Abstract
Mesenchymal stem cells (MSCs) respond to niche elasticity, which varies between and within tissues. Stiffness gradients result from pathological conditions but also occur through normal variation, e.g. muscle. MSCs undergo directed migration even in response to shallow stiffness gradients before differentiating. More refined gradients of both stiffness range and strength are needed to better understand mechanical regulation of migration in normal and disease pathologies. We describe polyacrylamide stiffness gradient fabrication using three distinct systems that generate stiffness gradients of physiological (1 Pa/µm), pathological (10 Pa/µm), and step (≥ 100Pa/um) strength spanning physiologically relevant stiffness for most soft tissue, i.e. 1–12 kPa. MSCs migrated to the stiffest region for each gradient. Time-lapse microscopy revealed that migration velocity scaled directly with gradient strength. Directed migration was reduced in the presence of the contractile agonist lysophosphatidic acid (LPA) and cytoskeletal-perturbing drugs nocodazole and cytochalasin; LPA- and nocodazole-treated cells remained spread and protrusive, while cytochalasin-treated cells did not. Untreated and nocodazole-treated cells spread in a similar manner, but nocodazole-treated cells had greatly diminished traction forces. These data suggest that actin is required for migration whereas microtubules are required for directed migration. The data also imply that in vivo, MSCs may have a more significant contribution to repairs in stiffer regions where they may preferentially accumulate.
Keywords: durotaxis, cell migration, stem cell, stiffness gradients, microfluidics
1. Introduction
In their native environment, cells are surrounded by extracellular matrix (ECM) which provides complex biochemical signals to cells as well as biophysical ones [1–3]. One example of a biophysical cue is the elastic modulus of the ECM, which varies dramatically between and within tissues [4, 5] (Figure 1A). By deforming their surroundings through cell-generated forces, cells can “feel” or sense this elastic modulus, often referred to as “stiffness” in the biological literature (measured in Pascals, or Pa). In the past two decades, hydrogel systems have been developed to more closely approximate the native ECM stiffness [6] in order to investigate mechanically coupled cellular functions such as cell morphology, spreading, maturation, and differentiation [6–14]. The majority of work to date has focused on cell responses to substrates of uniform stiffness, but cells may encounter dynamic environments where stiffness varies spatially, either naturally within tissues or as the result of a pathological condition such as the fibrotic lesions that develop after a myocardial infarct [15].
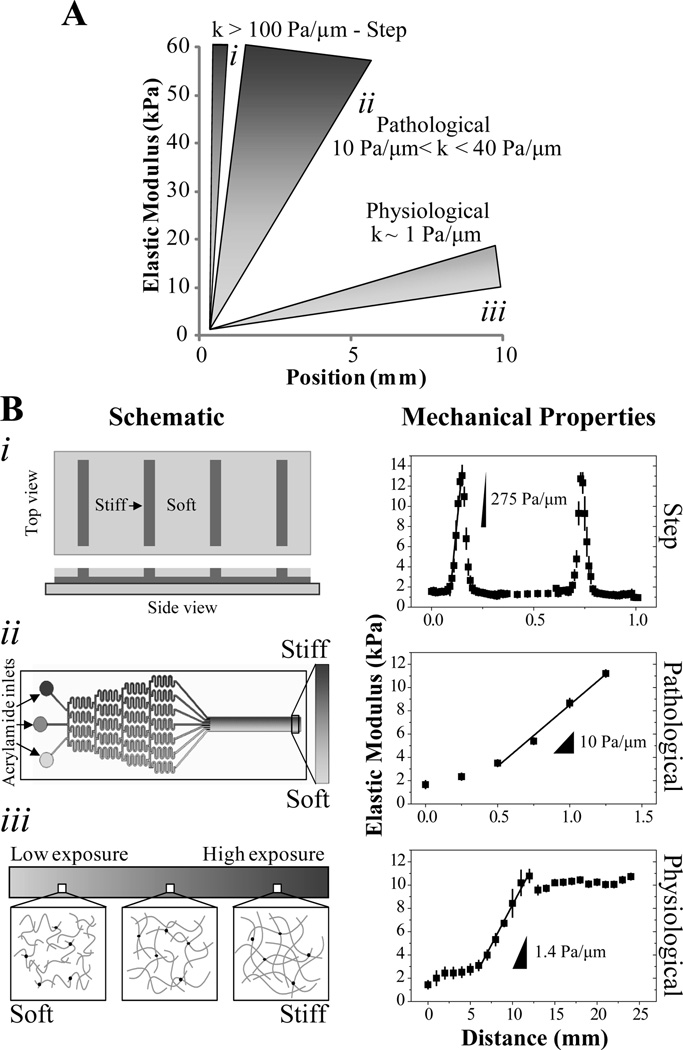
Figure 1.
(A) Schematic representation of step (i), pathological (ii), and physiological (i) stiffness gradients. (B) Three separate PA systems were developed to generate mechanical gradients of varying strength and of defined range spanning ~1–10 kPa. (i) At left is a schematic of a two-stiffness hydrogel where 500 µm wide regions of soft PA alternate with ~100 µm wide strips of stiff hydrogel producing a stripped stiffness profile. At right, there is a plot of stiffness with position, indicating that the sharp transitions between soft and stiff regions create gradients of >100 Pa/µm, n=4 gels. (ii) At left is a schematic of a microfluidic mixer that splits and recombines polymer solutions to generate a smooth gradient from discrete inputs. Photopolymerization of the solution in the outlet channel yields a PA hydrogel with a uniform, one-dimensional pathological stiffness gradient of 10 Pa/µm as indicated at right, n=3 gels. (iii) A photomask decreasing in transparency from 100 to 30% modulates the intensity of UV that reaches the polymer solution, leading to changes in polymer chain length as illustrated at left resulting in a UV transmission gradient. This results in PA substrate with a ~1 Pa/µm mechanical gradient as shown at right, n=3 gels.
Durotaxis, the directed migration of cells up a stiffness gradient, was originally observed in fibroblasts migrating across a soft-to-stiff interface of two juxtaposed polyacrylamide hydrogels [16]. While such sharp transitions clearly illustrate this behavior, most pathological conditions create gradients that are much less steep, e.g. myocardial infarction establishes gradients ~ 8 kPa/µm [15]. Indeed, preferential migration of vascular smooth muscle cells (VSMC) but not valvular interstitial cells has been documented on substrates ranging from ~2 to 40 kPa [17–19]. Both the range and the strength, or change in elastic modulus per unit length, of these mechanical gradients varies between studies, thus making it unclear as to how each parameter contributes to durotaxis. While VSMC polarization was found to increase with increasing gradient strength, differences in the gradient range require producing mechanical gradients of varying strength while keeping the stiffness value within a defined range to better understand the durotactic process [20]. Here, we explore this issue by using a defined range of physiological relevance.
Mesenchymal stem cells (MSCs) are also very migratory as they must egress from bone marrow, migrate through tissue, and hone in on an injury site, but unlike VSMCs, they also differentiate in response to stimuli, making their behavior more complex and difficult to predict. To migrate in vivo, local fibrotic tissue may serve as a homing signal encourage cells to preferentially accumulate, and while chemical signals are commonly implicated, ECM stiffness gradients may serve as a guide to MSC migration to ensure that the right cells can differentiate in the right place as part of the healing process [8, 21–23]. Such durotactic homing does occur for MSCs as we have previously observed MSCs undergoing directed migration even in response to shallow, physiological (1 Pa/µm) stiffness gradients. This process precedes differentiation [5], making its understanding more relevant to better address migration in therapeutic niches. Given the existence of stiffness variation between and within tissues, it remains to be seen whether MSC homing or durotaxis is influenced by the stiffness range or gradient strength. Such knowledge could help determine how stiffness gradients may be most effective at directing the therapeutic accumulation of MSCs.
