Abstract
Introduction
Mutations in EGFR and KRAS can impact treatment decisions for patients with NSCLC. The incidence of these mutations varies, and it is unclear whether there is a decreased frequency among African Americans (AfAs).
Methods
We performed a retrospective chart review of 513 NSCLC patients undergoing EGFR and KRAS mutational analysis at the Hospital of the University of Pennsylvania between May 2008 and November 2011. Clinical and pathologic data were abstracted from the patients’ electronic medical record.
Results
Of 497 patients with informative EGFR mutation analyses, the frequency of EGFR mutation was 13.9%. The frequency of EGFR mutations was associated with race (p<0.001) and was lower in AfA patients compared to Caucasian (C) patients but did not reach statistical significance (4.8% vs 13.7%, p=0.06). Mean Charlson Comorbidity Index and number of cigarette pack years were significantly lower in patients with EGFR mutations (p=0.01 and p<0.001, respectively). Multivariable logistic regression analysis showed a significant association between race and EGFR mutation (p=0.01), even after adjusting for smoking status (p<0.001) and gender (p=0.03). KRAS mutation (study frequency 28.1%) was not associated with race (p=0.08; p=0.51 for Afa vs C patients), but was more common among smokers (p<0.001) and females (p=0.01).
Conclusions
Based on multivariable analysis, even after adjusting for smoking status and gender, we found that race was statistically significantly associated with EGFR mutation, but not KRAS mutational status. To our knowledge, this is the largest single institution series to date evaluating racial differences in EGFR and KRAS mutational status among patients with NSCLC.
Keywords: EGFR, KRAS, Racial Disparity, NSCLC
Introduction
Non small cell lung cancer (NSCLC) is a common and deadly disease.(1) Recent data have shown that activating mutations in the epidermal growth factor receptor (EGFR) can be used to guide therapy with small molecule tyrosine kinase inhibitors.(2-4) This treatment modality has been evaluated in multiple trials in treatment-naïve patients and has been shown to improve both response rate and progression free survival compared to standard chemotherapy in patients whose tumors harbor this mutation.(5-8) Knowing a patient’s EGFR mutational status, therefore, is crucial when choosing a patient’s initial treatment regimen. KRAS is another mutation seen frequently in NSCLC, and though there is no established targeted therapy for this molecular abnormality, mutational analysis is often performed for this as well. There has been much debate whether such testing should happen automatically whenever a diagnosis of lung cancer is made, regardless of histology or demographic background, or if it should be an individualized decision.
The incidence of such mutations varies widely across different populations, with increased incidence of EGFR mutations in never smokers,(9) East Asian populations,(10, 11) women,(12) and those with adenocarcinoma.(2-4, 12) There have been conflicting data to date regarding the incidence of EGFR mutation in African American (AfA) patients with NSCLC. This is a particularly important population to study, as AfAs constitute 1/8th of the US population and AfA patients have the highest incidence and mortality from lung cancer.(1, 5-8, 13, 14) All prior analyses evaluating this issue combined multiple studies, compared banked samples to established cohorts and registries or had a relative paucity of AfA patients. Because we have a substantial AfA population at our institution, we felt we were well suited to address this question.
Materials and Methods
After acquiring approval from our institutional review board, in conjunction with our Department of Molecular Pathology (MP) we reviewed every EGFR mutation analysis for a diagnosis of NSCLC performed at Hospital of the University of Pennsylvania from May 2008 to November 2011. MP also provided us with the results of KRAS mutational analysis when performed on the same patients during this time period, although it should be noted that not all patients undergoing EGFR analysis underwent KRAS testing. Results that were indeterminate or inconclusive were excluded from further analysis.
Once the list of patient names was made available, a retrospective chart review was performed using our electronic medical record (EMR) to acquire demographic, treatment and tumor related data. Race was patient reported based upon a verbal discussion with the patient at the time of new patient registration. As a part of this chart review, we created a “disease status” variable, referring to disease extent at the time of EGFR testing. Localized disease referred to tumors resectable by traditional measures. Recurrent intrathoracic or extrathoracic disease referred to tumors previously treated definitively, which had subsequently recurred within or outside of the thorax. Metastatic intrathoracic or extrathoracic disease referred to metastatic disease within or outside the thorax at presentation. Although some EGFR testing was performed proximal to the time of diagnosis and thus correlated with a patient’s presenting stage, many other patients had EGFR testing performed later in their clinical course, usually at the time of recurrence. In these situations, the disease status variable was often discordant from true stage.
In addition, a Charlson Comorbidity Index as well as an age-weighted Index(15) were calculated for each patient using an online calculator (http://www.medal.org/OnlineCalculators/ch1/ch1.13/ch1.13.01.php) based on medical history data extracted from the EMR.
For mutation analysis, DNA was extracted from formalin fixed paraffin embedded tissue using conventional methods. EGFR mutation analysis for exon 19 deletions and the L858R point mutation (EGFR NM_005228.3:c.2573T>G) were performed as previously described.(10, 16) KRAS mutation analysis was performed using multiplex PCR coupled with analysis on a liquid bead array. Briefly, primers designed to detect the seven most common point mutations in nucleotides c.34G, c.35G and c.38G in codons 12 and 13 of KRAS (NM_004985.3) were used to amplify the target sequence. Amplified products were then hybridized to a liquid bead array and analyzed with a Luminex 100. The analytical sensitivity of both methods is approximately 5%.
