Abstract
Ubiquitin E3 ligase-mediated protein degradation promotes proteasomal degradation of key positive regulators of osteoblast functions. For example, the E3 ligases, Smurf1, Itch and Wwp1, promote degradation of Runx2, JunB, and CXCR4 proteins to inhibit their functions. However, the role of E3 ligases in age-associated bone loss is unknown. We found that the expression level of Wwp1, but not Smurf1 or Itch, was significantly increased in CD45− bone marrow-derived mesenchymal stem cells (MSCs) from 6- and 12-month-old WT mice. Wwp1−/− mice developed increased bone mass as they aged, associated with increased bone formation rates and normal bone resorption parameters. Bone marrow stromal cells (BMSCs) from Wwp1−/− mice formed increased numbers and areas of alkaline phosphatase+ and Alizarin Red+ nodules and had increased migration potential towards CXCL12 gradients. Runx2, JunB and CXCR4 protein levels were significantly increased in Wwp1−/− BMSCs. Wwp1−/−BMSCs had increased amount of ubiquitinated JunB protein, but Runx2 ubiquitination was no change. Knocking-down JunB in Wwp1−/− BMSCs returned Runx2 protein levels to that in WT cells. Thus, Wwp1 negatively regulates osteoblast functions by affecting both their migration and differentiation. Mechanisms designed to decrease Wwp1 levels in BMSCs may represent a new approach to prevent the decrease in osteoblastic bone formation associated with aging.
Keywords: Wwp1, mesenchymal stem cells, osteoblasts, bone formation
Introduction
Osteoporosis is a common skeletal disorder with an age-related reduction in bone mass and increased fracture risk that affects the quality of life of a growing population worldwide. Dysfunction of osteoblasts is a significant contributor to the pathogenesis of osteoporosis, but the mechanisms involved remain inadequately understood (1–4). Osteoblasts differentiate from mesenchymal stem cells (MSCs), a process that is tightly controlled by several transcription factors, including BMP-Smads and Runx2 (5, 6). The BMP-Smad-Runx2 pathway is regulated at both transcriptional and translational levels (7, 8) Ubiquitination and subsequent degradation of BMP-Smad-Runx2 proteins is an important regulatory mechanism in osteoblasts and their precursors and is carried out by members of the C2-WW-HECT family of ubiquitin E3 ligases. This class of E3 ligases consists of Nedd4-1, Nedd4-2, Itch, Smurf1, Smurf 2, Wwp1 and Wwp2 (9, 10) and promotes ubiquitination and subsequent proteasomal or lysosomal degradation of target proteins (11, 12). Previous studies have reported roles for Smurf1, Smurf2, Wwp1 and Wwp2 in bone cells. For example, Smurf1−/− mice develop age-related bone loss by promoting degradation of the MAP kinase, MEKK2 (13). The inflammatory cytokine, TNF, increases Smurf1 levels and this promotes ubiquitination and degradation of BMP-Smad-Runx2 proteins in mature osteoblasts (12, 14) Smurf2 negatively regulates chondrocyte function by regulating the stability of Smad3 and GSK3 proteins (15). Wwp2−/− mice develop malformations of the craniofacial region due to failure of Wwp2 to interact with the homeobox transcription factor, Goosecoid (16). Wwp1 negatively regulates osteoblast functions as revealed by the osteosclerotic phenotype of Schnurri-3 (Shn3)−/− mice; Shn3 enhances Runx2 ubiquitination and degradation via the recruitment of Wwp1 to Runx2 (17). We reported that Wwp1 is involved in TNF-induced bone loss by promoting proteasomal degradation of JunB proteins in osteoblasts and their precursors (18). JunB is a well known positive regulator of osteoblast differentiation (19, 20) and its degradation is mediated by Smurf1 and Wwp1 (18, 21). However, the role of HECT family of ubiquitin E3 ligases in age-associated osteoblast dysfunction in vivo has not been investigated in detail.
MSCs migrate to bone surfaces where they differentiate into osteoblasts. MSC migration is regulated by the CXCL12/CXCR4 axis (22, 23). Over-expression of CXCR4 in MSCs increases bone healing in mouse bone defects models because more CXCR4-expressing cells are recruited to the injury site (24). Depletion of CXCR4 in osteoblast precursors reduces postnatal bone formation (25). In cancer cells, CXCR4 is regulated partially through CXCL12-mediated lysosomal degradation, which involves Wwp1 (26). However, whether aging affects CXCL12-mediated cell migration and if this involves Wwp1 is unknown.
In this study, we investigated the role of Wwp1 in bone volume and osteoblast and osteoclast functions in young and aged mice. We demonstrate that Wwp1 negatively regulates bone mass by inhibiting MSC migration and osteoblast differentiation without affecting osteoclast formation. MSCs from Wwp1−/− mice have significantly elevated expression levels of the Wwp1 target proteins, Runx2, JunB and CXCR4. More importantly, Wwp1−/− mice are protected from age-related bone loss. Our findings identify Wwp1 as a new negative regulator of osteoblast functions and suggest that targeting Wwp1 may represent a novel therapeutic strategy to prevent age-related bone loss.
Materials and Methods
Animals
Mice used in this study were 1 to 12 months old. Wwp1+/− mice (on a C57/BL6 background) were obtained from Dr. L. Matesic (University South Carolina) and bred to generate Wwp1−/− mice. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Rochester Medical Center.
MSC-enriched CD45− cells and bone marrow stromal cells
Primary bone marrow stromal cells (BMSCs) were incubated with anti-CD45 antibody-conjugated microbeads (Miltenyi Biotec, Auburn, CA) and CD45-negative (CD45−) cells were isolated by negative selection, according to the manufacturer’s instructions. With this method, we can obtain 98% of the CD45− cells and these were used as MSC-enriched cells, as we described previously (18, 21). To generate a large quantity of MSC-enriched cells, we used 3rd passage bone marrow stromal cells and stained them with fluorescein isothiocyanate (FITC)-anti-CD45, FITC-anti-CD11b, FITC-anti-CD31, PE-anti-CD105, APC-anti-CD44, and PE-Cy5-Sca-1 (eBioscience, San Diego, CA, USA) and subjected to FACS analysis. The results were analyzed using FlowJo 7.6 software, indicating that this cell preparation contains enriched cells expressing MSC surface markers. Thus 3rd passage BMSCs were used in experiments requiring large numbers of cells. To examine frequency of cells expressing MSCs, bone marrow cells were stained with a panel of antibodies, including CD45, Scal-1, CD105, and Ter119, that distinguish mesenchymal cells from hematopoietic cells. MSCs were defined as CD45−Scal-1+CD105+Ter119− according to literatures (27).
Quantitative reverse transcriptase-PCR
Total RNA was extracted from the CD45− cells or osteoblast using TRIzol reagent (Invitrogen). cDNAs were synthesized using the iSCRIPT cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Quantitative RT-PCR amplifications were performed in an iCycler using iQ SYBR Green supermix (Bio-Rad). Gapdh was amplified on the same plates and used to normalize the data. Each sample was prepared in triplicate and each experiment was repeated at least three times. The relative abundance of each gene was calculated by subtracting the CT value of each sample for an individual gene from the corresponding CT value of Gapdh (ΔCT). ΔΔCT was obtained by subtracting the ΔCT of the reference point. These values were then raised to the power two (2ΔΔCT) to yield the fold-expression relative to the reference point. Expression levels of Smurf1, Runx2, alkaline phosphatase (ALP), Wwp1, Itch and Gapdh mRNA were examined using sequence specific primers, as described previously (18).
