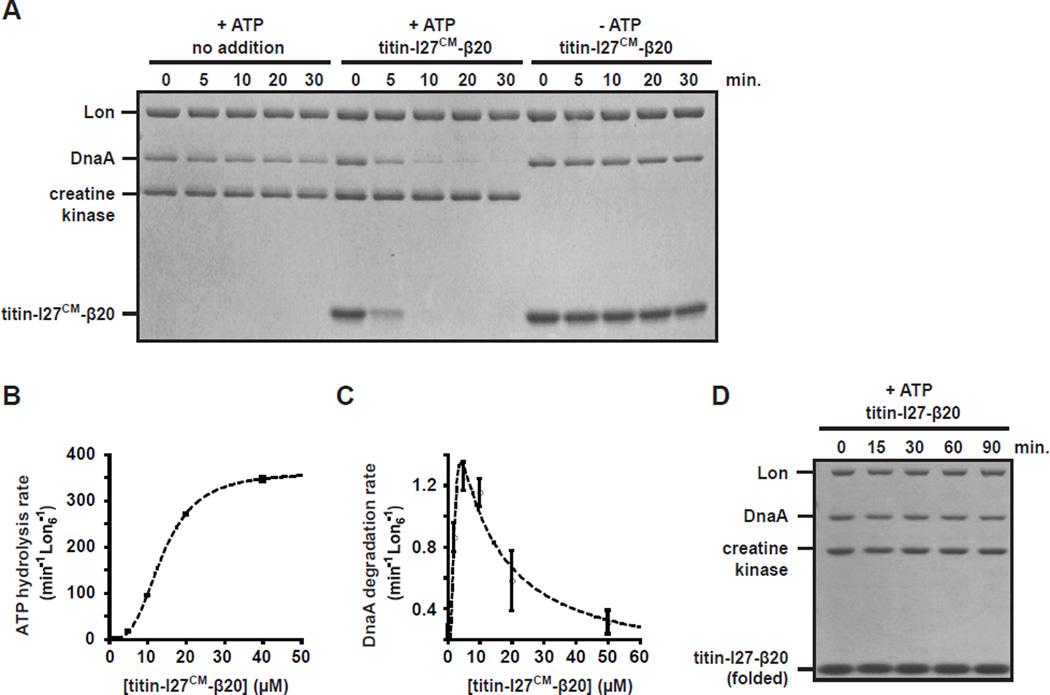
Fig. 5. Unfolded protein substrates stimulate Lon to degrade DnaA in vitro.
(A) Stimulatory effect of titin-I27CM-β20, an unfolded substrate, on DnaA degradation. DnaA (1.5 µM) was incubated with Lon6 (0.2 µM) in the absence or presence of titin-I27CM-β20 (10 µM). Creatine kinase is part of the ATP regeneration mix. (B) Interaction of titin-I27CM-β20 with Lon is positively cooperative. ATPase rate of Lon6 (0.1 µM) was measured in the presence of increasing concentration of titin-I27CM-β20. (C) Allosteric activation of Lon-mediated DnaA degradation. Degradation rates of DnaA (1.5 µM) by Lon6 (0.1 µM) are shown for increasing concentrations of titin-I27CM-β20. See methods for descriptions of curve fits. (D) Folded titin-I27-β20 does not stimulate DnaA degradation by Lon. DnaA (1.5 µM) was incubated with Lon6 (0.1 µM) in presence of native titin-I27-β20 (10 µM).