All of this information was entered into a password protected Excel® spreadsheet, which was then de-identified.
Statistical Analyses
Descriptive statistics were employed to describe patient characteristics of the study cohort. Each categorical variable was summarized by frequency and percentage. Each continuous variable was summarized by mean, standard deviation, median and range. Patients who were positive for either exon 19 mutation or L858R mutation were classified as EGFR mutation positive, while patients who were negative for both mutations were classified as EGFR mutation negative or wild type. Patients who were positive (or negative) for one mutation but were indeterminate or inconclusive for the other mutation were classified as non-informative. Prevalences of EGFR or KRAS mutations were described by frequencies and percentages based on informative cases. Bar graphs were used to visualize mutation rates for subgroups defined by gender, race and smoking status. Associations between patient characteristics and EGFR or KRAS mutations were tested by Fisher’s exact test for categorical variables and Student’s t test for continuous variables. Univariate logistic regression analysis was employed to estimate the magnitude of association with mutation status, using the odds ratio and 95% confidence interval. Multivariable logistic regression analysis was utilized to identify significant independent factors associated with EGFR mutation. The variables considered for model building exhibited univariate significance of p < 0.10. Backward selection was employed to construct the optimal multivariable model. Statistical significance was assessed by the Wald test. A p-value less than or equal to 0.05 was considered statistically significant. All statistical analyses were produced in SPSS 19 (SPSS Inc., Chicago, IL).
Results
Patient Characteristics
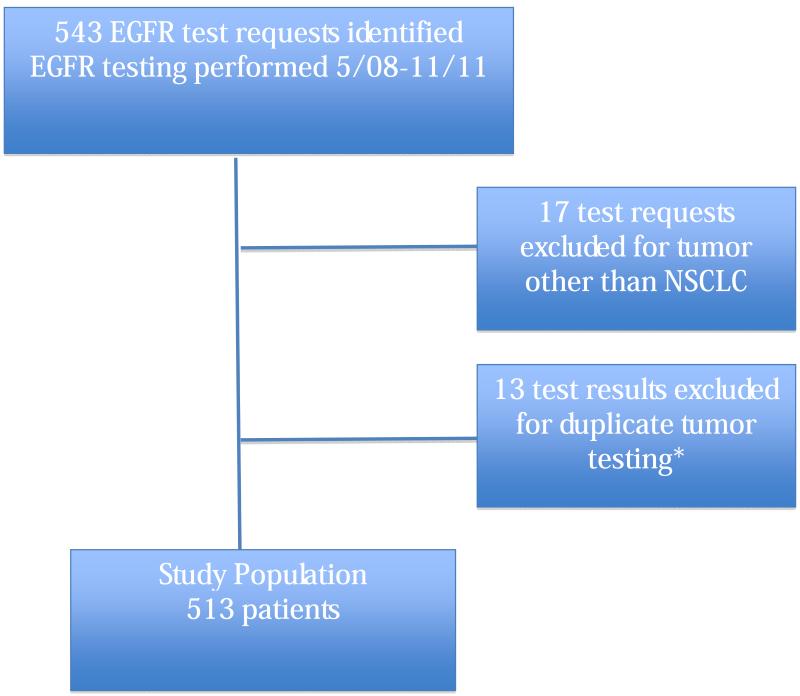
We identified 543 requests for EGFR testing. Of these, 17 were eliminated for a diagnosis other than NSCLC and 13 were eliminated because the same primary tumor was tested twice, leaving a total of 513 patients (CONSORT Diagram in Figure 1). When two tumors in a single patient had different histologies, they were regarded as separate primary cancers and both records were included (n=5 patients). When two records of the same primary tumor were noted, the tumor testing with the most complete information was included. This was usually due to previously indeterminate or inconclusive test results. Our cohort included 67 AfA patients, 399 C patients, 17 Asian American (AsA) patients, 3 Hispanic (H) patients and 19 patients who were identified as Other (O) in the EMR. The majority of patients (80%) were identified as either former or current smokers. (Table 1)
Figure 1.
Consort Diagram. *When the tumors had different histologies, they were regarded as separate primary cancers and both records were included. When two records of the same primary tumor were noted, the tumor testing with the most complete information was included. This usually occurred as a result of a previously indeterminate or inconclusive test result.
Table 1.