Here we sought to understand the response of MSCs on hydrogels that mimic natural tissue stiffness variations (1 Pa/µm), pathological conditions (10 Pa/µm), and tissue interfaces that present step changes in stiffness (>100 Pa/µm). By focusing on gradient ranges between 1–12 kPa, we can better determine how stiffness gradient strength dictates MSC migration versus differentiation. To this end, we first fabricated polyacrylamide (PA) hydrogels with gradients using three separate techniques (Figure 1B), each suited to create a specific gradient strength. Photopolymerization through a photomask yields shallow gradients due to polymer diffusion over the time scale required for polymerization [5, 17]. Microfluidic mixing chambers can create steeper gradients than photomasks, given that the input solutions and degree of mixing govern gradient strength [14, 24]. Two-step reverse cast polymerization techniques mirror previous juxtaposed polyacrylamide hydrogels [16] in such a way as to create reproducible, defined gradients. For example, Marklein and Burdick have recently created 500 µm-wide stripes of alternating stiffness in hyaluronic acid hydrogels [25] while Choi et al developed 100 and 500 µm-wide stripes of alternating stiffness in PA hydrogels [26]. We cultured MSCs on these gradients to ask whether stiffness-directed migration is influenced by gradient strength, and we provide the first evidence for MSC durotaxis as a function of gradient strength over a specified physiological range. We also suggest a potential cytoskeletal mechanism that could regulate durotaxis but not necessarily migration in general.
2. Materials and methods
2.1. Polyacrylamide hydrogels
Polyacrylamide (PA) hydrogels were prepared from acrylamide monomers and the crosslinker N,N’-methylene-bis-acrylamide (Fisher Scientific). PA stiffness gradients of approximately 1, 10, and 100 Pa per µm corresponding to physiological, pathological, and step gradients, respectively, were created using three distinct systems described below. To facilitate cell attachment, human plasma fibronectin was covalently attached to the hydrogel surface. Substrates were incubated in 0.2 mM sulfo-SANPAH (Pierce) in sterile 50 mM HEPES pH 8.5, treated with 4mW/cm2 350 nm UV light for 10 min, washed three times with HEPES, and incubated with 10 µg/mL human fibronectin overnight at 37°C. Samples were stored in PBS at 4°C and UV sterilized prior to use. All chemicals were obtained from Sigma unless otherwise noted.
2.2. Fabrication of substrates with step gradients
To create hydrogels with very steep stiffness gradients, we employed a two-step polymerization scheme recently developed by Choi and co-workers [26]. First, master Si wafers were patterned with 25 mm long by 100 µm wide by 20 µm high cuboids spaced 500 µm apart using soft photolithography like described above. SU-8 2015 was used instead of SU-8 2050, and the exposure, development, and pre- and post-bake times were adjusted according to manufacturer specification. To covalently attach substrates to glass, glass coverslips (Fisher) were cleaned of organics and oxidized by exposing both sides for 60 sec to UV/ozone (BioForce). Samples were immediately functionalized with 20 mM 3-(trimethoxysilyl)propyl methacrylate in ethanol, washed with ethanol, and dried. 20 µL of a polymer solution consisting of 4% acrylamide and 0.4% bis-acrylamide 1/100 volume of 10% ammonium persulfate (APS) and 1/1000 volume of N,N,N’,N’-tetramethylethylenediamine (TEMED) was pipetted onto the wafer and covered with a methacrylated coverslip and the solution allowed to polymerize for 15 min. The hydrogel was released from the wafer and placed face down onto a 20 µL drop of polymer solution consisting of 3.2% acrylamide and 0.4% bis-acrylamide, 1/100 volume of 10% APS, and 1/1000 volume of TEMED on top of a dichlorodimethylsilane treated glass slide. The second solution was allowed to polymerize for 15 minutes before soaking the resulting inter-penetrating hydrogel network in DI (deionized) water. Many gradients were fabricated simultaneously from the same polymer solutions by using multiple master wafers to limit batch to batch variability. Two hydrogels from each polymerization batch were checked by atomic force microscopy to verify substrate mechanical properties.
2.3. Fabrication of substrates with pathological stiffness gradients
Graded photoactivation is of insufficient resolution to achieve pathological stiffness gradients. Using the microfluidic mixing device developed by Zaari and coworkers [18] and detailed by Byfield and coworkers [27], we created gradients steep enough to mimic pathological stiffness variations [15]. Briefly, silicon wafers were cleaned with acetone, methanol, and ethanol prior to processing. Approximately 100 µm of SU-8 2050 negative photoresist (Microchem) was spin coated onto the wafer, prebaked at 65°C for 5 min and then 95°C for 20 min. The substrates were allowed to cool at room temperature before exposing with 300 mJ of 365 nm light through the transparency photomask designed in AutoCad depicting the microfluidic channels (CAD/Art Services). Exposure was performed on an MA-6 mask aligner (SUSS MicroTec). A postbake was performed for 1 min at 65°C followed 5 min at 95°C. The samples were once again allowed to cool to room temperature before submerging for 10 min in SU-8 developer to remove the uncrosslinked regions. Samples were washed with isopropanol and dried with ultrapure nitrogen. Feature dimensions were verified using a Dektak profilometer (Veeco). Master wafers were treated overnight with the fumes of (Tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane (United Chemical Technologies) to promote the subsequent polymer release. A 10:1 polydimethylsiloxane (PDMS) elastomer:curing agent solution (Sylgard 184, Dow Corning) was thoroughly mixed and degased under vacuum for 1 hour before pouring over the masters, baked at 60°C for an hour in an oven, and then released from the wafer. The resulting channels were prepped, treated for 30 sec under UV ozone along with methacrylate functionalized glass slides, and immediately reversibly bonded together. The microfluidic channels were then further processed before use by selectively wicking in a 10% solution bis(3-Triethoxysilylpropyl)disulfide (SCA 985, Struktol) in acetone into the outlet portion of the channel for 30 sec as indicated. The solution was suctioned out the outlet channel with vacuum and the photoinitiator solution consisting of 10% 2,2-diethoxyacetophenone (acetophenone) in acetone was wicked three times at 30 sec intervals into the outlet channel to remove residual SCA 985. The acetone swells the PDMS and the water insoluble hydrophobic initiator binds to the elastomer surface. Finally, the mixture was suctioned out and the channels used within the hour. Polymer solutions consisting of 10% acrylamide and either 0.05% (low) or 0.5% (high) bis-acrylamide in DI water were injected with a syringe pump (KD Scientific) at 30 µL/min into the three inlets in this order: low-high-low. The solutions split and recombined and after reaching steady state in the outlet portion of the channel the flow was turned off and the polymerization initiated by turning on the UV transilluminator for 6 min located directly beneath the outlet portion of the microchannel. After polymerization, the PDMS was gently removed and the resulting hydrogel stuck to the methacryalted glass, 1.7 mm in width and ~2 cm in length, immediately immersed in DI water.
2.4. Fabrication of substrates with physiological stiffness gradients
A strategy for making physiological stiffness gradients was modified from Tse and Engler [28]. Photomasks decreasing in transparency from 100 to 30% were designed in Photoshop (Adobe) over different lengths and were printed on transparency sheets using a 600 DPI printer. A stock polymer solution consisting of 10% acrylamide and 0.1% bis-acrylamide in DI water was prepared, stored at 4°C, and used for all experiments. Small aliquots of the polymer solution were mixed with 2,2′-azobis(2-methylpropionamidine) dihydrochloride (azobis), a photoinitiator, to a final concentratation of 0.5% w/v. 20 µL of the polymer solution was sandwiched between 25 mm square methacylate- and dichlorodimethylsilane-treated slides (the former for attachment and the latter for release). The glass-polymer-glass sandwich was aligned on top of the photomasks and the whole apparatus placed on the surface of a benchtop UV transilluminator equipped with 1 mW/cm2 305 nm tubes. A cutout was placed around and below the photomask to prevent stray light from influencing polymerization. After polymerization for 6 min, the substrate was removed and immediately immersed in water to remove unreacted species. Samples were created from the same polymer solution mixture, and two hydrogels from each polymerization batch were checked by atomic force microscopy to verify consistent substrate mechanical properties.