Prior studies of E. coli Lon indicated that the degradation of small peptides can be allosterically stimulated by other Lon substrates (Waxman and Goldberg, 1986) and that recognition motifs, or degrons, displayed by engaged substrates can convert Lon to a faster proteolysis state (Gur and Sauer, 2009). As Lon is the primary protease for degrading unfolded proteins following stress or DnaK depletion in E. coli (Tomoyasu et al., 2001; Tsilibaris et al., 2006), we speculated that the unfolded proteins that accumulate following DnaK/J depletion in Caulobacter might allosterically stimulate DnaA degradation. To test this hypothesis, we first investigated whether a carboxymethylated titin-I27 domain fused to a fragment of β-galactosidase (titin-I27CM-β20), a model unfolded protein substrate of E. coli Lon (Gur and Sauer, 2008) could stimulate DnaA degradation by Caulobacter Lon in vitro. Strikingly, addition of this substrate resulted in robust DnaA degradation with a half-life of < 5 minutes (Fig. 5A). DnaA degradation was ATP-dependent and likely did not result from a nonspecific activation of Lon as titin-I27CM-β20 did not increase degradation of CtrA or creatine kinase, a component of the reaction mix (Fig. 5A, S4A). Titration of titin-I27CM-β20 showed that increasing concentrations cooperatively activated Lon ATP hydrolysis (Fig. 5B) and that concentrations up to 5 µM titin-I27CM-β20 stimulated DnaA proteolysis, while a further increase reduced DnaA proteolysis (Fig. 5C). These findings support a model in which titin-I27CM-β20 stabilizes a conformation of Lon that promotes the binding and degradation of DnaA; however, at high concentrations, degradation of titin-I27CM-β20 itself competes with DnaA proteolysis.
In contrast to unfolded titin-I27CM-β20, the folded variant titin-I27-β20 did not stimulate DnaA proteolysis (Fig. 5D). However, folded titin-I27-β20 was poorly degraded by Lon itself, suggesting that only Lon substrates can stimulate DnaA proteolysis. Consistent with this notion, we found that casein, a natively unfolded protein and known Lon substrate (Charette et al., 1981), also stimulated Lon to degrade DnaA (Fig. S4B). Lon degradation of SciP was not stimulated by titin-I27CM-β20 and Lon degradation of CcrM, another Caulobacter Lon substrate (Wright et al., 1996), was only slightly enhanced by titin-I27CM-β20 (Fig. S4C,D), suggesting that the allosteric activation of Lon primarily affects a specific class of substrates that includes DnaA. DnaA degradation was not stimulated by higher temperature or upon denaturation of DnaA (Fig. S4E), indicating that a loss of DnaA´s native conformation is not sufficient to allow degradation by Lon in the absence of allosteric activators.
We conclude that unfolded proteins, which are themselves Lon substrates, allosterically stimulate Lon to recognize and robustly degrade DnaA. Together with our in vivo results, we propose that the depletion of DnaK/J results in a substantial accumulation of unfolded proteins that are substrates for Lon and that can also stimulate DnaA degradation, yielding a G1 arrest.
DnaK/J coordinates DNA replication with the heat shock response
Heat shock and other stress conditions can cause the accumulation of unfolded proteins, which bind DnaK/J and effectively sequester the chaperone, leading to release of σ32 and induction of the heat shock regulon (Arsene et al., 2000; Tomoyasu et al., 1998). Based on our finding that the depletion of DnaK/J triggers Lon to degrade DnaA, we hypothesized that heat shock might have a similar effect. To test this hypothesis, we examined how shifts in growth temperature affected DnaA and DNA replication. For wild-type cells shifted from 30 to 40°C we observed a rapid increase in DnaK levels (Fig. 6A, Fig. S5A), consistent with the notion that Caulobacter dnaK is activated by σ32 (Avedissian et al., 1995). The elevated level of DnaK likely prevented protein misfolding and a sustained activation of σ32. Consequently, and because Plon responds more slowly to σ32 than PdnaK (Fig. 6A) (Nonaka et al., 2006), Lon levels increased only mildly following the shift to 40°C, DnaA was not degraded, and DNA replication continued to initiate (Fig. 6B).