Patient Characteristics
| Variable | Levels | Frequency | Percentage |
|---|---|---|---|
| Gender | |||
| Male | 210 | 40.9% | |
| Female | 303 | 59.1% | |
| Race | |||
| African American | 67 | 13.3% | |
| Caucasian | 399 | 79.0% | |
| Asian American | 17 | 3.4% | |
| Hispanic | 3 | 0.6% | |
| Other | 19 | 3.8% | |
| Smoking Status | |||
| Never Smoker | 104 | 20.4% | |
| Former Smoker | 354 | 69.5% | |
| Current Smoker | 51 | 10.0% | |
| Histology | |||
| Adenocarcinoma | 405 | 79.6% | |
| Squamous | 46 | 9.0% | |
| Adenosquamous | 16 | 3.1% | |
| Poorly differentiated | 30 | 5.9% | |
| Large cell | 9 | 1.8% | |
| Sarcomatoid | 1 | 0.2% | |
| Pleomorphic | 1 | 0.2% | |
| NOS* | 1 | 0.2% | |
| Subcutaneous Mets | |||
| No | 495 | 98.6% | |
| Yes | 7 | 1.4% | |
| Surgery as part of treatment | |||
| No | 239 | 47.6% | |
| Yes | 263 | 52.4% | |
| Disease status | |||
| Localized | 138 | 27.3% | |
| Recurrent, extrathoracic | 33 | 6.5% | |
| Recurrent, intrathoracic | 46 | 9.1% | |
| Metastatic extrathoracic | 191 | 37.7% | |
| Metastatic intrathoracic | 98 | 19.4% |
NOS refers to NSCLC Not Otherwise Specified
EGFR Mutational Status
Of 497 patients with an informative EGFR mutational analysis (i.e., informative for both exon 19 and L858R), 69 (13.5%) had a detectable EGFR mutation (9.0% exon 19 deletion, 4.8% exon 21 L858R mutation, Table 2). The frequency of EGFR mutation was 4.8% in AfA patients, 13.7% in C patients, 65.2% in AsA patients, 0% in Hispanic and 10.5% in O patients (p<0.001, Table 3). On univariate analysis, mutation rates in AfA and C patients were not statistically significantly different (p= 0.06).
Table 2.
Mutational status
| Variable | Levels | Frequency | Percentage |
|---|---|---|---|
| EGFR Mutation (any) | |||
| Negative | 428 | 86.1% | |
| Positive | 69 | 13.9% | |
| Indeterminate/Inconclusive* | 16 | - | |
| EGFR Exon 19 Deletion | |||
| Negative | 456 | 91.0% | |
| Positive | 45 | 9.0% | |
| Indeterminate | 2 | - | |
| Inconclusive | 10 | - | |
| EGFR L858R Mutation | |||
| Negative | 475 | 95.2% | |
| Positive | 24 | 4.8% | |
| Indeterminate | 5 | - | |
| Inconclusive | 9 | - | |
| KRAS | |||
| Wild type | 269 | 71.9% | |
| Mutant | 105 | 28.1% | |
| Indeterminate | 4 | - | |
| Inconclusive | 4 | - |
Indeterminate or inconclusive for either or both Exon 19 or L858R Mutations
Table 3.
Associations between Patient Characteristics and EGFR Mutation, Categorical Variables
| Variable | #positive/# tested | Percentage | Fisher’s Exact p value |
|---|---|---|---|
| Gender | 0.002 | ||
| Male | 16/201 | 8.0% | |
| Female | 53/296 | 17.9% | |
| Race | <0.001 | ||
| African American | 3/63 | 4.8% | |
| Caucasian | 53/388 | 13.7% | |
| Asian American | 10/16 | 65.2% | |
| Hispanic | 0/3 | 0.0% | |
| Other | 2/19 | 10.5% | |
| Race, binary | 0.06 | ||
| African American | 3/63 | 4.8% | |
| Caucasian | 53/388 | 13.7% | |
| Smoking status | <0.001 | ||
| Never Smoker | 42/100 | 42.0% | |
| Ever Smoker | 26/393 | 6.6% | |
| Histology | 0.27 | ||
| Adenocarcinoma | 61/391 | 15.6% | |
| Squamous | 6/44 | 13.6% | |
| Adenosquamous | 0/16 | 0.0% | |
| Poorly Differentiated | 1/30 | 3.3% | |
| Large Cell | 0/9 | 0.0% | |
| Sarcomatoid | 0/1 | 0.0% | |
| Pleomorphic | 0/1 | 0.0% | |
| NOS* | 0/1 | 0.0% | |
| Disease Status | 0.53 | ||
| Localized | 21/137 | 15.3% | |
| Recurrent, extrathoracic | 7/32 | 21.9% | |
| Recurrent, intrathoracic | 6/43 | 14.0% | |
| Metastatic extrathoracic | 23/184 | 12.5% | |
| Metastatic intrathoracic | 10/95 | 10.5% | |
| Subcutaneous Mets | 0.60 | ||
| No | 67/480 | 14.0% | |
| Yes | 0/7 | 0.0% | |
| Surgery as part of treatment | 0.69 | ||
| No | 29/228 | 12.7% | |
| Yes | 37/259 | 14.3% |
NOS refers to NSCLC not otherwise specified
As expected, patients with EGFR mutations were significantly more likely to be female (p=0.002) and never smokers (p<0.001). In addition, we found that in current or former smokers, patients with EGFR mutations had a lower mean number of pack years than those without EGFR mutations (p<0.001, Table 3-4). Patients with EGFR mutations had lower Charlson Comorbidity Index (i.e., less comorbidity) than those without EGFR mutations (p=0.01) as well as a lower combined age-weighted Comorbidity Index (p=0.03).
Table 4.