2.5. Material stiffness and surface topography
Force-mode Atomic Force Microscope (AFM) was performed to determine the mechanical properties of the various hydrogels. Samples were mounted on glass slides using vacuum grease and then on the AFM stage (3DBio; Asylum Research). Samples were indented 300 nm using gold-coated, pyramid-shape SiN cantilevers (TR400PB; Olympus) with ~25 pN/nm nominal spring constants as determined from indentations on a silicon surface and thermal calibration. Measurements were taken every 500–1000 µm, 250–500 µm, and 10–50 µm for stiffness gradients of 1, 10, and >100 Pa/µm, respectively. Force curves were analyzed in Igor using a linearized Hertz model to determine the Young’s modulus [29]. For 100 Pa/µm gradients, the soft-stiff transition topography and modulus were obtained with 90×90 µm force maps.
2.6. Cell culture
Mesenchymal stem cells (MSCs, Lonza Walkersville) were cultured in low glucose DMEM supplemented 10% fetal bovine serum and 1% Penn/Strep (Gibco). For culture maintenance, media was changed every three days and cells passaged before reaching confluence to maintain multipotency. Stem cells between passages 4–8 were used for all experiments, seeded at 5×103 to 2.5×104 cells/ml and media changed every 2–3 days. For inhibitor studies, lysophosphatidic acid (LPA, Enzo Life Scienes), nocodazole and cytochalasin D were used at a final concentration of 20 µM, 0.5 µM, and 2 µM, respectively. Inhibitors were dissolved in DMSO, stored at −20°C and the final concentration of DMSO in the media did not exceed 0.1% v/v.
2.7. Immunofluorescent staining and imaging
Cells were fixed with 10% formalin for 15 min at room temperature. Actin cytoskeleton was stained with 1:500 rhodamine phalloidin (Invitrogen, Carlsbad, CA) in 1% bovine serum albumin and a wash buffer (1 mM MgCl2 in phosphate buffered saline) for 30 min at 37°C. After rinsing thrice with wash buffer, nuclei were stained with 1:5000 Hoechst 33342 for 10 min at room temperature. For surface protein visualization, fibronectin-coated hydrogels were incubated with 1:500 R457 rabbit polyclonal anti-rat antiserum against the amino-terminal 70 kDa fragment of fibronectin in staining solution for 30 min at 37°C, washed thrice with buffer, and then incubated with 1:500 Alexa fluor 488-conjugated secondary antibody (1:500; Invitrogen) for 30 min at 37°C. All samples were washed with DI water and mounted using Fluoromount-G (SouthernBiotech). Samples were imaged by a CARV II confocal (BD Biosciences) Nikon Eclipse Ti microscope equipped with a TE2000-U motorized, programmable stage using a Cool-Snap HQ camera (Photometrics) and controlled by Metamorph 7.6 (Molecuar Devices). For time-lapse measurements, cells were placed inside a temperature, CO2, and humidity controlled LiveCell chamber (Pathology devices) and custom Metamorph journals acquired, reconstructed, and processed multi-positional scan slides images. Post-processing was performed in Methamorph 7.6 and ImageJ (NIH).
2.8. Traction Force Microscopy (TFM)
TFM was performed as described elsewhere using in-house Matlab (Mathworks) routines [30]. 2% v/v of 0.5 µm diameter Fluoresbrite YG Microspheres (Polysciences, Inc) were added to the polymer solutions prior to gelation and the positions of the beads acquired over time using fluorescent and brightfield time-lapse microscopy. After cell trypsinization, bead positions were acquired again and displacement maps were generated using image correlation algorithms similar to particle image velocimetry [31]. Traction stress maps were determined from the measured displacement maps by solving the equation of elastic equilibrium for the substrate. The hydrogel’s spatially-varying mechanical properties were considered by performing a perturbation expansion of the solution in terms of the stiffness gradient [32]. Samples were again imaged by a CARV II confocal (BD Biosciences) Nikon Eclipse Ti microscope equipped with a TE2000-U motorized, programmable stage using a Cool-Snap HQ camera (Photometrics) and controlled by Metamorph 7.6 (Molecuar Devices). Cell velocities were computed with ImageJ and only migration along the gradient direction was measured. The error in migration velocities measurements due to the spatial resolution of the images is on the order of 1 µm/hr.
2.9. Statistical analyses
All data are expressed as mean ± standard deviation of experiments unless otherwise noted. Non-parametric Wilcoxon rank-sum tests were used to perform all statistical analysis. Differences were considered significant when p<0.005 and indicated for all comparisons. All experiments were performed in triplicate unless otherwise noted, and in such cases, the number of cells used in the measurement has been stated.
3. Results
Fabrication of hydrogels with stiffness gradients
To generate physiological, pathological, and step stiffness gradients of defined range corresponding to approximately 1 Pa/µm, 10 Pa/µm, and 100 Pa/µm respectively (Figure 1A), we utilized three distinct systems (Figure 1B). Since our goal was to investigate the migration behavior of human mesenchymal stem cells (MSCs) solely based on gradient strength, system parameters were chosen such that the stiffness range was identical across all systems, spanning 1 to 12 kPa.
For step gradients, a two-step polymerization mechanism was used resulting in mechanically-patterned matrix. The stiff, bottom hydrogel was first polymerized on top of a micropatterned Si wafer; a second polymer solution was subsequently added on top of the first where it could selectively polymerize into the ‘fingers’ of the first layer, making them even stiffer, or it could be retained in grooves where it formed a softer hydrogel (Figure 1Bi) [26]. To highlight this, top and bottom hydrogel composition was set to be relatively similar – 3.2% vs. 4.0% acrylamide, respectively– but the bottom hydrogel’s stiffness after a second polymerization was roughly 13 kPa (Figure 1Bi), four times the expected value when polymerized as a thin film (Figure S1). Changing the bottom hydrogel’s composition by increasing the acrylamide percentage lead to stiffer bottom hydrogels (Figure S1A), but also magnified the stiffening effect during the second polymerization step. For instance, increasing the bottom hydrogel’s stiffness to 7.4 kPa using a 6% acrylamide solution (Figure S1A) resulted in a mechanically-patterned matrix whose stiff regions consistently exceeded 100 kPa (Figure S1B). The transition distance from soft to stiff stripes occurred over approximately 40 µm, effectively creating a stiffness gradient of 275 Pa/µm whose range spans an order of magnitude, i.e. 1 to 13 kPa (Figure 1Bi). It is important to note that these hydrogels have a continuous top layer made with a high crosslinker-containing solution, i.e. 0.4% bis-acrylamide, which minimizes topographical differences [26].
For pathological stiffness gradients, a microfluidic mixing device made out of polydimethylsiloxane (PDMS) was used to generate hydrogels with varying mechanical properties [18, 27]. Solutions with different concentrations of bis-acrylamide crosslinker but similar acrylamide concentrations are flowed in the inlets of the microchannel (Figure 1Bii) after the outlets had been functionalized with SCA 985 to promote later release of the hydrogel from the mold and the photoinitiator acetophenone to induce polymerization only in the outlet channel (Fig 2A). As the solutions reach the branch points, they split and recombine, mixing in the process and generating a gradient of crosslinker in the outlet portion of the channel (Figure 2B and 2C). Photopolymerization of the solution produced a hydrogel with a defined stiffness gradient up to 10 Pa/µm (Figure 1Bii). Modulating crosslinker concentration in microchannel inlets changed the stiffness profile of the resulting hydrogel including the gradient range and magnitude (data not shown).