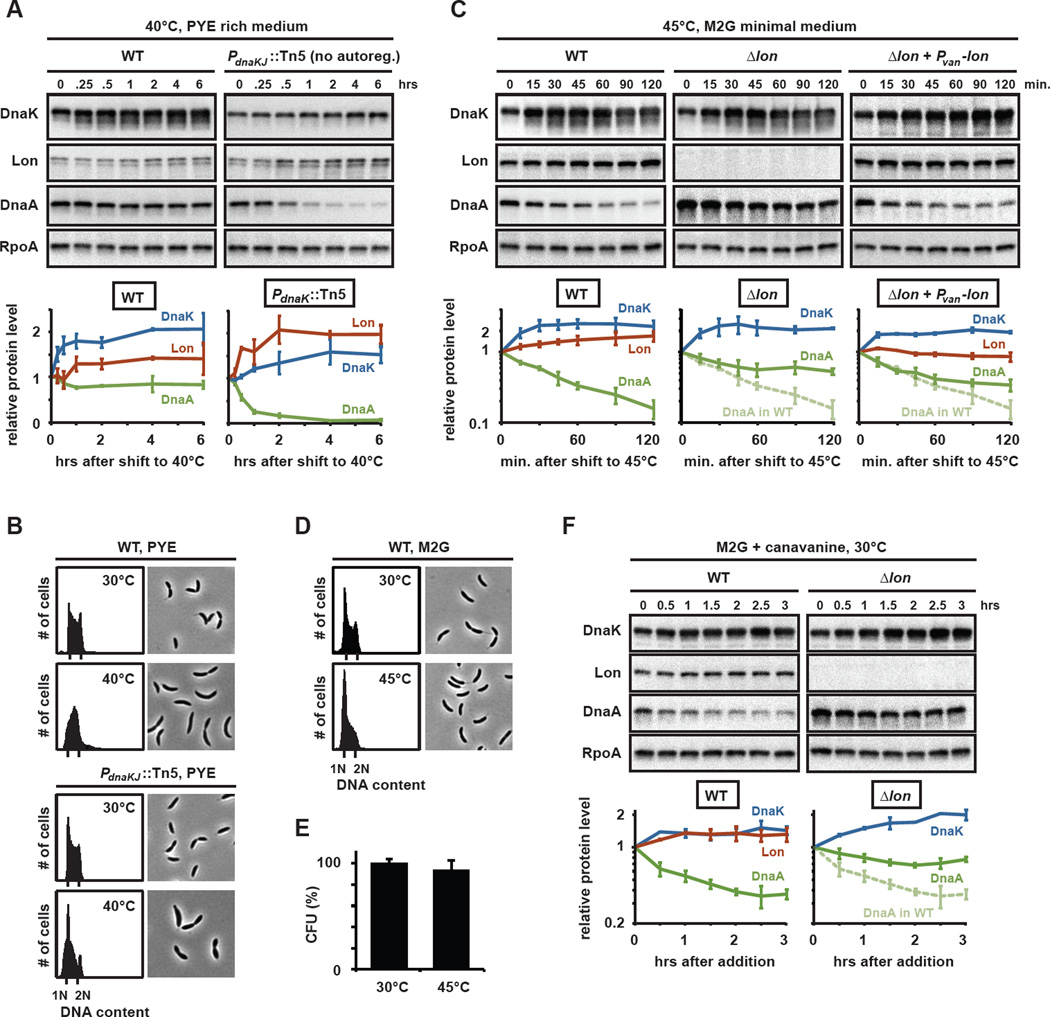
Fig. 6. Protein unfolding upon heat shock or canavanine treatment induces the rapid degradation of DnaA.
(A) DnaK, Lon, DnaA and RpoA levels in cells after shift to 40°C. Wild type or a mutant containing a transposon insertion in PdnaKJ and constitutively expressing dnaKJ were grown at 30°C and then shifted to 40°C. Protein levels were determined by Western blotting at the indicated time points after temperature shift (top) and quantified (bottom). Similar results were obtained in M2G (Fig. S5A). (B) Phase contrast images and flow cytometry profiles for wild type or PdnaKJ::Tn5 cells grown at 30°C or 40°C respectively. (C) DnaK, Lon, DnaA and RpoA levels measured by Western blots (top), with quantifications (bottom), in wild type, a Δlon mutant and in cells constitutively expressing lon (Δlon + Pvan-lon) grown in M2G medium and shifted to 45°C for the times indicated. Similar results were obtained in PYE (Fig. S5B). (D) Phase contrast images and flow cytometry profiles for wild type grown in M2G at 30°C or for two hours at 45°C. (E) Viability of cells grown in M2G at 30°C or for two hours at 45°C. (F) DnaK, Lon, DnaA and RpoA levels measured by Western blots (top), with quantifications (bottom), in wild type and Δlon after addition of canavanine to cultures grown in M2G. Means with standard deviations (n≥2) are shown.