Osteoblast and adipocyte differentiation
Bone marrow cells were flushed from femorae and tibiae. For CFU-F and CFU-ALP colony assays, total bone marrow cells were cultured in 10cm dishes at 106 cells/dish in 10ml of α-MEM containing 10% fetal calf serum (Hyclone Laboratories, Logan, UT) + 50 μg/mL ascorbic acid and 10 mM β-glycerophosphate for 28 days. At the end of the culture period, cells were stained for CFU-F or ALP activity. The stained CFU-F and ALP positive CFU-F, which contained >20 cells in a single colony were counted. Bone nodule formation was examined in Alizarin red-stained plates after 21 days in culture under these conditions. For adipocyte differentiation assay, cells were allowed to become confluent and then cultured in adipocyte-inducing medium (AIM, α-MEM containing 10% FBS, 1μM dexamethasone, and 0.5 mM methylisobutylxanthine, 10 μg/ml of insulin, and 100 μM indomethacin) for 3 days and then cultured in the adipocyte-maintaining medium (AMM, a-MEM containing 10% FBS and 10 μg/ml of insulin) for 2 days. After the first round of AIM and AMM treatment, cells were subjected to a second round of treatment with AIM and AMM until adipocytes formed. Adipocyte staining with oil red O was performed, as described previously (28).
Western blot analysis and Ubiquitination assay
Cells were lysed with mammalian protein extraction reagent (Pierce) containing a protease inhibitor mixture (Roche Applied Science). Samples were resolved by SDS-PAGE gel, transferred to a nitrocellulose membrane, and immunoblotting was carried out using antibodies to Wwp1 (Novus Biological), CXCR4 (Calbiochem), Smurf1 (Abcam), Runx2, JunB, and GAPDH (Santa Cruz Biotechnology Inc). Bands were visualized using ECL chemiluminescence (Amersham) and quantitated by Scion Image Beta 4.02 (Scion Corporation, NIH). For ubiquitination assay, BMSCs were treated with the proteasome inhibitor MG132 (10 μM) for 4 hours. Whole cell lysates (500 μg protein/sample) were incubated with UbiCapture-Q Matrix (Biomol) by gentle agitation at 4′ overnight to pull down all ubiquitinated proteins according to the manufacturer’s instructions. After washing three times, captured proteins were eluted with 2× SDS-PAGE loading buffer and analyzed by Western blotting using anti-JunB or anti Runx2 antibody, as described previously (18).
Small interfering RNAs (siRNA)
JunB siRNA and control siRNA were purchased from Santa Cruz. BMSCs from from Wwp1−/− mice and WT littermates were transfected with JunB siRNA or control siRNA using Santa Cruz Biotechnology’s siRNA Transfection Reagents (sc-29528 and sc-36868) according to the manufacturer’s instructions. Whole cell lysates were harvested xx hours after siRNA transfection and expression levels of JunB and Runx2 were examined by Western analysis.
Microcomputed tomography (Micro-CT)
Long bones were scanned on a VivaCT40 micro-CT scanner (Scanco Medical, Basserdorf, Switzerland) using 300 ms integration time, 55 kVp energy, and 145 μA intensity. A volume of interest for quantitative analysis of trabecular bone was defined, extending from the proximal to the distal end of the tibia. Three-dimensional images were generated using a constant threshold of 275 for all samples. For each sample, bone volume fraction, trabecular number, trabecular thickness, trabecular separation, and connectivity density were measured.
Histology
Bones were fixed in 10% formalin and decalcified in 10% EDTA solution for 14 days. Decalcified bones were dehydrated and embedded in paraffin, after which 5-μm sections were cut on a rotary microtome. Sections were stained with hematoxylin and eosin (H&E) or histochemically for tartrate-resistant acid phosphatase (TRAC) activity to help identify osteoclasts. For calcein labeling, mice were injected intraperitoneally with 10 mg calcein per gram of body weight (C-0875; Sigma) at 7 days and 1 day prior to sacrifice, as reported previously (18). Left femurs were embedded in LR white acrylic resin (L9774; Sigma), and 5-μm sections were viewed and imaged using fluorescence microscopy. Mineral apposition rate (MAR, μm/day), bone formation rate (BFR, μm3/μm2/day) and mineralizing surface per bone surface (MS/BS) were calculated according to the reported formula (18). TRAP staining was performed using a modification of a previously described protocol (29) In brief, dewaxed sections were preincubated in Naphthol AS-BI Phosphate Substrate (Sigma) solution and then incubated in Sodium Nitrite (Sigma) and Pararosaniline Dye (Sigma) solution. After washing with distilled water, sections were incubated in phosphomolybdic acid for 5 min, counterstained with 1% fast green, and mounted in Cytoseal 60 (Richard-Allan) mounting Medium.
Cell migration assay
C3H10T1/2 cells and BMSCs were labeled with Calcein AM (Molecular Probes, Invitrogen) at a final concentration of 2 μg/ml as we reported previously (26). Cells (106 cells/100 μl) were loaded into the upper chamber of a transwell insert (5 μm, Corning Costar). Transwell inserts were immediately moved to wells of a 24-well tissue culture dish containing different doses of the chemokine, CXCL12 (1, 10, 50 or 100 ng/ml). After various times, cells that had migrated to the underlying wells were collected, centrifuged and solubilized (in 100 μl Hank’s Buffered Salt Solution with 1% SDS/0.2 N NaOH). Calcein AM labeling was read in a 96-well FluoroNunc plate (Nalge Nunc International) and quantified in a Gemini XS microplate spectrofluorometer (Molecular Devices) at 485 nm/530 nm.
Statistical Analysis
Data are presented as mean ± SD, and all experiments were performed at least three times with similar results. Statistical analyses were performed with Stat view statistical software (SAS, Cary, NC, USA, www.sas.com). Differences between the two groups were compared using unpaired Student’s t test, while more than two groups were compared using one-way analysis of variance between groups (ANOVA), and p values less than 0.05 were considered to be statistically significant.
Results
Increased expression of ubiquitin E3 ligase Wwp1 in MSC-enriched CD45− cells from aged mice
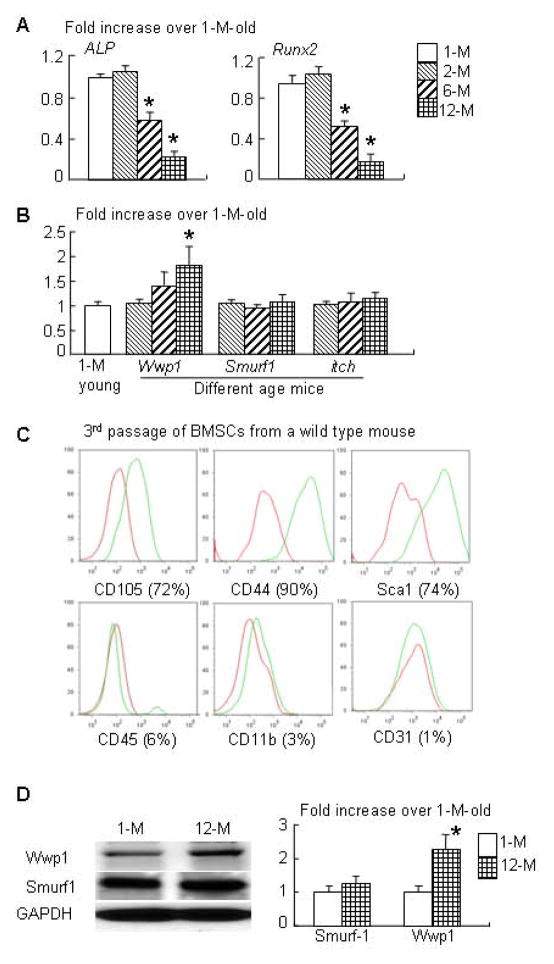
MSCs from aged mice have decreased osteogenic capacity via multiple mechanisms (30–32). The ubiquitin E3 ligases, Wwp1, Smurf1 and Itch, regulate the protein stability of the positive osteoblast regulators, Runx2, JunB and CXCR4 (12, 18, 33). To determine the role of E3 ligases in age-related osteoblast inhibition, we examined the expression levels of Wwp1, Smurf1 and Itch in MSC-enriched CD45− cells from 1-, 2-, 6-, and 12-month-old C57/B6 mice. As expected, the expression levels of the osteoblast markers, ALP and Runx2, were significantly decreased in cells from 6 and 12 month old WT mice (Fig. 1A). The expression levels of Wwp1 were significantly increased in cells from 12-month old mice, while Smurf1 and Itch levels were similar in cells from mice of different ages (Fig. 1B). In subsequent studies, we used 1-month old mice as young mice and 12-month-old mice as old mice.