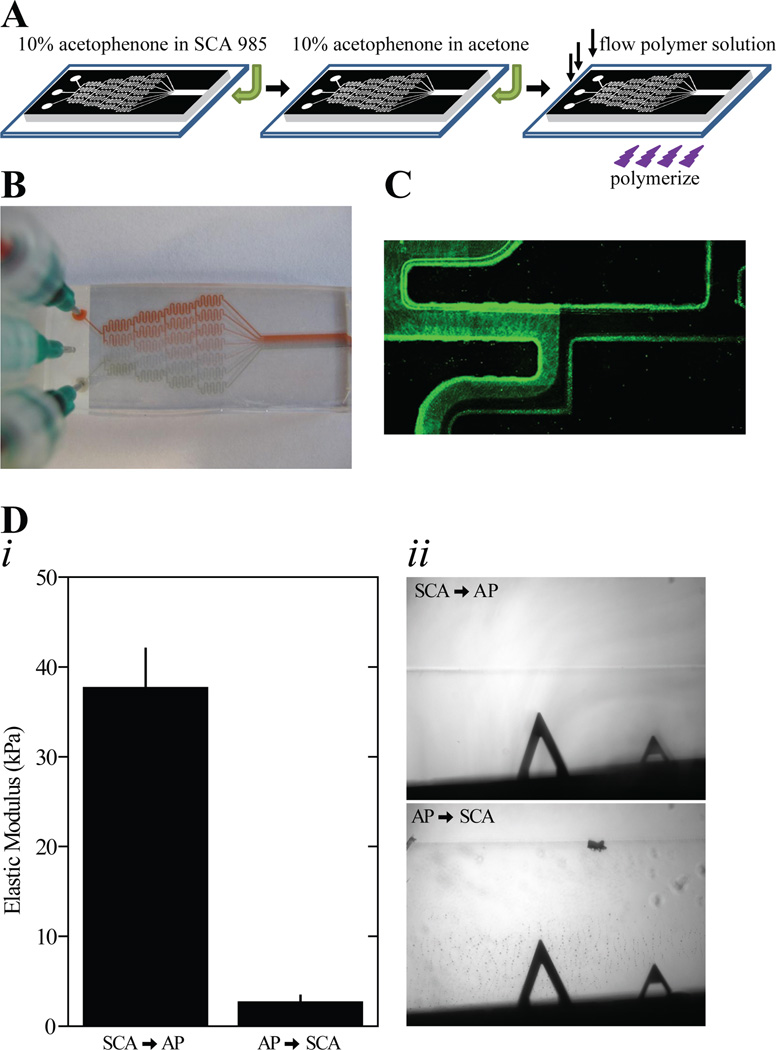
Figure 2.
(A) Schematic of the microfluidic gradient generator preparation process. (B) Image of the microfluidic gradient generator with inlets containing red, green, or no food coloring in water to visualize mixing in the channel. (C) Magnified fluorescent image of active flow in the microfluidic gradient generator at a branch point. The left inlet contains EosinY. (D) A solution was polymerized in the outlet of the gradient generator with acetophenone dissolved in SCA 985 or acetophenone dissolved in acetone and SCA 985 subsequently added. (i) Hydrogel stiffness is shown for the indicated order of adding acetophenone and SCA 985 to the gradient generator output channel. Error bars depict standard deviation. Measurements in triplicate were made at 6 distinct positions, n=3 gels. (ii) Phase contrast images are shown of the hydrogel edge when either SCA 985 or acetophenone was added first in the preparation process.
Several design changes from previous PDMS microfluidic mixing devices [18, 24, 33] were also introduced to ensure a consistent gradient with minimal swelling, limited topographical features, and a range of approximately 1 to 12 kPa, which were not controlled for in other methods and may have limited polymerization. For instance, using prior methods and a variety of UV wavelengths and exposure times, polymer solutions that normally yield 40 kPa substrates using APS/TEMED initiators produced less than 2 kPa here (Fig S2A) [18]. Moreover, a range of polymer concentrations and photoinitiators including irgacure 2959 and azobis when polymerized inside the PDMS microchannel could not create a gradient. Polymerization through fused quartz, which has superior UV transmission properties than glass [34], did not significantly increase gradient slope in the PDMS device either (Figure S2B). To address initiator concerns, acetophenone was adsorbed onto the PDMS surface to deliver it locally to the polymer solution [35], and photopolymerization of static hydrogels inside the outlet portion produced substrates with the same mechanical properties as thin films, e.g. 40 kPa (Fig 2Di). In contrast, acetophenone flowed through the channel prior to the releasing agent SCA 985 resulted in softer hydrogels with numerous surface defects that may interfere with cell adhesion (Figure 2D).
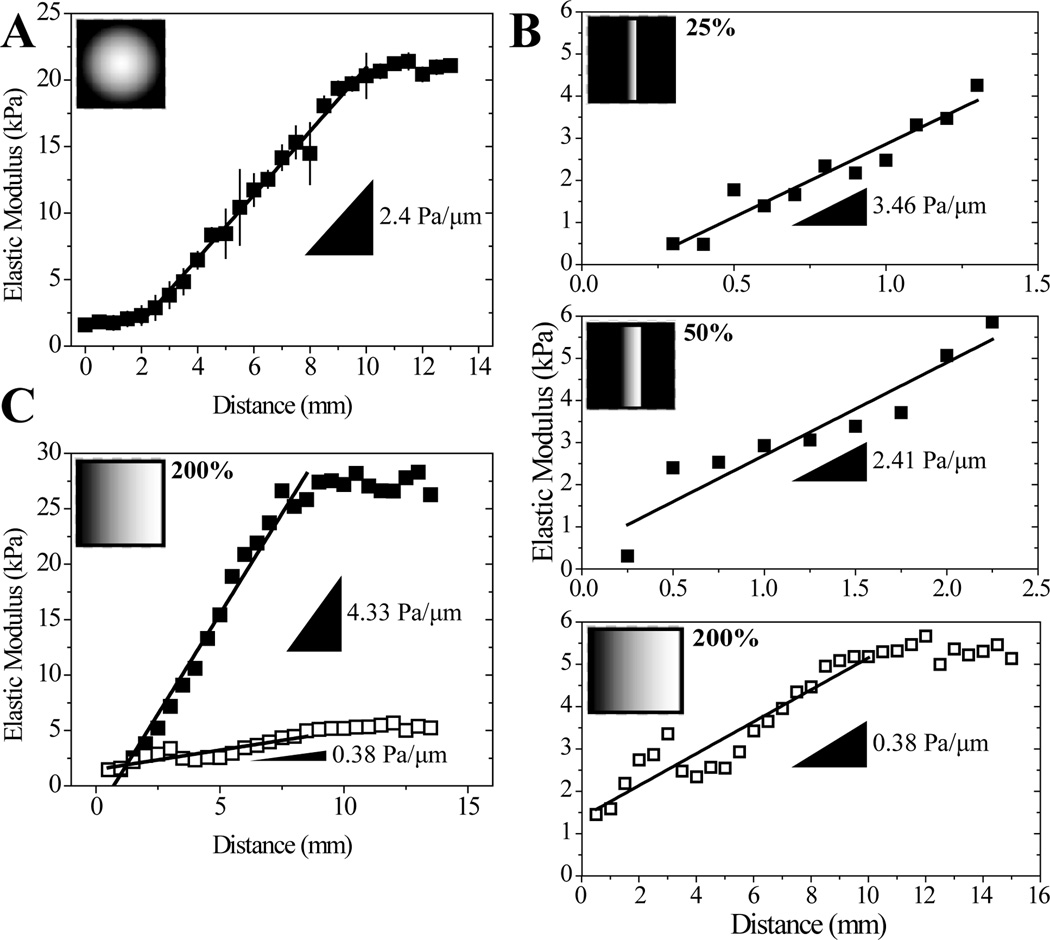
Photomasks allow one to spatially control the amount of UV light reaching the PA solution to modulate photopolymerization kinetics (Figure 1Biii). To achieve physiological gradients, we adapted a photoactivated polymerization procedure used by Tse and Engler where irgacure and a radial photomask are used to generate a shallow gradient (Figure 3A) [5]. Substituting in a linear photomask to more easily change gradient distance and using the more water-soluble initiator azobis, we were able to modulate gradient strength 10-fold, e.g. from 0.38 to 3.46 Pa/µm, even for gradient hydrogels with narrow stiffness ranges, i.e. 1–5 kPa (Figure 3B). For any given photomask, changing solution concentration, and thus the gradient hydrogel’s stiffness range, also changes the gradient strength from ~0.4 to 4 Pa/µm (Figure 3C). Tailoring this system to our specifications, we are able to span the 1 to 12 kPa range at a gradient strength of ~1 Pa/µm (Figure 1Biii) using a 12 mm opacity gradient photomask and a 10% acrylamide/0.1% bis-acrylamide solution.