To better assess the impact of heat shock on Lon, we used a strain harboring a transposon in the promoter of dnaK, such that the levels of DnaK remained nearly constant upon shift from 30 to 40°C (Fig. 6A, Fig. S5A). These cells have a reduced capacity to cope with misfolded proteins and should, therefore, mount a more sustained heat shock response, including the stimulation of Lon synthesis by σ32 and the allosteric activation of Lon by misfolded protein substrates. Consistent with these predictions, Lon levels increased and DnaA was cleared rapidly from these cells (Fig. 6A, Fig. S5A), causing a G1 arrest (Fig. 6B). Together, these experiments suggest that wild type cells cope with the protein misfolding induced at 40°C by adjusting the cellular concentration of DnaK. Cells that cannot upregulate DnaK accumulate unfolded proteins and degrade DnaA.
We hypothesized that a more severe heat shock that causes extensive protein misfolding might exceed the buffering capacity of DnaK in wild type cells leading to DnaA degradation by Lon. Indeed, when wild type cells were shifted to 45°C, we found that DnaA levels decreased, despite a rapid increase in DnaK levels (Fig. 6C, S5B). In Δlon cells DnaA levels did not decrease significantly after a shift to 45°C demonstrating that the elimination of DnaA upon heat shock largely depended on Lon-mediated degradation. Importantly, we found that DnaA levels also dropped after a shift to 45°C in a strain constitutively expressing lon, and thus deficient in upregulating Lon synthesis after heat shock. This finding is consistent with a model in which DnaA degradation is triggered through direct stimulation of Lon even without increased synthesis of the protease. Consistent with the decline in DnaA levels, we found that many cells arrested in G1 within two hours of growth at 45°C (Fig. 6D). Cell viability, however, remained largely unaffected (Fig. 6E).
To more directly test that unfolded proteins are the trigger for Lon-dependent degradation of DnaA in vivo, we treated wild-type cells with the amino acid canavanine which can be incorporated into proteins in place of arginine, preventing them from assuming normal conformations and consequently destabilizing them (Goff and Goldberg, 1985; Jubete et al., 1996). Addition of canavinine led to a significant decrease in DnaA, but only to a mild increase in DnaK or Lon (Fig. 6F). Moreover, the decrease in DnaA was dependent on Lon.
In sum, our data suggest that the depletion of DnaK or the sequestration of DnaK by severe, stress-induced protein unfolding results in the destabilization of DnaA and a consequent inhibition of DNA replication initiation. We conclude that DnaK-mediated regulation of DnaA stability is critical for cells to coordinate DNA replication and cell cycle progression with environmental conditions, such as heat shock, that lead to proteotoxic stress.
Discussion
In virtually all bacteria, cell cycle progression requires the accumulation of active DnaA to trigger the initiation of DNA replication. Despite decades of study, our understanding of how DnaA and replication initiation are regulated remains incomplete. Here, we presented evidence that a sophisticated regulatory network involving the DnaK/Hsp70 chaperone and the protease Lon controls DnaA stability in C. crescentus (Fig. 7A). We find that DnaK is required for the accumulation of DnaA and, consequently, for replication initiation. Loss of the chaperone system triggers the protease Lon to degrade DnaA. Degradation is stimulated both by increased Lon synthesis and through direct activation of the protease by misfolded proteins, which accumulate during certain stress conditions or upon chaperone loss.