Figure 1. MSC-enriched CD45− cells from old mice express decreased osteoblast marker genes and increased E3 ligase Wwp1.
Different ages of WT mice were used. CD45− cells were isolated from bone marrow cells via negative selection using micro-beads conjugated with anti-mouse CD45 antibody. The gene expression levels of osteoblast markers (ALP and Runx2, A) and E3 ligases (Wwp1, Smurf1 and itch, B) were determined by qPCR. Values are the means ± SD of 3 culture dishes. (C) 3rd passage of bone marrow stromal cells (BMSCs) were stained with MSC and non-MSC surface markers and subjected to FACS analysis. (D) Protein expression of Wwp1 and Smurf1 in BMSCs from young (1-month-old) and old (12-month-old) mice was determined by Western blot analysis. The fold changes were calculated from the intensity of bands on Western blot image using Scion Image Beta 4.02 (Scion Corporation, NIH). Values are the means ± SD of 3 separate experiments. *, p<0.05 vs data from 1-month-old mice.
Generating large numbers of CD45− MSC-enriched cells for in vitro mechanistic studies is technically challenging. Bone marrow stromal cells are often used in osteoblast studies. We found that 3rd passage bone marrow stromal cells contained a high percentage of cells expressing MSC surface markers, including CD105, CD44 and Scal1 and very low levels of non-MSC markers CD45, CD11b and CD31 (Fig. 1C). Thus, 3rd passage BMSCs (called BMSCs in this study) contains enriched MSCs and therefore were used. We confirmed that BMSCs from old mice expressed increased levels of Wwp1 protein, but not Smurf1 (Fig. 1D), showing a change similar to that of CD45− cells in Fig. 1B.
Increased bone volume and bone formation in Wwp1−/− mice
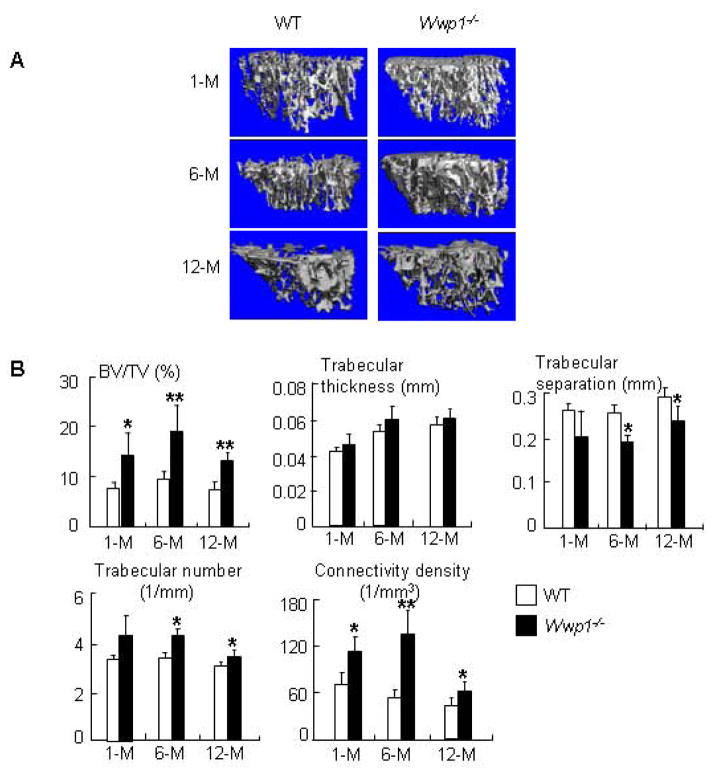
Wwp1 knockout (Wwp1−/−) mice have been generated, but their bone phenotype has not been investigated. We examined tibial bone volumes of 1-, 6- and 12-month-old Wwp1−/− mice and WT littermates. Micro-CT analyses (Fig. 2A&B) showed that 1-month-old Wwp1−/− mice had significantly increased trabecular bone volume, which became more obvious in 6- and 12-month-old Wwp1−/− mice and was accompanied by increased trabecular number and connectivity density, and reduced trabecular separation. Tibial cortical thickness was not increased in Wwp1−/− mice (data not shown).
Figure 2. Increased trabecular bone volume in Wwp1−/− mice.
Femurs isolated from 1-month-, 6-month- and 12-month-old Wwp1−/− mice and WT littermates were subjected to micro-CT analysis. (A) Representative 3D reconstructed images show significantly increased trabecular structure parameters in bones of Wwp1−/− mice. (B) Trabecular bone parameters from microCT analysis. Values are the means ± SD of 5 mice. *, p<0.05 vs data from WT mice.
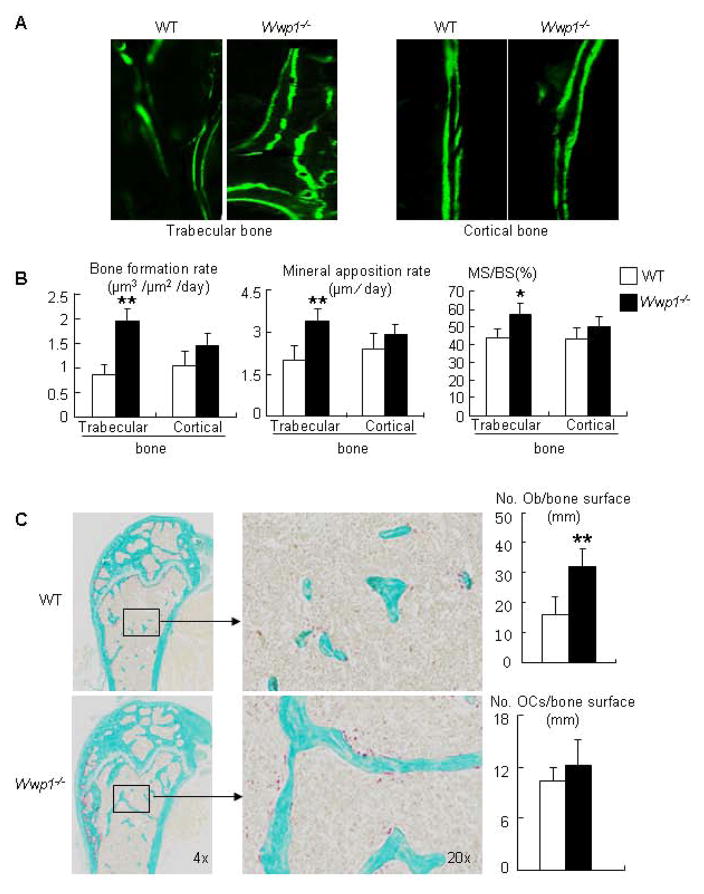
To determine if the increased bone volume in Wwp1−/− mice is associated with enhanced osteoblastic bone formation, we examined double calcein-labeled plastic-embedded sections and found that 6-month-old Wwp1−/− mice have significantly increased bone formation rate (BFR), mineral apposition rate (MAR), and mineralized surface (MS/BS) (Fig. 3A&B). No significant difference in any of these variables was detected in cortical bone endosteal surfaces. Histomorphometric analysis indicated that long bone sections from 6-month-old Wwp1−/− mice had increased osteoblast numbers, but normal osteoclast numbers (Fig. 3C). These in vivo data demonstrate that the increased bone mass in Wwp1−/− mice are due to increased osteoblast-mediated bone formation.