Associations between Patient Characteristics and EGFR Mutation, Continuous Variables
| Variable | EGFR mutation | Negative | EGFR mutation | Positive | t test p value |
|---|---|---|---|---|---|
| N | Mean ± SE | N | Mean ± SE | ||
| Pack years* | 362 | 36.7 ± 1.4 | 26 | 14.3 ± 4.3 | <0.001 |
| Age | 428 | 64.2 ± 0.6 | 69 | 64.5 ± 1.3 | 0.60 |
| Charlson Index | 428 | 5.6 ± 0.1 | 67 | 4.9 ± 0.2 | 0.01 |
| Charlson & age | 428 | 7.6 ± 0.1 | 67 | 6.9 ± 0.3 | 0.03 |
Former or current smokers only
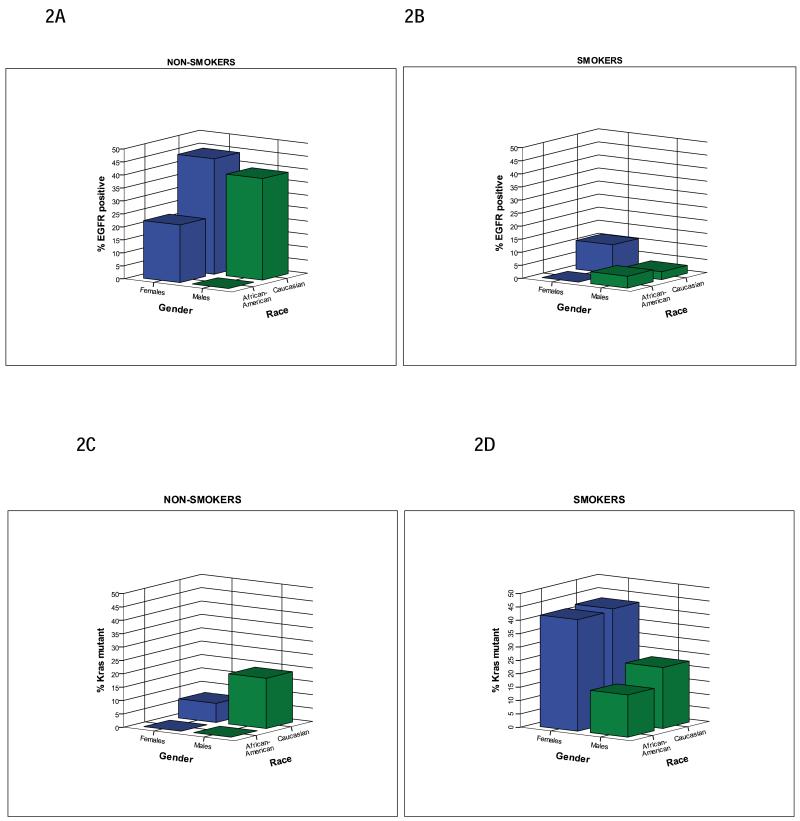
We analyzed 3-way interactions of smoking status, gender and race on frequency of EGFR mutations (Figure 2A and 2B). Overall, the rate of never smokers was identical in AfA and C patients (p=0.96), although the absolute number in AfA patients was very small (n=12). We found that the EGFR mutational frequency was lower in AfA female smokers than in their C counterparts (p=0.05, Figure 2B), but that this was not the case when comparing AfA and C male smokers (p=0.54). There was no significant difference detected in the frequency of EGFR mutation between the two races in never smokers (Figure 2A), but small numbers (9 AfA female never smokers and 1 AfA male never smoker) limit any conclusions drawn from this subgroup. Multivariable logistic regression determined that even after adjusting for smoking status (p<0.001) and gender (p=0.03), race was statistically significantly associated with EGFR mutations (p=0.01). Moreover, the 95% CI of the odds ratio for EGFR mutations for AfAs relative to Cs, did not include 1.0, indicating a significant difference between these groups. (Table 5)
Figure 2.
Association of Gender, Race and EGFR or KRAS positivity by Smoking Status. Females are represented by blue bars and males are represented by green bars. 2A. Caucasian female non-smokers had higher rate of EGFR positivity compared to African American female non-smokers, which did not meet statistical significance (44.4% (20/45) vs. 22.2% (2/9), p=0.28). Caucasian male non-smokers had a higher rate of EGFR positivity compared to African American male non-smokers; this difference did not meet statistical significance due to small number of African American never-smokers (39.1% (9/23) vs. 0% (0/1), p=1.00). 2B. Caucasian female smokers had a significantly higher rate of EGFR positivity than African American female smokers (11% (20/182) vs. 0% (0/31), p=0.05). Caucasian male smokers had a similar rate of EGFR positivity as African American male smokers (3% (4/135) vs. 4.5% (1/22), p=0.54). 2C. Caucasian female never-smokers had higher rate of KRAS mutant positivity compared to African American female never-smokers; this difference did not meet statistical significance (7.1% (2/28) vs. 0% (0/8), p=1.0). Caucasian male non-smokers had a higher rate of KRAS mutant positivity compared to African American male never-smokers, which did not meet statistical significance due to small number of African American never-smokers (18.8% (3/16) vs. 0% (0/1), p=1.00). 2D. Caucasian female smokers had a similar rate of KRAS mutant positivity as African American female smokers (42.5% (57/134) vs. 41.7% (10/24), p=1.0). Caucasian male smokers had a similar rate of KRAS mutant positivity as African American male smokers (22.8% (23/101) vs. 15.8% (3/19), p=0.76).