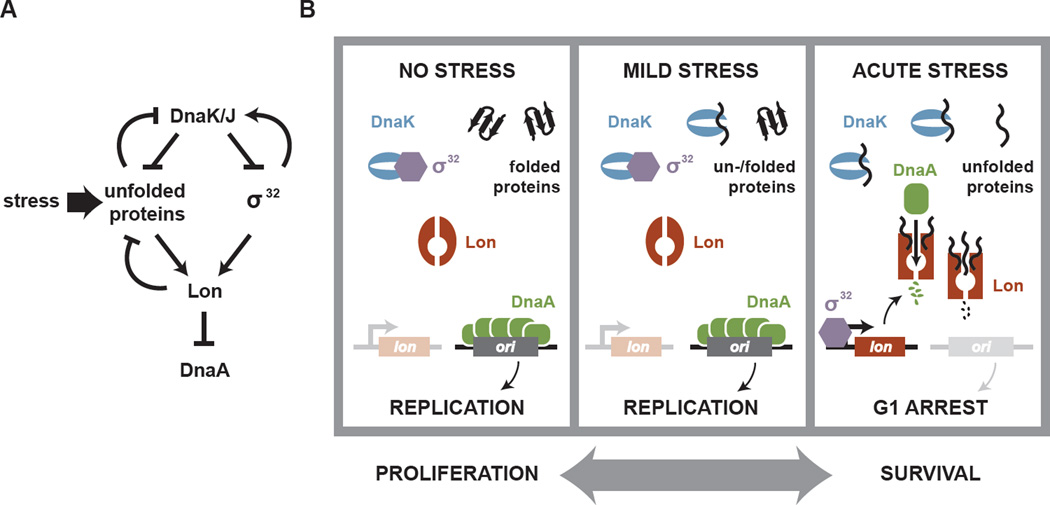
Fig. 7. Regulation of DnaA proteolysis by the DnaK/J chaperone and the Lon protease.
(A) Summary of the genetic relationship of DnaK/J, σ32, Lon and DnaA. (B) Model for the coordination of DNA replication with stress-induced protein misfolding. In the absence of stress DnaK promotes the folding of proteins and destabilizes σ32, allowing the stable accumulation of DnaA and DNA replication. With mild stress, DnaK is rapidly upregulated, preventing protein misfolding and a sustained activation of σ32. Under acute or severe stress, the dramatic increase in misfolded proteins exceeds DnaK capacity, leading to a sustained activation of σ32 and an upregulation of Lon synthesis. Additionally, unfolded proteins allosterically activate Lon to rapidly degrade DnaA. Consequently, DNA replication can no longer initiate until the stress subsides. The reciprocal coordination of DNA replication and the heat shock response allows cells to promote either a proliferative or survival mode.
Although best known for their roles in maintaining protein homeostasis (Hartl and Hayer-Hartl, 2009), molecular chaperones can have additional regulatory functions. DnaK has a clear role in regulating σ32 in E. coli (Arsene et al., 2000) and in Caulobacter (Fig. 4). Previous studies demonstrated that DnaK is also important for mini-F’ plasmid and λ phage replication in E. coli (Kawasaki et al., 1990; Yochem et al., 1978). However, in those cases, DnaK appears to directly influence plasmid-encoded replication factors and to promote release of the replicative helicase from the λ-OP preinitiation complex, respectively (Zylicz, 1993). It has long been suggested that DnaK also affects bacterial chromsome replication; in fact, the 'Dna' designation for DnaK and DnaJ was based on this idea (Yochem et al., 1978). However, a precise role for DnaK/J in controlling chromosomal replication has, until now, never been elucidated.
Coordination of DNA replication with stress-induced protein misfolding
The finding that DnaA stability is regulated by Lon and DnaK leads to a model for how DNA replication and the cell cycle are impacted by stress-induced protein unfolding (Fig. 7B). In the absence of stress, DnaK destabilizes σ32 thereby restraining lon expression. Additionally, DnaK directly promotes protein folding, which reduces the level of unfolded proteins that are both substrates for Lon and would otherwise stimulate the protease to degrade DnaA. Hence, by enforcing a low level of unstimulated Lon, DnaK promotes the accumulation of DnaA needed for replication initiation. Following mild heat shock, cells transiently upregulate DnaK in a σ32-dependent manner. These higher levels of DnaK prevent, or reverse, the accumulation of misfolded proteins such that Lon remains uninduced and DnaA remains stable. However, following a more severe heat shock, pervasive protein unfolding likely exceeds the buffering capacity of DnaK, leading to the sequestration of essentially all cellular DnaK, thereby freeing σ32 to induce expression of lon and other heat shock genes. The excess unfolded proteins also allosterically activate Lon, leading to the rapid degradation of DnaA. Consequently, during a severe or sustained heat shock, cells cannot initiate DNA replication and arrest in G1. While we have focused on heat shock, many other conditions can disrupt protein homeostasis (see Fig. 6F) and likely also trigger Lon-dependent degradation of DnaA and a cell cycle arrest.