Figure 3. Increased bone formation in Wwp1−/− mice.
Six-month-old Wwp1−/− mice and WT littermates were labeled with calcein, and femurs were subjected to histologic examination. (A) Representative images of calcein-labeled trabecular and cortical bone. (B) Histomorphometric analyses of calcein-labeled sections. (C) Right panels: representative TRAP-stained sections. Left panels: histomorphometric analyses of TRAP-stained sections. Values are the means ± SD of 5 mice. **, p<0.01 vs data from WT mice.
Wwp1 negatively regulates osteoblast differentiation and migration
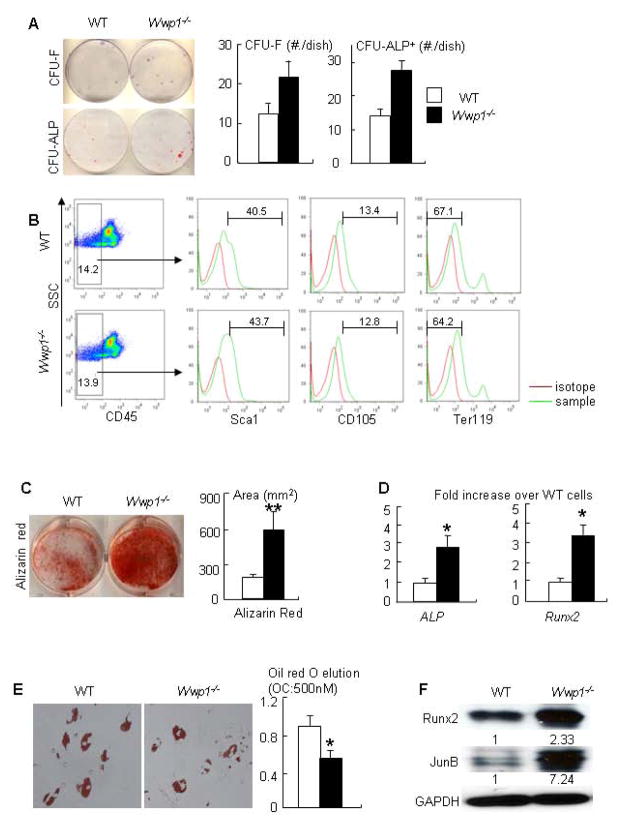
To determine the role of Wwp1 in osteoblast differentiation, we first examined colony forming potential of Wwp1-null bone marrow cells. BMSCs from Wwp1−/− mice were cultured in basal or in osteoblast-inducing medium. Compared with WT cells, Wwp1−/− BMSCs formed significantly increased numbers of CFU-F and CFU-ALP+ colonies (Fig. 4A). FACS of primary bone marrow cells showed the frequency of cells expressing MSC markers (CD45−/Scal+/CD105+/Ter119−) was similar among cells from Wwp1−/− mice and WT littermates (Fig. 4B). We then examine the osteoblast differentiation potential of Wwp1−/− BMSCs and found that they formed significantly increased area of calcified nodules (Fig. 4C), and expressed increased mRNA levels of ALP and Runx2 (Fig. 4D). In contrast to the increased osteoblast differentiation, adipocyte differentiation was significantly reduced in Wwp1−/− BMSCs (Fig. 4E). Wwp1 is known to mediate proteasomal degradation of the positive osteoblast regulators, Runx2 (17) and JunB (18). We found that Wwp1−/− BMSCs had a more than 2-fold increased level of Runx2 and JunB proteins (Fig. 4F), suggesting that Wwp1 controls osteoblast differentiation by changing the expression levels of these proteins.
Figure 4. MSC-enriched bone marrow stromal cells from Wwp1−/− mice have increased osteoblast differentiation.
BMSCs from 6-month-old Wwp1−/− mice and WT littermates were used. (A) Cells were cultured in the basal (CFU-F) or in the osteoblast-inducing medium (CFU-ALP) for 14 or 21 days respectively. Cells were stained with H&E or ALP. The number of colonies was counted (upper panels). The bone marrow cells from Wwp1−/− mice and WT littermates were stained with a panel of antibodies for MSC marker and subjected to FACS analysis (lower panels). (B) Primary bone marrow cells were stained with MSC and non-MSC surface markers and subjected to FACS analysis. CD45− cells were examined the frequency of cells expressing Sca1, CD105 and Ter119. (C) Cells were cultured in osteoblast-inducing medium for 21 days. Cells were stained with Alizarin Red and the area of calcified nodules was measured. (D) The expression levels of osteoblast markers (ALP and Runx2) were determined by qPCR. (E) Cells were cultured in adipocyte-inducing medium for 6 days and subjected to Oil red O staining. (F) Expression levels of Runx2 and JunB proteins by Western blot analysis. The fold changes were calculated as Fig. 1D. Values are the means ± SD of 3 separate dishes. *, p<0.05 or **, p<0.01 vs WT cells.
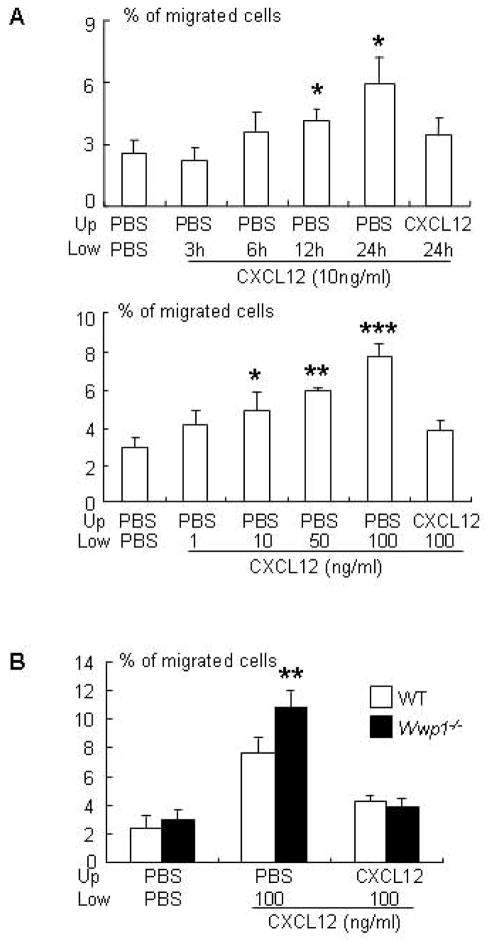
MSCs that differentiate into osteoblasts reside mainly within the bone marrow cavity and and osteoblast precursors that arise from them migrate to bone surfaces. The CXCL2/CXCR4 axis controls cell migration (22). In the bone marrow, CXCL12 is produced mainly by CXCL12-abundant reticular (CAR) cells on sinusoids (34) and perhaps also by osteoblastic cells on trabecular and cortical bone surfaces (35, 36). We recently found that Wwp1 negatively regulates the migration of cancer cells to CXCL12 by regulating degradation of CXCR4, the CXCL12 receptor (26). This finding led us to speculate that Wwp1−/− BMSCs may also have increased migration to CXCL12 as another mechanism for the high osteoblast-mediated bone mass in Wwp1−/− mice. To determine if Wwp1 affects cell migration, we first established an optimal protocol for CXCL12-mediated migration of WT BMSCs in a transwell migration assay. Time- and dose-dependent experiments demonstrated that 100ng/ml of CXCL12 induced maximal cell migration after 24 hours (Fig. 5A) and that Wwp1−/− BMSCs had significantly increased CXCL12-medicated migration (Fig. 5B).