Table 5.
Multivariable analysis of EGFR mutation positivity (Exon 19 or L858R Mutation)
| Univariate Logistic regression* | Multivariable Logistic regression# | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI | Wald test P value |
OR | 95% CI | Wald test P value |
|
| Gender | 0.002 | 0.03 | ||||
| Male | 1.00 | 1.00 | ||||
| Female | 2.52 | 1.40 - 4.55 | 2.13 | 1.10 - 4.15 | ||
| Race | <0.001 | 0.01 | ||||
| Caucasian | 1.00 | 1.00 | ||||
| African American | 0.32 | 0.10 - 1.04 | 0.26 | 0.07 - 0.93 | ||
| Asian American | 10.54 | 3.68 - 30.19 | 5.10 | 1.41 - 18.41 | ||
| Hispanic | ND | ND | ||||
| Other | 0.74 | 0.17 - 3.31 | 0.22 | 0.03 - 1.92 | ||
| Smoking status | <0.001 | <0.001 | ||||
| Ever smoker | 1.00 | 1.00 | ||||
| Never smoker | 10.22 | 5.83 - 17.93 | 9.66 | 5.27 - 17.71 | ||
| Charlson index, continuous | 0.86 | 0.76 - 0.97 | 0.01 | |||
| Charlson & age, continuous | 0.89 | 0.81 - 0.99 | 0.03 | |||
N = 484 with data on all 5 candidate variables seleced by univariate significance (p < 0.10)
Best model determined by backward elimination OR = Odds Ratio, CI = Confidence Interval ND = not determined, none of 3 Hispanic patients had EGFR mutations
KRAS Mutational Status
Of 374 patients with an informative KRAS mutational analysis, 105 (28.1%) had a KRAS mutation detected (Table 2). Of 366 patients with informative EGFR and KRAS mutational analyses, only 1 (0.3%) patient exhibited both mutations. The relative frequency of KRAS mutations was 25% in AfAs, 30.6% in Cs, 50% in Hs, 0% of AsAs and 18.8% in Os (p=0.08, Table 6). When comparing only AfA and C patients, no significant difference was found (p=0.51). KRAS mutations were more frequent among females (p=0.01). Patients with intrathoracic recurrent disease had a higher rate of KRAS mutation (51.5%) than any other disease status (p=0.03)
Table 6.
Associations between Patient Characteristics and KRAS Mutation
| Variable | #positive/#tested | Percentage | Fisher’s Exact p value |
|---|---|---|---|
| Gender | 0.01 | ||
| Male | 32/154 | 20.8% | |
| Female | 73/220 | 33.2% | |
| Race | 0.09 | ||
| African American | 13/52 | 25.0% | |
| Caucasian | 86/281 | 30.6% | |
| Asian American | 0/15 | 0.0% | |
| Hispanic | 1/2 | 50.0% | |
| Other | 3/16 | 18.8% | |
| Race, binary | 0.51 | ||
| African American | 13/52 | 25.0% | |
| Caucasian | 86/281 | 30.6% | |
| Smoking status | <0.001 | ||
| Never Smoker | 6/72 | 8.3% | |
| Ever Smoker | 98/300 | 32.7% | |
| Histology | 0.68 | ||
| Adenocarcinoma | 88/294 | 29.9% | |
| Squamous | 5/31 | 16.1% | |
| Adenosquamous | 4/12 | 33.3% | |
| Poorly Differentiated | 5/24 | 20.8% | |
| Large Cell | 2/8 | 25.0% | |
| Sarcomatoid | 0/1 | 0.0% | |
| Pleomorphic | 0/1 | 0.0% | |
| NOS* | 0 | -- | |
| Disease Status | 0.03 | ||
| Localized | 23/92 | 25.0% | |
| Recurrent, extrathoracic | 6/18 | 33.3% | |
| Recurrent, intrathoracic | 17/33 | 51.5% | |
| Metastatic extrathoracic | 35/147 | 23.8% | |
| Metastatic intrathoracic | 24/83 | 28.9% | |
| Subcutaneous Mets | 0.02 | ||
| No | 101/365 | 27.7% | |
| Yes | 4/5 | 80.0% | |
| Surgery as part of treatment | 0.05 | ||
| No | 43/183 | 23.5% | |
| Yes | 62/187 | 33.2% |
NOS refers to NSCLC not otherwise specified
While a history of smoking was associated with the presence of a KRAS mutation (p<0.001), the mean number of pack years did not differ significantly between patients with or without KRAS mutations (p=0.56, Table 7). We analyzed 3-way interactions of smoking status, gender and race on frequency of KRAS mutations and discovered no significant differences in mutation rates were observed between groups (Figure 2C and 2D).