More generally, our findings shed new light on the sophisticated mechanisms used by cells to regulate their behavior in response to changing environmental conditions. Under favorable conditions cells often strive to grow and proliferate. However, during stressful conditions, cells must stop replicating to maintain genome integrity and ensure cellular survival. This trade-off between proliferation and survival strategies requires the reciprocal coordination of two competing modes or cellular states, and often involves extensive transcriptional changes. For example, a suite of regulatory mechanisms ensure that bacteria employ different sigma factors during exponential growth and upon exposure to stress to ensure that global gene expression is appropriately geared to either growth or survival (Nystrom, 2004).
Our work highlights the importance of regulated proteolysis in the switch between proliferative and survival modes in Caulobacter. Conditions that lead to severe protein unfolding trigger an induction of chaperones that help mitigate the proteotoxic stress; simultaneously, these conditions lead to the rapid degradation of DnaA thereby preventing DNA replication in adverse conditions. Because of Lon's intrinsic ability to recognize many substrates, we suspect that additional regulatory proteins promoting growth and cell cycle progression are also degraded upon protein unfolding stress. Indeed, loss of the DnaK chaperone in E. coli also appears to trigger Lon to degrade the translation elongation factor-Tu (Bruel et al., 2012).
Arresting the cell cycle in response to proteotoxic stress is likely a broadly conserved phenomenon critical to the survival of most organisms. For example, in yeast, stress-induced protein misfolding, including growth at high temperature, induces a G1 arrest by triggering proteolysis of a critical cell cycle regulator, the cyclin Cln3 (Rowley et al., 1993; Trotter et al., 2001). Interestingly, both bacterial and eukaryotic responses appear to arrest replication initiation rather than elongation, presumably because an arrest during replication could result in incomplete replication and fork collapse.
Lon as a sensor of intracellular protein folding status
Lon is likely the primary protease for DnaA in Caulobacter. DnaA has a half-life of ~45 minutes under non-stress conditions and Lon was the only protease that affected steady-state levels of DnaA in rich media (Fig. 3A). Stressful conditions that lead to protein unfolding, including the depletion of DnaK/J or heat shock, trigger Lon to degrade DnaA at a faster rate. As noted, this increased rate of proteolysis stems from a combination of increased Lon synthesis and the allosteric activation of Lon. The transcriptional activation of lon by σ32 has been described in other bacteria (Tsilibaris et al., 2006), and we speculate that the allosteric activation mechanism uncovered here may also be widely conserved.
Hsp70 has long been recognized as a direct molecular sensor of misfolded proteins (Craig and Gross, 1991). Based on our finding that Lon is directly stimulated by unfolded proteins, we propose that Lon also acts as a molecular sensor that directly responds to changes in the cellular protein milieu by altering degradation of selected substrates. An allosteric mechanism to regulate Lon activity was suggested previously based on in vitro studies with E. coli Lon (Gur and Sauer, 2009; Waxman and Goldberg, 1986). However, the in vivo consequence of this regulation had been unclear as native targets affected by such an activation of Lon had not been identified. Our work highlights DnaA as one such target of Lon, but other proteins may also be degraded by Lon in a similar manner.
Our results indicate that the allosteric activators of DnaA degradation by Lon must themselves be Lon substrates. In addition to unfolded proteins, the folded Lon substrate SciP also stimulates DnaA degradation in vitro (data not shown). Folded Lon substrates may drive some degradation of DnaA in vivo, although the increased rate of proteolysis seen following heat shock and DnaK/J depletion suggests that allosteric activators of Lon may have to accumulate to high levels to fully stimulate DnaA proteolysis. Importantly, our results suggest that unfolded proteins do not simply stabilize Caulobacter Lon in a fast proteolysis conformation (Fig. S4), but instead selectively promote Lon's ability to recognize and degrade a specific set of substrates that includes DnaA. The exact biochemical mechanism by which unfolded proteins affect substrate recognition or processing remains to be elucidated. Understanding what features of DnaA are specifically recognized by Lon also remains an important challenge for the future.