Figure 5. MSC-enriched bone marrow stromal cells from Wwp1−/− mice migrate faster to a CXCL12 gradient.
(A) BMSCs from WT mice were labeled with calcein AM and seeded in the upper chamber of a transwell dish. CXCL12 (10 ng/ml) was added to the lower chamber and cells were incubated for different times (upper panel) or different doses of CXCL12 were added to the lower chamber and cells were incubated for 24 hrs (lower panel). The % of cells that migrated to the lower chamber was determined by measuring calcein intensity. (B) BMSCs from Wwp1−/− mice and WT littermates were labeled and seeded as above. CXCL12 was added to the lower chamber and cells were incubated for 24hrs. The % of cells that migrated to the lower chamber was determined. Values are the means ± SD of 3 well/group. *, p<0.05, **, p<0.01, and ***, p<0.001 vs PBS or WT group.
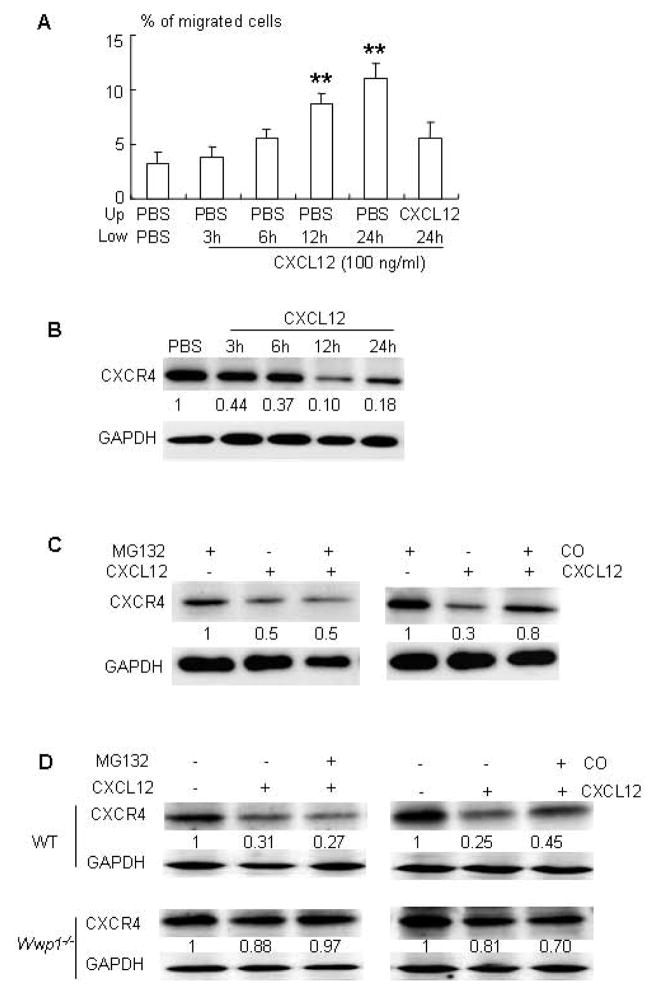
We reported that Wwp1 regulates CXCL12-induced CXCR4 lysosomal degradation in breast cancer cells, thereby affecting cell migration (26). To investigate if Wwp1 affects CXCR4 degradation in MSCs, we first used C3H10T1/2 cells, an embryonic mesenchymal cell line. C3H10T1/2 cells migrated toward a CXCL12 gradient (Fig. 6A). CXCL12 treatment markedly reduced CXCR4 protein levels, which peaked at 12 hours (Fig. 6B). To determine if CXCL12-induced CXCR4 degradation is through a proteasome or lysosome pathway, we treated cells with the proteasome inhibitor, MG132, or the lysosome inhibitor, chloroquine (CQ), in the presence of CXCL12. CQ partially prevented CXCL12-induced CXCR4 degradation, but MG132 had no effect (Fig. 6C). We then examined CXCR4 protein levels in BMSCs from Wwp1−/− mice in the presence of CXCL12 plus CQ or MG132. In WT cells, CXCL12 markedly decreased CXCR4 protein levels, an effect that was partially prevented by CQ, but not by MG132 (Fig. 6D, upper panels). However, CXCL12-induced CXCR4 degradation was significantly reduced in cells from Wwp1−/− mice (Fig. 6D, lower panels).
Figure 6. MSC-enriched bone marrow stromal cells from Wwp1−/− mice have decreased CXCL12-induced CXCR4 degradation.
(A–C) C3H10T1/2 cells were used. (A) Cells were labeled and seeded as Fig. 5A. CXCL12 was added to the lower chamber and cells were incubated for different times. The % of cells that migrated to the lower chamber was determined. Values are the means ± SD of 3 wells/group. **, p<0.01 vs PBS group. (B) Expression of CXCR4 protein in CXCL12-treated cells was determined by Western blot analysis. (C) Cells were pre-treated with the proteasome inhibitor, MG132, or the lysosome inhibitor, chloroquine (CQ), and then treated with CXCL12 for 12 hours. Expression of CXCR4 protein was determined. (D) BMSCs from Wwp1−/− mice and WT littermates were pre-treated with MG132 or CQ and then with CXCL12 as above. Expression of CXCR4 protein was determined by Western blot analysis and the fold changes were calculated as Fig. 1D.
Age-related decreased osteoblast function is rescued in Wwp1−/− mice
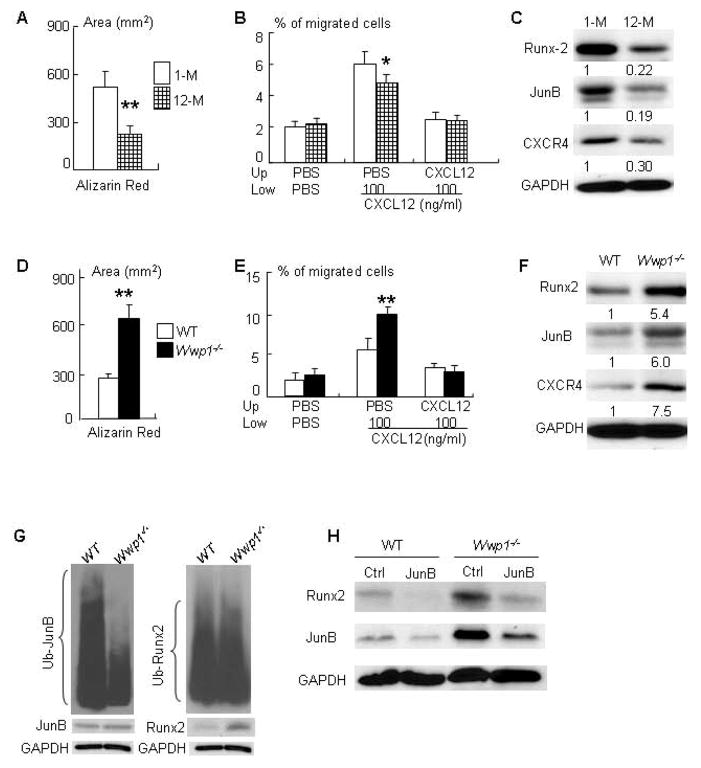
If elevated levels of Wwp1 in MSCs (Fig. 1) are responsible for reduced osteoblast function in old WT mice, the phenotypes in cells from old WT mice should be different from those from young WT and old Wwp1−/− mice. To test this, we first compared osteoblast differentiation, CXCL12-mediated migration, and expression of Wwp1 target proteins using BSMCs from young (1-month-old) and old (12-month-old) mice. BSMCs from old WT mice formed decreased Alizarin Red-stained calcified nodules (Fig. 7A), migrated less to a CXCL12 gradient (Fig. 7B), and expressed lower levels of Runx2, JunB and CXCR4 proteins (Fig. 7C) compared cells from young mice. However, in contrast to cells from old WT littermates, BMSCs from Wwp1−/− mice had increased Alizarin Red-stained calcified nodules (Fig. 7D), greater migration to a CXCL12 gradient (Fig. 7E), and higher expression levels of Runx2, JunB and CXCR4 proteins (Fig. 7F).