Figure 3.
(A) Gradients generated using a radially symmetric mask and a solution containing 10% acrylamide, 0.3% bis-acrylamide, and 0.5% irgacure as the initiator. n=4 gels (B) Gradients produced with the same polymer solution (10% acrylamide, 0.1% bis-acrylamide) but using photomasks where the opacity gradient was scaled to 25%, 50%, or 200% of the distance used in Figure 1Bii, n=1. (C) Two different gradients made with the same photomask but different polymer solutions. Closed squares: 10% acrylamide and 0.3% bis-acrylamide, open squares: 10% acrylamide, 0.1% bis-acrylamide, n=1. Insets (A)–(C): Photomask images used for gradient fabrication with indicated photomask gradient distance relative to the photomask in Figure 1Bii.
Directed cell migration on stiffness gradients
Prior to cell adhesion, each PA hydrogel was functionalized with fibronectin to promote cell attachment to an otherwise inert substrate. With different PA hydrogel concentrations made by different methods, it was important to verify that the protein coating was consistent across individual gels, i.e. independent of substrate stiffness. Confocal cross-sections of fluorescently-labeled fibronectin indicated that there were no qualitative differences in protein attachment as a function of fabrication method, gradient strength, or absolute stiffness (Figure 4). Thus subsequent cell behavior differences should not be the result of spatial changes in surface ligand density.
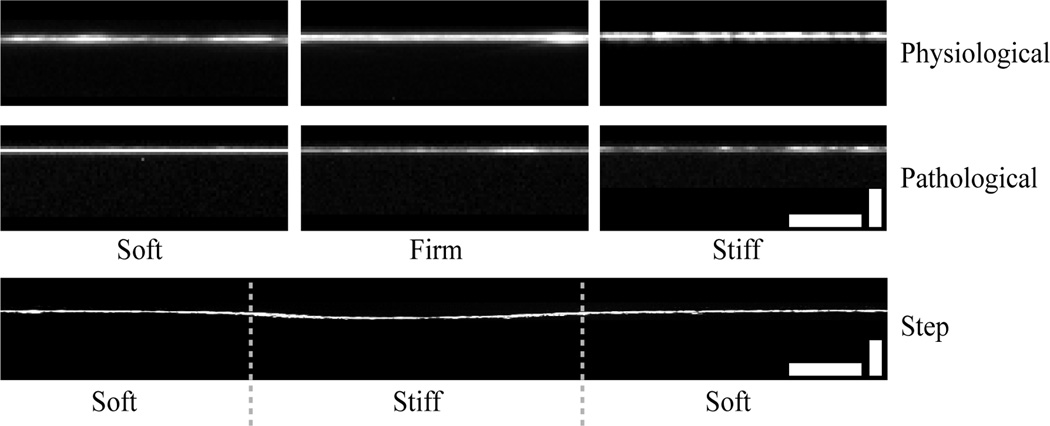
Figure 4.
Representative confocal cross-sections of each hydrogel system with fluorescently-labeled human plasma fibronectin. Each physiological gradient and pathological gradient image was averaged over 13 overlapped fluorescent cross-sections and repeated at least twice. Scale bars are 25 µm (horizontal) and 5 µm (vertical) for the physiological and pathological gradients and 25 µm (horizontal) and 20 µm (vertical) for step gradient.
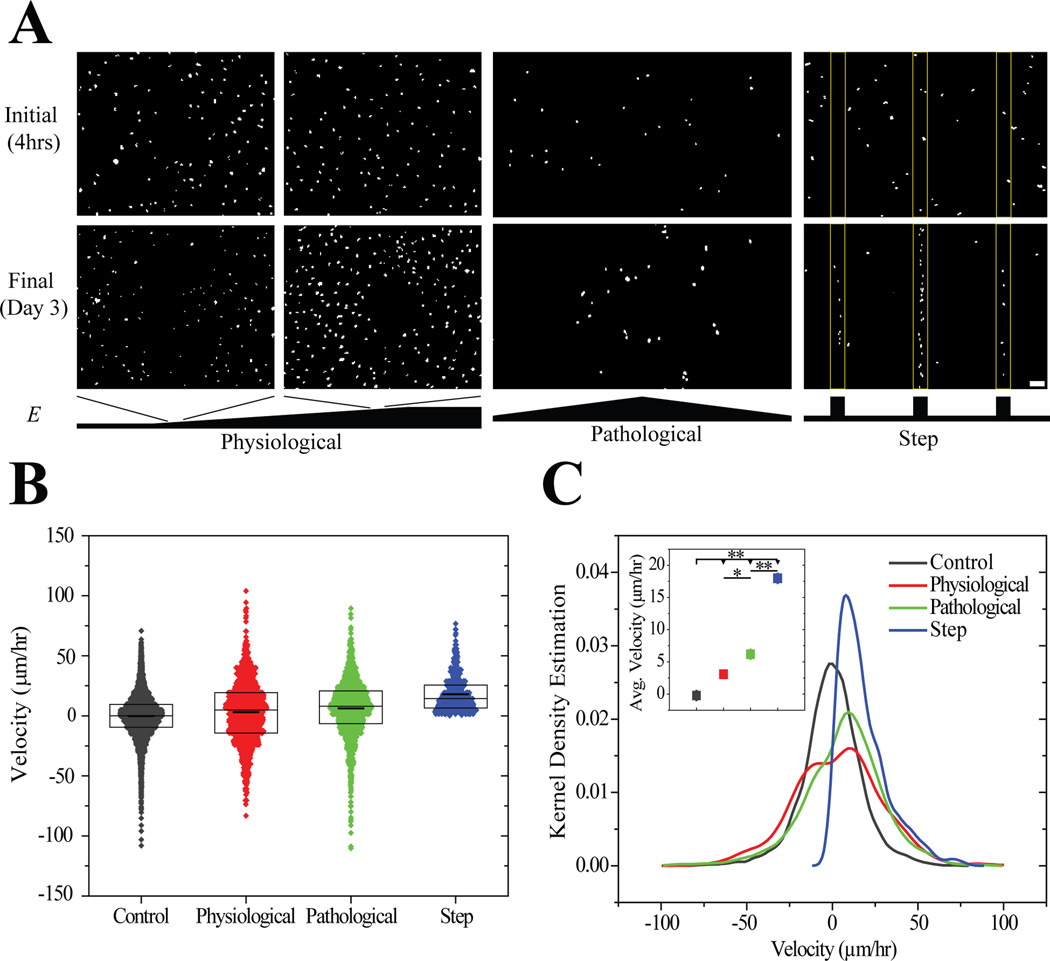
MSCs attached and spread independent of gradient strength or stiffness within hours of seeding, and after 3 days, cells migrated to stiffer portions of the substrates. Migration was most evident on step and pathological gradients (Figure 5A). It is important to note that MSCs durotax on physiological gradients although this is less evident in short-term experiments [28]. To verify that cells durotax and that the spatial differences in cell density were due to migration and not preferential proliferation on stiffer regions [13], durotactic migration velocity, i.e. migration in the direction of the gradient, was measured from time-lapse video microscopy. The distribution of instantaneous migration speeds was broad for cells on physiological and pathological gradients (Figure 5B), yet the population average indicated a net biased migration in the direction of the gradient at a rate of 3.0 ± 0.7 and 6.2 ± 0.6 µm/hr for physiological and pathological gradients, respectively (Figure 5C). Cells on step gradients migrated at 18.0 ± 0.7 µm/hr, more than 6-fold faster than on other gradient strengths, and it should be noted that negative velocities were not observed for cells migrating up step gradients. These data imply that biased migration velocity scales directly with gradient strength over two orders of magnitude, i.e. 1 to >275 Pa/µm (Figure 5C inset).
Figure 5.