Finally, we note that although our work does not point to a direct interaction between DnaK/J and DnaA, we do not rule out the possibility that DnaK/J may also influence the folding of all or part of DnaA, particularly during heat shock conditions. The folding of DnaA by DnaK/J could, in principle, help bury a region recognized by Lon and so further stabilize DnaA from degradation.
Protein homeostasis and cell cycle control
In sum, our work demonstrates that the status of intracellular protein folding plays a pivotal role in the regulation of DnaA and the Caulobacter cell cycle. Conditions that disrupt protein homeostasis result in the proteolysis of DnaA and a G1 arrest. The ability to block DNA replication in conditions that cause protein unfolding is likely a critical adaptive response that helps bacterial cells survive acute stress. Cells that attempt to replicate their chromosome in adverse conditions could compromise genome integrity and long-term survival. Finally, we note that the chaperone DnaK and Lon are among the most highly conserved proteins in bacteria; Hsp70 and AAA+ proteases are also found in almost all eukaryotic organisms. The use of unfolded proteins as a trigger for the proteolysis of specific substrates may be a common and powerful mechanism for coordinating cell cycle progression, or other cellular activities, with the onset of acute stress.
Experimental Procedures
A detailed description of all experimental procedures can be found in the Supplemental Information. Strains, plasmids, and primers used are listed in Tables S2 and S3.
Supplementary Material
Highlights.
The chaperone DnaK is required for DNA replication and DnaA stability in Caulobacter.
Loss of DnaK causes an upregulation of the protease Lon that can degrade native DnaA.
Unfolded proteins that are Lon substrates can directly stimulate Lon to degrade DnaA.
Stress-induced protein unfolding results in reduced DnaA levels and a G1 arrest.
Acknowledgments
We thank J. Modell for help analyzing Illumina data and members of the Laub lab and R.T. Sauer for critical discussion and comments on the manuscript. We acknowledge S. Gomes, R. T. Sauer, G.T. Marczynski, C. Aakre, N. Hickey, and D. Curran for reagents. M.T.L. is an Early Career Scientist at the Howard Hughes Medical Institute. This work was supported by a National Institutes of Health grant (5R01GM082899) to M.T.L, a National Institutes of Health grant (R00GM084157) to P.C. and a research fellowship (JO 925/1-1) from the German Research Foundation (DFG) to K.J. J.L. was supported in part by a Fellowship from the University of Massachusetts as part of the Chemistry-Biology Interface Training Program (T32 GM08515).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arsene F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Avedissian M, Lessing D, Gober JW, Shapiro L, Gomes SL. Regulation of the Caulobacter crescentus dnaKJ operon. J Bacteriol. 1995;177:3479–3484. doi: 10.1128/jb.177.12.3479-3484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Bruel N, Castanie-Cornet MP, Cirinesi AM, Koningstein G, Georgopoulos C, Luirink J, Genevaux P. Hsp33 controls Elongation Factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and Trigger Factor chaperones. J Biol Chem. 2012 doi: 10.1074/jbc.M112.418525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette MF, Henderson GW, Markovitz A. ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1981;78:4728–4732. doi: 10.1073/pnas.78.8.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, Shapiro L. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol. 2009;191:5706–5716. doi: 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Gross CA. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- da Silva AC, Simao RC, Susin MF, Baldini RL, Avedissian M, Gomes SL. Downregulation of the heat shock response is independent of DnaK and sigma32 levels in Caulobacter crescentus. Mol Microbiol. 2003;49:541–553. doi: 10.1046/j.1365-2958.2003.03581.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Fernandez C, Gonzalez D, Collier J. Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus. PLoS One. 2011;6:e26028. doi: 10.1371/journal.pone.0026028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli : a paradigm for the repartition of chaperone functions. Mol Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- Goff SA, Goldberg AL. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- Gora KG, Cantin A, Wohlever M, Joshi KK, Perchuk BS, Chien P, Laub MT. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2013;87:1277–1289. doi: 10.1111/mmi.