Figure 7. Decreased bone volume and osteoblast function in old mice is rescued in Wwp1−/−mice.
(A–C) BMSCs from young (1-month-old) and old (12-month-old) WT mice were used. (A) The area of alizarin-red-stained calcified nodules. (B) The % of CXCL12-mediated migration. Values are the means ± SD of 3 wells/group. *, p<0.05, **, p<0.01 vs data from young mice. (C) Expression of Runx2, JunB and CXCR4 proteins was determined by Western blot analysis and the fold changes were calculated as Fig. 1D. (D–F) BMSCs from old Wwp1−/− mice and WT littermates were used. (D) The area of alizarin-red-stained calcified nodules. (E) The % of CXCL12-mediated migration. Values are the means ± SD of 3 wells/group. *, p<0.05, **, p<0.01 vs data from WT littermates. (F) Expression of Runx2, JunB and CXCR4 proteins was determined by Western blot analysis and the fold changes were calculated as Fig. 1D. (G) BMSCs derived from Wwp1−/− mice and WT littermates were treated with MG 132 (10 μM) for 4 hours and then lysed for ubiquitination assays to detect endogenous ubiquitinated JunB and Runx2 proteins. (H) BMSCs from Wwp1−/− mice and WT littermates were transfected with JunB siRNA or control siRNA. Protein expressions of Runx2 and JunB were determined by Western blot analysis.
Wwp1 directly targets JunB protein for degradation in bone marrow stromal cells
Both JunB and Runx2 proteins positively regulate osteoblast differentiation and are Wwp1 substrates (17, 18). However, we found that cells from Wwp1−/− mice expressed increased levels of runx2 mRNA (Fig. 4D). If lack of Wwp1 mainly stabilized Runx2 protein from degradation, one would expect that protein levels would increase without major changes in mRNA. This raises an interesting question, e.g. are levels of Runx2 ubiquination actually lower in Wwp1−/− cells and it is possible that Runx2 is not the major substrate of this ligase and instead it is destabilizing another factor that regulates Runx2 expression? To address this question, we performed ubiquitination assay using BMSCs from Wwp1−/− mice to examine Runx2 and JunB ubiquitination because JunB regulates Runx2 expression in osteoblast/stromal cells (37). We found that a significantly reduced JunB ubiquitination in Wwp1−/− cells, but Runx2 ubiquitination is similar between WT and Wwp1−/− cells (Fig. 7G). Furthermore, knockdown of JunB in Wwp1−/− or WT cells resulted in significantly decreased Runx2 expression (Fig. 7H).
Discussion
We have demonstrated that MSC-enriched CD45− cells from aged mice have increased expression of Wwp1, an ubiquitin E3 ligase that promotes the degradation of target proteins. Wwp1−/− mice have increased bone mass. MSC-enriched bone marrow stromal cells (BMSCs) from Wwp1−/− mice have increased osteoblast differentiation, CXCL12-mediated migration, and expression of the known Wwp1 target proteins, Runx2, JunB and CXCR4. Consistent with increased Wwp1 expression, BMSCs from old mice have decreased osteoblast differentiation, CXCL12-mediated migration and Runx2, JunB and CXCR4 protein levels. Wwp1 deficiency prevented the bone loss and osteoblast inhibition observed in old mice. Thus, Wwp1 is a new negative regulator of osteoblast function and contributes to age-related bone loss by reducing the expression levels of key positive osteoblast proteins in MSCs.
Wwp1 promotes ubiquitination and degradation of multiple proteins including p53, KLFs (38, 39), Smad4 (40), Runx2 (41), JunB (18) and CXCR4 (26). Among Wwp1 target proteins, Runx2 and JunB play a critical role in osteoblast differentiation (19, 42) CXCR4 is essential for retention of hematopoietic stem cells in the bone marrow cavity (43) and controls MSC homing to injury sites in bones as well as osteoblast differentiation (44, 45). Elevated levels of Runx2, JunB and CXCR4 proteins in cells from Wwp1−/− mice suggest that these are potential endogenous targets of Wwp1 and are likely responsible for increased osteoblast-mediated bone formation in Wwp1−/− mice. However, our ubiquitination assay clearly indicates that Runx2 is not a direct substrate of Wwp1 because the amount of ubiquitinated Runx2 protein did not change in Wwp1−/− cells. Rather, the amount of ubiquitinated JunB was reduced in Wwp1−/− cells (Fig. 7G). JunB promotes Runx2 expression during bone formation (37) and JunB knocking-down in Wwp1−/− cells decreased Runx2 levels (Fig. 7H). These findings indicate that Runx2 is not the major substrate of Wwp1 and its primary endogenous target in osteo-progenitor cells is likely JunB. This finding is similar to the finding obtained from calvarial-derived osteo-progenitor from Smurf1−/− mice (13) in which Runx2 also is not considered as an endogenous target of Smurf1. Since early studies on Runx2 degradation are mainly carried out in cell lines with over-expression system, more experiments need to be done to elucidate the importance of controlling Runx2 degradation as a critical mechanism for osteoblast regulation.
One of important functions of E3 ligases is to promote ubiquitination and degradation of target proteins via proteasomal pathway. For example, Smurf1 promotes the proteasomal degradation of Smad1/5 (12), Runx2 (12, 46), JunB (21) and MEKK2 (13) proteins, all of these proteins are positive osteoblast regulators. However, ubiquitinated proteins also can undergo lysosomal degradation. E3 ligase itch-mediated CXCR4 degradation in lysosomal pathway has been reported (47). We reported that Wwp1 promotes CXCR4 lysosomal degradation in MDA231 breast cancer cells (48). Thus, Wwp1 may affect osteogenic function by regulating protein stability through both proteasomal and lysosomal pathways. In this study, we demonstrated that Wwp1-null cells migrate to a CXCL12 gradient faster as a result of reduced CXCR4 lysosomal degradation (Fig. 6). We suspect that increased migration of MSCs and osteo-progenitors in Wwp1−/− mice to CXCL-12 enriched bone surface serves as a mechanism for increased osteoblast function by providing more precursor cells. This speculation is based on finding that cells on the endosteal surface of the bone are stained positively for CXCL12 (36) and that systemic delivery of CXCR4-over-exressing cells prevents OVX-reduced bone loss (49). However, currently we do not have an in vivo cell migration assay that will allow us to examine if cells from WWp1−/− mice migrate to CXCL12 faster. Thus if CXCL12/CXCR4-mediated cell migration that we found using in vitro assay contribute to general bone remodeling needs further investigation.
Wwp1 is a member of C2-WW-HECT family E3 ligases and several of them regulate osteoblast functions and target some of the same proteins (refs). It will be important to study the specificity of individual E3 ligase in bone. We suspect that different E3 ligases regulate osteoblast function at different stages of differentiation and/or in different physiologic or pathologic settings. Wwp1 likely plays a more critical role in MSCs or osteoblast precursors in disease conditions, such as chronic inflammation (18) and aging. Ww1p may not play an important role in bone homeostasis because it is expressed at very low levels physiologically. Inflammation or the aging process up-regulate Wwp1 expression in MSCs and osteoblast precursors, leading to inhibition of bone formation as a consequence of increased degradation of positive osteoblast regulatory proteins.