(A) Thresholded images of Hoechst-stained MSCs on physiological (1.4 Pa/µm), pathological (10 Pa/µm), and step gradients (275 Pa/µm) 4 hours and 3 days after plating. Scale bar is 100 µm. (B) Velocities of migrating MSCs in the direction of the gradient determined from tracking live cells using time-lapse microscopy on physiological, pathological, and step gradients. Boxes indicate median, 25th, and 75th percentile and the thicker line indicates the average. (C) Kernel density estimation of cell velocities on the three gradient systems and average ± standard error of cell migration velocity for each system (inset). * p-value < 10−2, ** p-value <10−5. For step gradient, n=450 independent velocities. For physiological and pathological gradients, n>1300 independent velocities. Data were obtained from 3 biological replicates.
Regulating directed cell migration
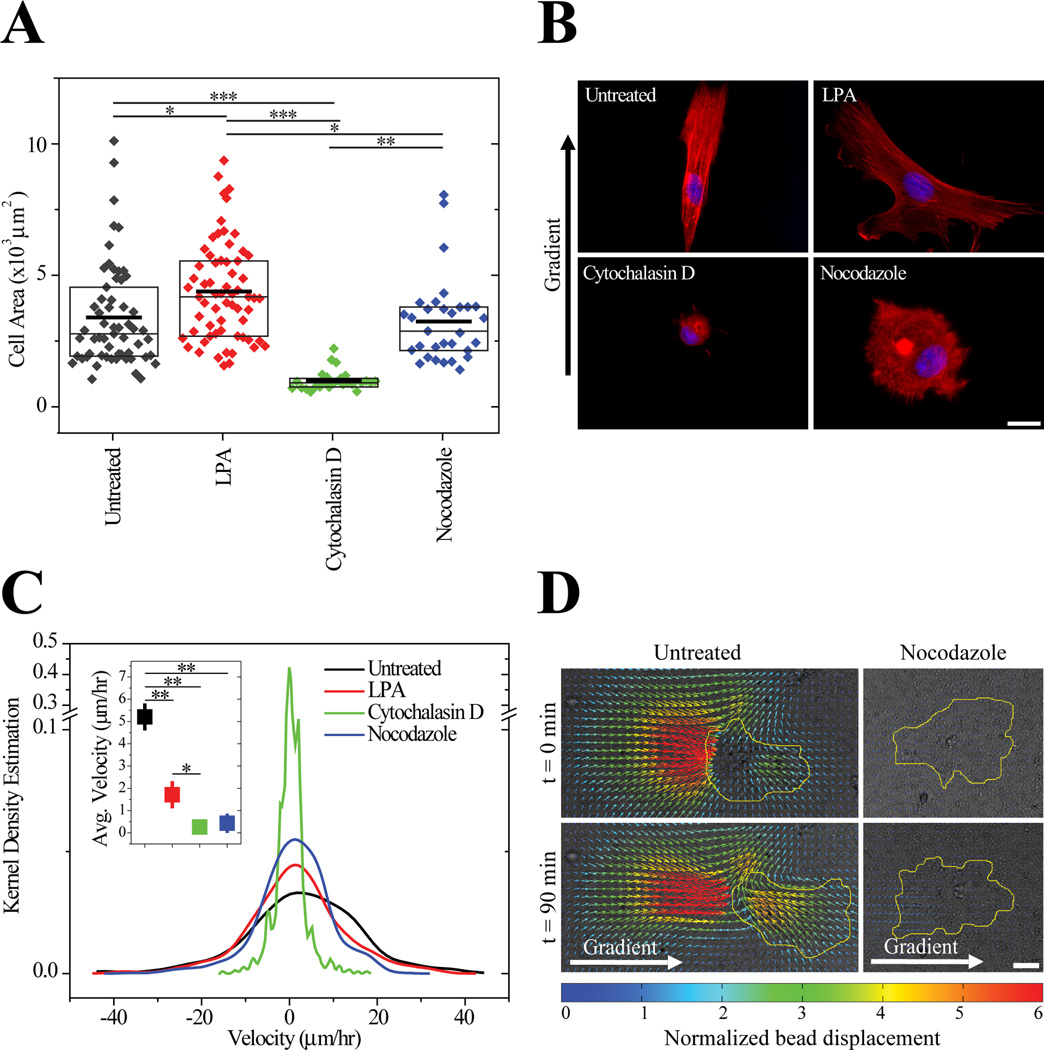
Since cell migration results from the coordination of cytoskeleton assembly and disassembly in both space and time, we sought to understand which elements were crucial for durotaxis. Untreated cells were less spread that cells treated with lysophosphatidic acid (LPA), a contractile agonist, equally spread compared to nocodazole-treated cells that cannot form stable microtubules, and more spread than cytochalasin D-treated cells which cannot polymerize an actin cytoskeleton (Figure 6A). Untreated cells on gradient substrates did not differ in spread area compared to cells on static matrices (3570±340 µm2 vs. 3400±260µm2). After 3 days in culture, untreated cells polarized their actin cytoskeleton in the direction of a pathological gradient, e.g. 8.7 ± 1.9 Pa/µm, whereas LPA-treated cells were randomly polarized and nocodazole- and cytochalasin D-treated cells maintained a rounded morphology that could not polarize (Figure 6B). All treatments impaired durotaxis significantly (Figure 6C), but while cytochalasin D- and nocodazole-treated MSCs remained largely stationary, LPA-treated MSCs remained spread, protrusive and migrated randomly (Supplemental videos 1–4). Together these data suggest that a stable actin cytoskeleton under appropriate tension is essential for MSC spreading but that microtubules are required for MSC polarization to initiate directed migration.
Figure 6.
(A) Spread area of MSCs on gradients either untreated (gray) or treated with lysophosphatidic acid (red), cytochalasin D (green), or nocodazole (blue) after 3 days. Boxes indicate median, 25th, and 75th percentile and the thicker line indicates the average. n>27 cells for each condition. (B) Nuclei (blue) and actin (red) of MSCs stained after 3 days in culture with inhibitors. Scale bar is 20 µm. (C) Migration of untreated and inhibitor treated MSCs on hydrogel with a pathological gradient of 8.7 ± 1.9 Pa/µm and range of 1 to 12 kPa. Inset depicts average ± standard error of cell migration velocity for each condition. For each condition, n>460 independent velocities. Data were obtained from 2 biological replicates. (D) Displacement maps of fluorescent particles embedded in the hydrogel obtained using particle image velocimetry for untreated and nocodazole-treated cells. Brightfield images and cells contours in yellow are overlaid with the displacement maps. Gradient is from left to right. Scale bar is 30 µm. * p-value < 10−2, ** p-value <10−5.
To determine if the directed migration of MSCs was due to contractile differences as a result of cytoskeletal changes caused by drug treatments, we performed traction force microscopy (TFM) on durotaxing MSCs plated on these gradients. Since MSCs were plated onto gradients, TFM software used to calculate traction stresses was specifically modified to adjust for a spatial stiffness gradient, and these changes were detailed elsewhere [32]. Over time, deformations on the stiffer, right side of the image for an untreated cell increased while cell deformations on the softer side decreased, resulting in MSC directed migration. In contrast, nocodazole-treated cells only slightly deformed the hydrogel despite changing morphology (Figure 6D). Converting displacements to traction forces and taking into account the graded mechanics of the hydrogel, untreated migrating MSCs are also better at forming force-generating protrusions than nocodazole-treated cells over time (Supplemental videos 5 and 6). These data indicate that stable microtubules are crucial to generate directed traction forces that encourage durotaxis but not essential for some modes of spreading.
4. Discussion
To study durotaxis over a range of gradient strengths, i.e. 1 to >100 Pa/µm, but defined stiffness range, i.e. 1 to 12 kPa, we developed three individual hydrogel systems, each of which was optimally suited for a specific gradient strength. Within that context, it is also critical to appreciate basic mechanistic reasons why cells would undergo directed migration. Thus here we put these systems, their troubleshooting, and the subsequent MSC behavior and mechanism in a broader context.