Table 7 Continuous Variable Associations between Patient Characteristics and KRAS Mutation
| Variable | KRAS | Wild type | KRAS | Mutant | t test p value |
|---|---|---|---|---|---|
| N | Mean ± SE | N | Mean ± SE | ||
| Pack years* | 200 | 34.6 ± 1.9 | 97 | 36.6 ± 2.6 | 0.56 |
| Age | 269 | 63.9 ± 0.7 | 105 | 63.1 ± 1.0 | 0.52 |
| Charlson Index | 268 | 5.6 ± 0.1 | 105 | 5.7 ± 0.2 | 0.66 |
| Charlson & age | 268 | 7.6 ± 0.2 | 105 | 7.7 ± 0.2 | 0.92 |
Former or current smokers only
Discussion
Available data for the incidence of EGFR mutation among AfA patients with NSCLC are conflicting. While Yang et al(17) and Leidner et al(18) have found a lower frequency of EGFR mutation among African Americans, Riely et al(19), Cote et al(20) and Reinersman et al(21) did not find a statistically significant difference between the two groups. All of the studies that included more than 20 AfAs combined data from multiple clinical trials, which is methodologically questionable. The largest and most recent evaluation, by Reinersman et al(21) compared archived tissue at 3 different institutions to the tumor registry at Memorial Sloan Kettering Cancer Center (MSKCC). The use of multiple institutions to increase the number of AfAs would have been more appropriate if Cs were also evaluated at all institutions. As it is, the referral patterns to MSKCC make such a conflation of patients problematic, particularly because they did not perform a multivariable analysis and only evaluated resected tumor specimens. The former is particularly relevant since statistical significance between AfAs and Cs in our series was only revealed on multivariable analysis. Our series was the largest to date analyzing this question in contemporaneous populations of C and AfA patients with all stages of NSCLC treated by the same clinicians at the same institution.
The frequency of EGFR mutation in our cohort was consistent with prior data. The frequency in North American cohorts usually ranges between 10% and 20% as referenced above; so our overall frequency of 13.5% is comparable. The statistical significance of smoking as a binary and continuous variable in relation to EGFR mutational frequency is not surprising.(22, 23) Within our cohort, the frequency of EGFR mutation among AfA patients was decreased compared to C patients, but was not statistically significant on univariate analysis. On multivariable analysis, however, EGFR mutational frequency was significantly affected by race. In addition, the 95% CI of the odds ratio for EGFR mutations for AfAs relative to Cs no longer crossed 1.0 when adjusted for gender and smoking status. This is supported by our analysis of differential mutational frequency in specific subgroups, most notably women. There was no significant difference in the frequency of L858R mutations, and male smokers also had a similar frequency between the two racial groups. This is interesting, since Cote et al observed that the EGFR mutations found in AfA patients were exclusively within exon 19. While this study did not find a significant difference between individual mutation frequencies by race, recent data indicate that AfA patients may harbor different mutations than those commonly seen in C patients. These mutations may still be sensitive to EGFR directed therapies.(24) Our data support this theory, and our large data set is able to show a significant difference between certain mutations in EGFR between the AfA and C patients. The lack of a difference between male smokers of AfA and C ancestry is, to our knowledge, a novel finding.
The association of EGFR mutational status with Charlson Comorbidity Index as well as the age-weighted Index is the first time such a relationship has been described. As referenced above, it is well known that patients with EGFR mutation are less likely to be smokers. Given that six of the conditions evaluated in the Index (cerebrovascular disease, chronic pulmonary disease, congestive heart failure, myocardial infarction, peripheral vascular disease, and malignant solid tumor) are frequently associated with smoking, it is certainly possible that tobacco exposure is acting as a confounding variable. That being said, this result supports the general medical impression that the EGFR mutated patient population tends to be healthier than the EGFR wild type population. We evaluated the association of EGFR mutation with Charlson Comorbidity Index in smokers and non-smokers to attempt to eliminate this bias (data not shown). In all groups, the Index was lower for patients with EGFR mutation, though this did not reach statistical significance. We feel this is likely secondary to the decreased power of the subgroup analysis. The consistent numerical difference supports a true distinction.
The multivariable analysis of EGFR mutational frequency confirms the association with race. Of note, however, the frequency of EGFR mutation was much more strongly associated with smoking status than any other variable. As such, while there may be a lower frequency of EGFR mutation among AfA patients, it would not be appropriate to make decisions about whom to test solely on the basis of race. This is consistent with the recent work of D’Angelo et al(9), who reported that if only female never smokers were tested for EGFR mutation, 57% of all EGFR mutations would be missed. Within our cohort, if only female, never smokers of Caucasian or Asian ancestry with adenocarcinoma were tested, 45.5% of EGFR mutations would be missed.
The lack of significant distinction between EGFR frequency among squamous and adenocarcinoma NSCLC stands in stark contrast to available data on the incidence of EGFR mutations as it relates to lung cancer histology,(12) which are the basis for the NCCN guidelines recommendation that testing for EGFR mutation in patients with squamous cell histology not be pursued.(25) Our clinical practice has been to test patients with squamous histology only if their smoking history or other clinical factors raise our suspicion that their tumors may more likely harbor an EGFR mutation. Indeed, of the patients with squamous NSCLC harboring EGFR mutation, 67% were never smokers, as opposed to 21.7% of all squamous NSCLC in the cohort. Our cohort was composed of all patients for whom EGFR mutational testing was performed; there is selection bias inherent in this comparison. The similar incidence in squamous NSCLC, however, supports a rationale for testing in appropriate patients, including selected patients with squamous histology. This type of individualized decision-making has not been analyzed formally in the literature and could be an opportunity for further research.