Currently, we do not know why Wwp1 levels are increased in MSCs from old mice. Ubiquitin E3 ligases are largely activated constitutively and are regulated at the level of target protein interaction (50). However, they can also be regulated by phosphorylation (51), utilization of adaptor proteins (52) and at the transcription level. For example, LPS induces muscle atrophy by stimulating the expression of atrogin-like-muscle-specific E3 ligase through NF-κB (53). TNF rapidly (within 24 hours) increases Wwp1 mRNA expression in primary murine and human MSCs. This is strikingly faster than TNF-induced Smurf1 expression (3 days) (12), suggesting that TNF directly regulates Wwp1 transcription. We identified a putative NF-κB response element in the murine Wwp1 promoter. The role of NF-κB in osteogenesis of MSCs has not been extensively investigated. However, mice over-expressing an NF-κB inhibitor (54) in osteoblasts or with osteoblast-specific NF-κB knockout (55) have increased bone formation, indicating that NF-κB is a negative regulator of osteoblasts (56). Aging is a complicated process associated with low-grade chronic inflammation, including increased TNF levels (57, 58) and NF-κB activity (59). Thus, it is likely that these factors up-regulate Wwp1 expression, but this will require further investigation.
A previous study indicated that Wwp1 contributes to the high bone mass phenotype of Shn3−/− mice by promoting Runx2 ubiquitination in osteoblasts (17). A recent report demonstrated that the high bone mass phenotype of Shn3−/− mice is also due to decreased osteoclast formation. Shn3−/− MSCs express low levels of RANKL, resulting in reduced numbers of osteoclasts in the diaphyseal region of long bones (60). We found that Wwp1−/− mice have normal osteoclast numbers in long bones and that their MSCs express comparable levels of RANKL (data not shown), indicating that Wwp1 regulates bone mass mainly through the regulation of osteoblast function.
In summary, our results demonstrate that Wwp1 is a new negative regulator of osteoblast function by reducing the protein levels of multiple positive osteoblast regulators. Wwp1−/− mice exhibit high bone mass due to increased osteoblast precursor differentiation and migration. Wwp1 expression is low in MSCs from young mice and it increases gradually as mice age, while Wwp1 deficiency in mice prevents age-related bone loss and osteoblast inhibition. Wwp1 inhibitors may represent a new class of bone anabolic agents to treat patients with osteoporosis.
Acknowledgments
The authors thank Dr. Lydia E. Matesic for providing Wwp1−/− mice and Ms. Evelyn Henderson for manuscript editing. This work was supported by research grants from National Institute of Health PHS awards (AR48697 to LX and AR43510 to BFB). MicroCT was supported by P30AR0613007 to Edward M. Schwarz.
Footnotes
Disclosure: The authors have nothing to disclose.
Conceived and designed the experiments: Xing, Lianping and Boyce, Brendan. Performed the experiments: Shu, Lei and Zhang, Hengwei. Analyzed the data: Shu, Lei, Zhang, Hengwei, Boyce, Brendan and Xing, Lianping. Wrote the paper: Shu, Lei, Zhang, Hengwei, Boyce, Brendan and Xing, Lianping.
References
- 1.McCarthy TL, Ji C, Centrella M. Links among growth factors, hormones, and nuclear factors with essential roles in bone formation. Crit Rev Oral Biol Med. 2000;11:409–22. doi: 10.1177/10454411000110040201. [DOI] [PubMed] [Google Scholar]
- 2.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 3.Zuo C, Huang Y, Bajis R, Sahih M, Li YP, Dai K, Zhang X. Osteoblastogenesis regulation signals in bone remodeling. Osteoporos Int. 2012;23:1653–63. doi: 10.1007/s00198-012-1909-x. [DOI] [PubMed] [Google Scholar]
- 4.Marie PJ, Kassem M. Osteoblasts in osteoporosis: past, emerging, and future anabolic targets. Eur J Endocrinol. 2011;165:1–10. doi: 10.1530/EJE-11-0132. [DOI] [PubMed] [Google Scholar]
- 5.Xing L, Zhang M, Chen D. Smurf control in bone cells. J Cell Biochem. 2010;110:554–63. doi: 10.1002/jcb.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazono K, Maeda S, Imamura T. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 2005;16:251–63. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O’Keefe RJ, Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem. 2006;281:3569–76. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8:272–88. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhi X, Chen C. WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci. 2012;69:1425–34. doi: 10.1007/s00018-011-0871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 11.Durrington HJ, Upton PD, Hoer S, Boname J, Dunmore BJ, Yang J, Crilley TK, Butler LM, Blackbourn DJ, Nash GB, Lehner PJ, Morrell NW. Identification of a lysosomal pathway regulating degradation of the bone morphogenetic protein receptor type II. J Biol Chem. 2010;285:37641–9. doi: 10.1074/jbc.M110.132415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo R, Yamashita M, Zhang Q, Zhou Q, Chen D, Reynolds DG, Awad HA, Yanoso L, Zhao L, Schwarz EM, Zhang YE, Boyce BF, Xing L. Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins. J Biol Chem. 2008;283:23084–92. doi: 10.1074/jbc.M709848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–13. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, Boyce BF, Xing L. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281:4326–33. doi: 10.1074/jbc.M509430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Chen D, Zuscik MJ, O’Keefe RJ, Rosier RN. Overexpression of Smurf2 stimulates endochondral ossification through upregulation of beta-catenin. J Bone Miner Res. 2008;23:552–63. doi: 10.1359/JBMR.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou W, Chen X, Shim JH, Huang Z, Brady N, Hu D, Drapp R, Sigrist K, Glimcher LH, Jones D. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat Cell Biol. 2011;13:59–65. doi: 10.1038/ncb2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–7. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Huang J, Zhang H, Wang Y, Matesic LE, Takahata M, Awad H, Chen D, Xing L. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells. 2011;29:1601–10. doi: 10.1002/stem.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenner L, Hoebertz A, Beil FT, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, Angel P, Wagner EF. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol. 2004;164:613–23. doi: 10.1083/jcb.200308155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess J, Hartenstein B, Teurich S, Schmidt D, Schorpp-Kistner M, Angel P. Defective endochondral ossification in mice with strongly compromised expression of JunB. J Cell Sci. 2003;116:4587–96. doi: 10.1242/jcs.00772. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Huang J, Guo R, Wang Y, Chen D, Xing L. Smurf1 inhibits mesenchymal stem cell proliferation and differentiation into osteoblasts through JunB degradation. J Bone Miner Res. 2010;25:1246–56. doi: 10.1002/jbmr.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Duan B, Cheng Z, Jia X, Mao L, Fu H, Che Y, Ou L, Liu L, Kong D. SDF-1/CXCR4 axis modulates bone marrow mesenchymal stem cell apoptosis, migration and cytokine secretion. Protein Cell. 2011;2:845–54. doi: 10.1007/s13238-011-1097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu MH, Li CZ, Hu CJ, Fan YH, Wang SM, Wu YY, Liang GP, Yang SM. microRNA-27b suppresses mouse MSC migration to the liver by targeting SDF-1alphain vitro. Biochem Biophys Res Commun. 2012;421:389–95. doi: 10.1016/j.bbrc.2012.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Lien CY, Chih-Yuan Ho K, Lee OK, Blunn GW, Su Y. Restoration of bone mass and strength in glucocorticoid-treated mice by systemic transplantation of CXCR4 and cbfa-1 co-expressing mesenchymal stem cells. J Bone Miner Res. 2009;24:837–48. doi: 10.1359/jbmr.081257. [DOI] [PubMed] [Google Scholar]