Challenges of Fabricating Reproducible Gradient Hydrogel Systems of Varying Gradient Strength and Stiffness Range
A variety of methods have been used to achieve spatial stiffness gradients, but each has a limited range of stiffness and gradient strength. Each method also requires standardizing methods to ensure that the gradient is reproducible and that there are no other mitigating factors that could unduly influence cell behavior. We have provided an overview of three gradient hydrogel systems noting their range and strength, and here describe their similarities and differences with similar systems used previously to fabricate stiffness gradients. For photolithographically patterned gradients, modulating light intensity to change radical polymerization kinetics has previously been used with photomasks [5, 17] or a sliding mask to vary hydrogel UV exposure time [36, 37]. This technique is well suited to generate physiological to pathological gradients, and we show its versatility in precisely controlling both the range and strength of gradients between 0.4 and 8.7 Pa/µm and 1 to 12 kPa. Here azobis was used as the photoinitiator due to its increased solubility in aqueous solutions and activation with long wave UV. Silica glass does not transmit below 300 nm, and since irgacure’s peak absorption occurs at 276 nm, its reaction is always suboptimal [34, 38]. Light diffraction and lateral diffusion of radicals also limit gradient resolution, but photomasks with high resolution have been used to control stiffness at the micron lengthscale of pre-polymerized hydrogels [25].
To achieve higher resolution and gradient strength, microfluidic channels more tightly constrain hydrogel dimensions and provide superior control over stiffness at the micron lengthscale based on channel design. In this type of system, stiffness was modulated by changing crosslinker concentration instead of modulating initiator activation along the width of the hydrogel [18, 24, 27]. Changing input solutions is simple, and allows for systematic spatial control over the mechanics in the outlet channel. However, PDMS is the common material used in these devices; it is porous and allows molecular oxygen to diffuse through the surface which reacts with radicals generated by the photoinitiator, effectively terminating the polymerization reaction [39, 40]. This effect has been minimal in quickly polymerizing systems such as poly(ethylene glycol) and even advantageous as the non-polymerized layer at the interface of the hydrogel and PDMS serves as a lubricating layer and promotes hydrogel release [24, 40]. For radical-based polymerization that takes several minutes, e.g. PA, oxygen radical scavengers in PDMS prevented the synthesis of hydrogels above 2 kPa over a range of water-soluble initiators, initiator concentrations, and UV treatments here despite studies showing otherwise [18, 20]. Absorbing hydrophobic acetophenone to the surface of PDMS allows for the polymerization of stiff PA hydrogels [27, 35], presumably because the initiator-generated radicals saturate at the PDMS surface and the rest diffuse into the polymer solution. However, PA polymerization with this method lacks the lubricating layer, and thus the PA grafts into the PDMS [27]. This prevents release and may cause the substrate to rip. SCA 985 covalently bound to the PDMS surface likely maintains the lubricating layer, and by coating it prior to absorbing acetophenone, stiff hydrogels were released from the PDMS mold without damaging the substrate’s surface.
There are also several challenges associated with step stiffness gradients from mechanically-patterned substrates. For example, the original method of polymerizing adjacent droplets of distinct acrylamide concentrations [16] forms a gradient from uncontrolled mixing, and while microfluidic gradients are well controlled, they often cannot achieve the steepest gradients, i.e. >100 Pa/µm [18, 24, 27]. Controlling the transition from soft to stiff was recently described by using a 2-step polymerization method to make mechanically-patterned hydrogels [25, 26]; here, by increasing the soft region width, we allowed cells to spread and randomly migrate before feeling the gradient near the interface and undergoing directed migration to the stiffer region. Though we used this system to understand migration up an exceedingly steep gradient, this platform has also been used to study how aligning cells improves their function, e.g. muscle cell fusion [26]. There are specific technical challenges worth noting; differential swelling between layers in microfabricated hydrogels have been reported and could introduce contract guidance cues similar to topographical patterns. By using substrates with high crosslinker content however [41], polymer chains become less free to slide across each other, dramatically reducing differential swelling between layers, i.e. < 2 µm, as well as roughness changes between stripes on the hydrogel, i.e. < 200 nm [26]. Polymer depletion effects in the soft stripes formed for the second layer’s polymerization also confounds predicting layer stiffness. For example the small changes in bottom hydrogel stiffness dramatically changed the final stiffness after the second acrylamide solution was polymerized and significantly deviated from monolayer hydrogel stiffness [5]. Materials that do not undergo such mixing may form more predictable layered materials, e.g. PDMS [42]. Despite both of these challenges, it should be noted that across all three systems, protein coating appeared uniform and initial cell adhesion was similar independent of polymer concentration and crosslink density.
The Origins of Durotaxis
Cells plated on the gradient systems migrated to the stiffer end of the hydrogels at different speeds, indicating that MSC durotaxis velocity depends on gradient strength as previously suggested with other cell types [20] but over a wider range of gradients here. It is important to note that the migration velocities reported here were obtained over the same physiological stiffness range of 1 to 12 kPa, indicating that gradient strength and not absolute stiffness drove directed MSC migration on gradient substrates. While these velocities were time-averaged, MSCs did not always feel the gradient, as with step gradients for the mechanically-patterned hydrogels; MSCs that approached the interface migrated faster than the time-averaging used here and thus their 18 µm/hr speed could be an underestimation. Despite that consideration, it is clear that durotactic speed increases but is not linear with respect to gradient strength. In addition to speed, cell traction forces enable MSCs to feel stiffness gradients and then migrate, but as of yet, this process remains unclear. Neutrophils, which undergo amoeboid-like migration, form rearward contractile centers, which subsequently squeeze the cell forward in the direction of a chemotactic gradient [43, 44]. This process, though different from the multi-step mesenchymal migration of MSCs [45], also requires myosin II [46], and interestingly, MSC traction distribution in cells migrating up the gradient seemingly mirror the rearward contractile stresses observed in neutrophils. One plausible explanation may be that cells feel their environment by sensing the strain they can impart on their matrix [47]. When oriented into a gradient, displacements were highest in the more compliant rear of the cells, where strain sensors could be most active and positively feeding signals to the cell. A variety of strain sensors have been proposed [48–50] and likely require some minimum level of signaling to encourage cell behaviors such as spreading and migration [51, 52].
The sensors required for this process, and thus their signaling capabilities, are likely connected to the cells’ cytoskeleton, since altering the cell’s ability to assemble a stable cytoskeleton significantly reduced MSC migration though these observations did not necessarily correlate with changes in cell spread area. The loss of microtubule architecture inhibited cell polarity with the cells remaining spread yet non-migratory. Similar to a previous report on nocodazole-treated fibroblasts [53], MSCs appear to require assembled microtubules to transmit forces to their surrounding environment but an actin cytoskeleton is necessary to adopt a spread morphology. Interestingly, cells treated with the multifunctional phospholipid messenger LPA, which promotes activation of the Rho and Ras GTP-ases [54] and thus cellular tension [55], did not undergo directed migration despite their heightened ability to ‘feel’ matrix stiffness [56]. LPA signaling is thought to contribute to cancer initiation, progression, and metastasis [57] though our results also suggest the inhibition of durotaxis. This observation is particularly relevant since the migration of untreated cells up the gradient relies on their ability to deform the matrix and generate traction stresses on the order of a few hundred Pascals.
Together these data imply that durotaxis mechanism(s) are force dependent, require an assembled microtubule network, and also require precise coordination of contractility in time and space since both increased and decreased cell contractility abolished directed migration. More importantly through the development of these three platforms, it was possible to fabricate gradients of the necessary range and strength to answer these questions.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Somyot Chirasatitsin and Alexander Fuhrmann for technical assistance with atomic force microscopy and quantification of cell spread areas, respectively. This work was supported by grants from the NIH (DP02OD006460 to A.J.E., and 1R01GM084227 to J.C.A), the Human Frontiers Science Foundation (RGY0064/2010 to A.J.E.), Graduate Research Fellowship from the National Science Foundation (to L.G.V), and Achievement Rewards for College Scientists Fellowship (to L.G.V).