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol Microbiol. 2001;40:485–497. doi: 10.1046/j.1365-2958.2001.02404.x. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc Natl Acad Sci U S A. 2009;106:18503–18508. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Ishida T, Akimitsu N, Kashioka T, Hatano M, Kubota T, Ogata Y, Sekimizu K, Katayama T. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J Biol Chem. 2004;279:45546–45555. doi: 10.1074/jbc.M402762200. [DOI] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Chen YE, Laub MT. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr Biol. 2011;21:1092–1101. doi: 10.1016/j.cub.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubete Y, Maurizi MR, Gottesman S. Role of the heat shock protein DnaJ in the lon-dependent degradation of naturally unstable proteins. J Biol Chem. 1996;271:30798–30803. doi: 10.1074/jbc.271.48.30798. [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem Cell Biol. 2010;88:49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Wada C, Yura T. Roles of Escherichia coli heat shock proteins DnaK, DnaJ and GrpE in mini-F plasmid replication. Mol Gen Genet. 1990;220:277–282. doi: 10.1007/BF00260494. [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: taking directions from the genome. Annu Rev Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley JA, Shapiro L. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol. 2008;190:6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski GT. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J Bacteriol. 1999;181:1984–1993. doi: 10.1128/jb.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Grossman AD. Control of the replication initiator DnaA by an anti-cooperativity factor. Mol Microbiol. 2011;82:434–446. doi: 10.1111/j.1365-2958.2011.07821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P. An expanded view of bacterial DNA replication. Proc Natl Acad Sci U S A. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- Phillips TA, VanBogelen RA, Neidhardt FC. lon gene product of Escherichia coli is a heat-shock protein. J Bacteriol. 1984;159:283–287. doi: 10.1128/jb.159.1.283-287.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol Cell. 2008;32:347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Rowley A, Johnston GC, Butler B, Werner-Washburne M, Singer RA. Heat shock-mediated cell cycle blockage and G1 cyclin expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1034–1041. doi: 10.1128/mcb.13.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin MF, Baldini RL, Gueiros-Filho F, Gomes SL. GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J Bacteriol. 2006;188:8044–8053. doi: 10.1128/JB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JA, Ouimet MC, Wargachuk R, Marczynski GT. The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol Microbiol. 2011;82:312–326. doi: 10.1111/j.1365-2958.2011.07785.x. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Ogura T, Tatsuta T, Bukau B. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol Microbiol. 1998;30:567–581. doi: 10.1046/j.1365-2958.1998.01090.x. [DOI] [PubMed] [Google Scholar]
- Trotter EW, Berenfeld L, Krause SA, Petsko GA, Gray JV. Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:7313–7318. doi: 10.1073/pnas.121172998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Tsokos CG, Laub MT. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol. 2012;15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman L, Goldberg AL. Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La) Science. 1986;232:500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- Wright R, Stephens C, Zweiger G, Shapiro L, Alley MR. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- Yochem J, Uchida H, Sunshine M, Saito H, Georgopoulos CP, Feiss M. Genetic analysis of two genes, dnaJ and dnaK necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol Gen Genet. 1978;164:9–14. doi: 10.1007/BF00267593. [DOI] [PubMed] [Google Scholar]
- Zawilak-Pawlik A, Kois A, Stingl K, Boneca IG, Skrobuk P, Piotr J, Lurz R, Zakrzewska-Czerwinska J, Labigne A. HobA - a novel protein involved in initiation of chromosomal replication in Helicobacter pylori. Mol Microbiol. 2007;65:979–994. doi: 10.1111/j.1365-2958.2007.05853.x. [DOI] [PubMed] [Google Scholar]
- Zylicz M. The Escherichia coli chaperones involved in DNA replication. Philos Trans R Soc Lond B Biol Sci. 1993;339:271–277. doi: 10.1098/rstb.1993.0025. discussion 277–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.