- 25.Zhu W, Liang G, Huang Z, Doty SB, Boskey AL. Conditional inactivation of the CXCR4 receptor in osteoprecursors reduces postnatal bone formation due to impaired osteoblast development. J Biol Chem. 2011;286:26794–805. doi: 10.1074/jbc.M111.250985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subik K, Shu L, Wu C, Liang Q, Hicks D, Boyce B, Schiffhauer L, Chen D, Chen C, Tang P, Xing L. The ubiquitin E3 ligase WWP1 decreases CXCL12-mediated MDA231 breast cancer cell migration and bone metastasis. Bone. 2012;50:813–23. doi: 10.1016/j.bone.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung JH, Yang HM, Park JB, Choi GS, Joh JW, Kwon CH, Chun JM, Lee SK, Kim SJ. Isolation and characterization of mouse mesenchymal stem cells. Transplant Proc. 2008;40:2649–54. doi: 10.1016/j.transproceed.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–64. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao Z, Xing L, Boyce BF. NF-kappaB p100 limits TNF-induced bone resorption in mice by a TRAF3-dependent mechanism. J Clin Invest. 2009;119:3024–34. doi: 10.1172/JCI38716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeman E. Bone modeling and remodeling. Crit Rev Eukaryot Gene Expr. 2009;19:219–33. doi: 10.1615/critreveukargeneexpr.v19.i3.40. [DOI] [PubMed] [Google Scholar]
- 31.Robling AG, Turner CH. Mechanical signaling for bone modeling and remodeling. Crit Rev Eukaryot Gene Expr. 2009;19:319–38. doi: 10.1615/critreveukargeneexpr.v19.i4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng X, McDonald JM. Disorders of bone remodeling. Annu Rev Pathol. 2011;6:121–45. doi: 10.1146/annurev-pathol-011110-130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhandari D, Robia SL, Marchese A. The E3 ubiquitin ligase atrophin interacting protein 4 binds directly to the chemokine receptor CXCR4 via a novel WW domain-mediated interaction. Mol Biol Cell. 2009;20:1324–39. doi: 10.1091/mbc.E08-03-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–88. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Otsuru S, Tamai K, Yamazaki T, Yoshikawa H, Kaneda Y. Circulating bone marrow-derived osteoblast progenitor cells are recruited to the bone-forming site by the CXCR4/stromal cell-derived factor-1 pathway. Stem Cells. 2008;26:223–34. doi: 10.1634/stemcells.2007-0515. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, Guo R, Schwarz EM, Boyce BF, Xing L. TNF inhibits production of stromal cell-derived factor 1 by bone stromal cells and increases osteoclast precursor mobilization from bone marrow to peripheral blood. Arthritis Res Ther. 2008;10:R37. doi: 10.1186/ar2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lian JB, Stein GS. Runx2/Cbfa1: a multifunctional regulator of bone formation. Curr Pharm Des. 2003;9:2677–85. doi: 10.2174/1381612033453659. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Kruppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316:139–48. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 39.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–61. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 40.Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280:22115–23. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 41.Jones DC, Wein MN, Glimcher LH. Schnurri-3: a key regulator of postnatal skeletal remodeling. Adv Exp Med Biol. 2007;602:1–13. doi: 10.1007/978-0-387-72009-8_1. [DOI] [PubMed] [Google Scholar]
- 42.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 43.Moll NM, Ransohoff RM. CXCL12 and CXCR4 in bone marrow physiology. Expert Rev Hematol. 2010;3:315–22. doi: 10.1586/ehm.10.16. [DOI] [PubMed] [Google Scholar]
- 44.Georgiou KR, Foster BK, Xian CJ. Damage and recovery of the bone marrow microenvironment induced by cancer chemotherapy - potential regulatory role of chemokine CXCL12/receptor CXCR4 signalling. Curr Mol Med. 2010;10:440–53. doi: 10.2174/156652410791608243. [DOI] [PubMed] [Google Scholar]
- 45.Sordi V. Mesenchymal stem cell homing capacity. Transplantation. 2009;87:S42–5. doi: 10.1097/TP.0b013e3181a28533. [DOI] [PubMed] [Google Scholar]
- 46.Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J Biol Chem. 2004;279:12854–9. doi: 10.1074/jbc.M313294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003;5:709–22. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- 48.Subik K, Shu L, Wu C, Liang Q, Hicks D, Boyce B, Schiffhauer L, Chen D, Chen C, Tang P, Xing L. The ubiquitin E3 ligase WWP1 decreases CXCL12-mediated MDA231 breast cancer cell migration and bone metastasis. Bone. 2012;50:813–23. doi: 10.1016/j.bone.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho SW, Sun HJ, Yang JY, Jung JY, An JH, Cho HY, Choi HJ, Kim SW, Kim SY, Kim D, Shin CS. Transplantation of mesenchymal stem cells overexpressing RANK-Fc or CXCR4 prevents bone loss in ovariectomized mice. Mol Ther. 2009;17:1979–87. doi: 10.1038/mt.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143:677–81. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 51.Lu K, Yin X, Weng T, Xi S, Li L, Xing G, Cheng X, Yang X, Zhang L, He F. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat Cell Biol. 2008;10:994–1002. doi: 10.1038/ncb1760. [DOI] [PubMed] [Google Scholar]
- 52.Malik R, Soh UJ, Trejo J, Marchese A. Novel roles for the E3 ubiquitin ligase atrophin-interacting protein 4 and signal transduction adaptor molecule 1 in G protein-coupled receptor signaling. J Biol Chem. 2012;287:9013–27. doi: 10.1074/jbc.M111.336792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langen RC, Haegens A, Vernooy JH, Wouters EF, de Winther MP, Carlsen H, Steele C, Shoelson SE, Schols AM. NF-kappaB Activation Is Required for the Transition of Pulmonary Inflammation to Muscle Atrophy. Am J Respir Cell Mol Biol. 2012;47:288–97. doi: 10.1165/rcmb.2011-0119OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alles N, Soysa NS, Hayashi J, Khan M, Shimoda A, Shimokawa H, Ritzeler O, Akiyoshi K, Aoki K, Ohya K. Suppression of NF-kappaB increases bone formation and ameliorates osteopenia in ovariectomized mice. Endocrinology. 2010;151:4626–34. doi: 10.1210/en.2010-0399. [DOI] [PubMed] [Google Scholar]
- 55.Chang J, Wang Z, Tang E, Fan Z, McCauley L, Franceschi R, Guan K, Krebsbach PH, Wang CY. Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat Med. 2009;15:682–9. doi: 10.1038/nm.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krum SA, Chang J, Miranda-Carboni G, Wang CY. Novel functions for NFkappaB: inhibition of bone formation. Nat Rev Rheumatol. 2010;6:607–11. doi: 10.1038/nrrheum.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fagiolo U, Cossarizza A, Scala E, Fanales-Belasio E, Ortolani C, Cozzi E, Monti D, Franceschi C, Paganelli R. Increased cytokine production in mononuclear cells of healthy elderly people. Eur J Immunol. 1993;23:2375–8. doi: 10.1002/eji.1830230950. [DOI] [PubMed] [Google Scholar]
- 58.Arnaud CD. An integrated view of the role of the endocrine system in the genesis of the osteoporosis associated with aging. Osteoporos Int. 1993;3 (Suppl 1):37–9. doi: 10.1007/BF01621859. [DOI] [PubMed] [Google Scholar]
- 59.Tilstra JS, Robinson AR, Wang J, Gregg SQ, Clauson CL, Reay DP, Nasto LA, St Croix CM, Usas A, Vo N, Huard J, Clemens PR, Stolz DB, Guttridge DC, Watkins SC, Garinis GA, Wang Y, Niedernhofer LJ, Robbins PD. NF-kappaB inhibition delays DNA damage-induced senescence and aging in mice. J Clin Invest. 2012;122:2601–12. doi: 10.1172/JCI45785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wein MN, Jones DC, Shim JH, Aliprantis AO, Sulyanto R, Lazarevic V, Poliachik SL, Gross TS, Glimcher LH. Control of bone resorption in mice by Schnurri-3. Proc Natl Acad Sci U S A. 2012;109:8173–8. doi: 10.1073/pnas.1205848109. [DOI] [PMC free article] [PubMed] [Google Scholar]