Our study confirms prior work(18, 21) regarding the lack of racial variation among patients with KRAS mutation between AfA and C patients. It is, however, the first paper, to our knowledge, expanding the comparison to other races as well. The increased incidence of KRAS mutations among female patients in our cohort adds to a mixed body of literature. Some studies have shown increased incidence of KRAS mutation among women(26) while others report equal frequencies in both men and women.(27-29) Our study is the largest series to date to address this question, and the results are in agreement with the report by Nelson et al, the second largest series.
There are several limitations to our study. First, mutational testing was not tested at a fixed time point. This could introduce bias if mutations arise or alter throughout the disease course. Previous data, however, suggest that tumor heterogeneity for EGFR is very rare.(30) Second, the retrospective nature limits control of biases. We attempted to account for this by including a large series of patients presenting over a long period of time. That being said, our cohort included 59.1% females while the lung cancer group at our hospital had only 48% female representation. This indicates a potential selection bias to test more females for EGFR mutation. There does not seem to have been a selection bias for race in our cohort, however; while our cohort included 13.3% AfAs the proportion of AfAs in the cancer center clinics was 16%. Lastly, our data were confined exclusively to L858R point mutations and exon 19 deletions in EGFR; less common mutations including resistance mutations were not evaluated. However, deletions in exons 19 and L858R point mutations have been reported to account for up to 90% of known mutations in EGFR.(31) We had a largely complete set of data, with only 16 patients having indeterminate or inconclusive EGFR mutational status in our entire cohort. Moreover, our study of KRAS in 374 patients with informative analysis is the largest single center series, to our knowledge, analyzing the impact of race on KRAS.
In conclusion, our study demonstrates that while the frequency of EGFR mutation was not statistically significantly lower among AfA patients than among C patients on univariate analysis, multivariable modeling revealed a significant difference in mutational frequency. Patients with EGFR mutation were more likely to be female, never or less extensive smokers, and have fewer comorbid conditions than their wild-type counterparts. Within our cohort, the incidence of EGFR mutation was not significantly different between adenocarcinoma and squamous NSCLC, thus supporting an individualized approach to mutational testing. Patients with KRAS mutation were more likely to be female and either former or current smokers. To our knowledge, this is the largest series to date evaluating molecular mutational status among patients with NSCLC as a function of race.
Acknowledgements
This project is funded, in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. This project is supported, in part, by GO grant 5UC2CA148310-02.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement:
None Declared
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62(1):10–29. doi: 10.3322/caac.20138. doi: papers2://publication/doi/10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C, Wu Y-L, Chen G, Feng J, Liu X-Q, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. The lancet oncology. 2011;12(8):735–42. doi: 10.1016/S1470-2045(11)70184-X. doi: papers2://publication/doi/10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Wu Y-L, Thongprasert S, Sunpaweravong P, Leong S-S, Sriuranpong V, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(21):2866–74. doi: 10.1200/JCO.2010.33.4235. doi: papers2://publication/doi/10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. doi: papers2://publication/doi/10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, Park K, Kim SW. A randomized phase III study of gefitinib versus standard chemotherapy (gemcitabine plus cisplatin) as a first-line treatment fornever-smokers with advanced or metastatic adenocarcinoma of the lung; World Conference on Lung Cancer; 2009; doi: papers2://publication/uuid/6B768E63-9B38-45C6-BEA3-C3BCB9556889. [Google Scholar]
- 6.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine. 2010;362(25):2380–8. doi: 10.1056/NEJMoa0909530. doi: papers2://publication/doi/10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The lancet oncology. 2010;11(2):121–8. doi: 10.1016/S1470-2045(09)70364-X. doi: papers2://publication/doi/10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The lancet oncology. 2012;13(3):239–46. doi: 10.1016/S1470-2045(11)70393-X. doi: papers2://publication/doi/10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 9.D’Angelo SP, Pietanza MC, Johnson ML, Riely GJ, Miller VA, Sima CS, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(15):2066–70. doi: 10.1200/JCO.2010.32.6181. doi: papers2://publication/doi/10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo E, Baselga J. Ethnic differences in response to epidermal growth factor receptor tyrosine kinase inhibitors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24(14):2158–63. doi: 10.1200/JCO.2006.06.5961. doi: papers2://publication/doi/10.1200/JCO.2006.06.5961. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(36):13306–11. doi: 10.1073/pnas.0405220101. doi: papers2://publication/doi/10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23(4):857–65. doi: 10.1200/JCO.2005.08.043. doi: papers2://publication/doi/10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 13.Underwood JM, Townsend JS, Tai E, Davis SP, Stewart SL, White A, et al. Racial and regional disparities in lung cancer incidence. Cancer. 2012;118(7):1910–8. doi: 10.1002/cncr.26479. doi: papers2://publication/doi/10.1002/cncr.26479. [DOI] [PubMed] [Google Scholar]
- 14.Berger M, Lund MJ, Brawley OW. Racial disparities in lung cancer. Current problems in cancer. 2007;31(3):202–10. doi: 10.1016/j.currproblcancer.2007.02.002. doi: papers2://publication/doi/10.1016/j.currproblcancer.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. Journal of chronic diseases. 1987;40(5):373–83. doi: 10.1016/0021-9681(87)90171-8. doi: papers2://publication/uuid/EBD7B5DE-6D4F-49BD-836D-C55E8103B0D8. [DOI] [PubMed] [Google Scholar]
- 16.Aisner DL, Deshpande C, Baloch Z, Watt CD, Litzky LA, Malhotra B, et al. Evaluation of EGFR mutation status in cytology specimens: An institutional experience. Diagnostic cytopathology. 2011 doi: 10.1002/dc.21851. doi: papers2://publication/doi/10.1002/dc.21851. [DOI] [PubMed] [Google Scholar]
- 17.Yang SH, Mechanic LE, Yang P, Landi MT, Bowman ED, Wampfler J, et al. Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(6):2106–10. doi: 10.1158/1078-0432.CCR-04-1853. doi: publicationpapers2://publication/doi/10.1158/1078-0432.CCR-04-1853. [DOI] [PubMed] [Google Scholar]
- 18.Leidner RS, Fu P, Clifford B, Hamdan A, Jin C, Eisenberg R, et al. Genetic abnormalities of the EGFR pathway in African American Patients with non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(33):5620–6. doi: 10.1200/JCO.2009.23.1431. doi: papers2://publication/doi/10.1200/JCO.2009.23.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12(3 Pt 1):839–44. doi: 10.1158/1078-0432.CCR-05-1846. doi: papers2://publication/doi/10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 20.Cote ML, Haddad R, Edwards DJ, Atikukke G, Gadgeel S, Soubani AO, et al. Frequency and type of epidermal growth factor receptor mutations in African Americans with non-small cell lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6(3):627–30. doi: 10.1097/JTO.0b013e31820a0ec0. doi: papers2://publication/doi/10.1097/JTO.0b013e31820a0ec0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinersman JM, Johnson ML, Riely GJ, Chitale DA, Nicastri AD, Soff GA, et al. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in African Americans. J Thorac Oncol. 2011;6(1):28–31. doi: 10.1097/JTO.0b013e3181fb4fe2. doi: 10.1097/JTO.0b013e3181fb4fe2. PubMed PMID: 21107288; PubMed Central PMCID: PMC3337520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sequist LV, Joshi VA, Jänne PA, Muzikansky A, Fidias P, Meyerson M, et al. Response to treatment and survival of patients with non-small cell lung cancer undergoing somatic EGFR mutation testing. The oncologist. 2007;12(1):90–8. doi: 10.1634/theoncologist.12-1-90. doi: papers2://publication/uuid/7112082D-247A-474B-A8D4-86C0B9E6C204. [DOI] [PubMed] [Google Scholar]
- 23.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(3):1167–73. doi: papers2://publication/uuid/F6D01F68-5DAC-4BD9-BA36-8C0530A5159B. [PubMed] [Google Scholar]
- 24.Harada T, Lopez-Chavez A, Xi L, Raffeld M, Wang Y, Giaccone G. Characterization of epidermal growth factor receptor mutations in non-small-cell lung cancer patients of African-American ancestry. Oncogene. 2011;30(15):1744–52. doi: 10.1038/onc.2010.545. doi: papers2://publication/doi/10.1038/onc.2010.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettinger DS, Akerley W, Bepler G, Blum MG, Chang A, Cheney RT, et al. Non-small cell lung cancer. Journal of the National Comprehensive Cancer Network: JNCCN. 2010;8(7):740–801. doi: 10.6004/jnccn.2010.0056. doi: papers2://publication/uuid/32AC9037-9D65-4D59-B024-E6857CBFFF18. [DOI] [PubMed] [Google Scholar]
- 26.Nelson HH, Christiani DC, Mark EJ, Wiencke JK, Wain JC, Kelsey KT. Implications and prognostic value of K-ras mutation for early-stage lung cancer in women. Journal of the National Cancer Institute. 1999;91(23):2032–8. doi: 10.1093/jnci/91.23.2032. doi: papers2://publication/uuid/D6404CFC-0A9F-4954-A516-9D0DED69CAE8. [DOI] [PubMed] [Google Scholar]
- 27.Bacchi CE, Ciol H, Queiroga EM, Benine LC, Silva LH, Ojopi EB. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics (São Paulo, Brazil) 2012;67(5):419–24. doi: 10.6061/clinics/2012(05)03. doi: papers2://publication/uuid/F04AA1B0-F128-493A-A47B-F45F7553371A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouquette I, Lauwers-Cances V, Allera C, Brouchet L, Milia J, Nicaise Y, et al. Characteristics of lung cancer in women: importance of hormonal and growth factors. Lung cancer (Amsterdam, Netherlands) 2012;76(3):280–5. doi: 10.1016/j.lungcan.2011.11.023. doi: papers2://publication/doi/10.1016/j.lungcan.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clinical cancer research: an official journal of the American Association for Cancer Research. 1996;2(2):411–8. doi: papers2://publication/uuid/2191341A-2C0B-4BFC-988A-5EAF30CD76E1. [PubMed] [Google Scholar]
- 30.Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(22):2972–7. doi: 10.1200/JCO.2010.33.3906. doi: papers2://publication/doi/10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 31.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nature reviews Cancer. 2007;7(3):169–81. doi: 10.1038/nrc2088. doi: papers2://publication/doi/10.1038/nrc208. [DOI] [PubMed] [Google Scholar]