Abbreviations
- APS
ammonium persulfate
- AFM
atomic force microscope
- azobis
2,2′-azobis(2-methylpropionamidine) dihydrochloride
- Da
Dalton
- DI
deionized
- DMEM
Dulbecco’s modified eagle medium
- DMSO
dimethyl sulfoxide
- DPI
dots per inch
- ECM
extracellular matrix
- LPA
lysophophatidic acid
- MSCs
Mesenchymal stem cells
- Pa
pascal
- PA
polyacrylamide
- PDMS
polydimethylsiloxane
- TEMED
N,N,N’,N’-tetramethylethylenediamine
- UV
ultraviolet
- VSMC
vascular smooth muscle cells
Footnotes
Conflict-of-Interest Statement
The authors declare no commercial or financial conflict of interest.
Citations
- 1.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badylak SF. Regenerative medicine and developmental biology: the role of the extracellular matrix. Anat Rec B New Anat. 2005;287:36–41. doi: 10.1002/ar.b.20081. [DOI] [PubMed] [Google Scholar]
- 3.Hay ED. Extracellular matrix. J Cell Biol. 1981;91:205s–223s. doi: 10.1083/jcb.91.3.205s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent L, Engler AJ. 5.504 - Effect of Substrate Modulus on Cell Function and Differentiation. In: Paul D, editor. Comprehensive Biomaterials. Oxford: Elsevier; 2011. pp. 51–63. Editor-in-Chief. [Google Scholar]
- 5.Tse JR, Engler AJ. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saha K, Keung AJ, Irwin EF, Li Y, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 9.Huebsch N, Arany PR, Mao AS, Shvartsman D, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong JY, Leach JB, Brown XQ. Balance of chemistry, topography, and mechanics at the cell–biomaterial interface: Issues and challenges for assessing the role of substrate mechanics on cell response. Surface Science. 2004;570:119–133. [Google Scholar]
- 15.Berry MF, Engler AJ, Woo YJ, Pirolli TJ, et al. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am J Physiol Heart Circ Physiol. 2006;290:H2196–H2203. doi: 10.1152/ajpheart.01017.2005. [DOI] [PubMed] [Google Scholar]
- 16.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong JY, Velasco A, Rajagopalan P, Pham Q. Directed Movement of Vascular Smooth Muscle Cells on Gradient-Compliant Hydrogels†. Langmuir. 2003;19:1908–1913. [Google Scholar]
- 18.Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Photopolymerization in Microfluidic Gradient Generators: Microscale Control of Substrate Compliance to Manipulate Cell Response. Advanced Materials. 2004;16:2133–2137. [Google Scholar]
- 19.Kloxin AM, Benton JA, Anseth KS. In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials. 2010;31:1–8. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isenberg BC, Dimilla PA, Walker M, Kim S, Wong JY. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama Y, Battista M, Kao WM, Hidalgo A, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 22.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 23.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol Cell Physiol. 2008;295:C1037–C1044. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 24.Burdick JA, Khademhosseini A, Langer R. Fabrication of Gradient Hydrogels Using a Microfluidics/Photopolymerization Process. Langmuir. 2004;20:5153–5156. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 25.Khetan S, Burdick JA. Patterning network structure to spatially control cellular remodeling and stem cell fate within 3-dimensional hydrogels. Biomaterials. 2010;31:8228–8234. doi: 10.1016/j.biomaterials.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 26.Choi YS, Vincent LG, Lee AR, Kretchmer KC, et al. The alignment and fusion assembly of adipose-derived stem cells on mechanically patterned matrices. Biomaterials. 2012;33:6943–6951. doi: 10.1016/j.biomaterials.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byfield FJ, Wen Q, Levental I, Nordstrom K, et al. Absence of filamin A prevents cells from responding to stiffness gradients on gels coated with collagen but not fibronectin. Biophys J. 2009;96:5095–5102. doi: 10.1016/j.bpj.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. Chapter 10, Unit 10 16. [DOI] [PubMed] [Google Scholar]
- 29.Kaushik G, Zambon AC, Fuhrmann A, Bernstein SI, et al. Measuring passive myocardial stiffness in Drosophila melanogaster to investigate diastolic dysfunction. J Cell Mol Med. 2012;16:1656–1662. doi: 10.1111/j.1582-4934.2011.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Alamo JC, Meili R, Alonso-Latorre B, Rodriguez-Rodriguez J, et al. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc Natl Acad Sci U S A. 2007;104:13343–13348. doi: 10.1073/pnas.0705815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willert CE, Gharib M. Digital particle image velocimetry. Experiments in Fluids. 1991;10:181–193. [Google Scholar]
- 32.Alonso-Latorre B. University of California, San Diego, United States -- California. 2010:153. [Google Scholar]
- 33.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, et al. Generation of Solution and Surface Gradients Using Microfluidic Systems. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 34.Kitamura R, Pilon L, Jonasz M. Optical constants of silica glass from extreme ultraviolet to far infrared at near room temperature. Appl. Opt. 2007;46:8118–8133. doi: 10.1364/ao.46.008118. [DOI] [PubMed] [Google Scholar]
- 35.Hu S, Ren X, Bachman M, Sims CE, et al. Surface-Directed, Graft Polymerization within Microfluidic Channels. Analytical Chemistry. 2004;76:1865–1870. doi: 10.1021/ac049937z. [DOI] [PubMed] [Google Scholar]
- 36.Sunyer R, Jin AJ, Nossal R, Sackett DL. Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLoS One. 2012;7:e46107. doi: 10.1371/journal.pone.0046107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marklein RA, Burdick JA. Spatially controlled hydrogel mechanics to modulate stem cell interactions. Soft Matter. 2010;6:136–143. [Google Scholar]
- 38.Ciba Specialty Chemicals Inc. 2003:8. [Google Scholar]
- 39.Decker C, Jenkins AD. Kinetic approach of oxygen inhibition in ultraviolet- and laser-induced polymerizations. Macromolecules. 1985;18:1241–1244. [Google Scholar]
- 40.Dendukuri D, Pregibon DC, Collins J, Hatton TA, Doyle PS. Continuous-flow lithography for high-throughput microparticle synthesis. Nat Mater. 2006;5:365–369. doi: 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- 41.Charest JM, Califano JP, Carey SP, Reinhart-King CA. Fabrication of substrates with defined mechanical properties and topographical features for the study of cell migration. Macromol Biosci. 2012;12:12–20. doi: 10.1002/mabi.201100264. [DOI] [PubMed] [Google Scholar]
- 42.Gray DS, Tien J, Chen CS. Repositioning of cells by mechanotaxis on surfaces with micropatterned Young's modulus. J Biomed Mater Res A. 2003;66:605–614. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 43.Jannat Risat A, Dembo M, Hammer Daniel A. Traction Forces of Neutrophils Migrating on Compliant Substrates. Biophysical Journal. 2011;101:575–584. doi: 10.1016/j.bpj.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alonso-Latorre B, Meili R, Bastounis E, del Alamo JC, et al. Engineering in Medicine and Biology Society, 2009. EMBC 2009. Annual International Conference of the IEEE. 2009:3346–3349. doi: 10.1109/IEMBS.2009.5333191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harb Perspect Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meili R, Alonso-Latorre B, del Alamo JC, Firtel RA, Lasheras JC. Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Mol Biol Cell. 2010;21:405–417. doi: 10.1091/mbc.E09-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saez A, Buguin A, Silberzan P, Ladoux B. Is the Mechanical Activity of Epithelial Cells Controlled by Deformations or Forces? Biophysical Journal. 2005;89:L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holle AW, Engler AJ. More than a feeling: discovering, understanding, and influencing mechanosensing pathways. Current Opinion in Biotechnology. 2011;22:648–654. doi: 10.1016/j.copbio.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 50.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, et al. Stretching Single Talin Rod Molecules Activates Vinculin Binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity Sensing at the Leading Edge through αvβ3 Integrins and RPTPα. Biophysical Journal. 2006;90:1804–1809. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhee S, Jiang H, Ho CH, Grinnell F. Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc Natl Acad Sci U S A. 2007;104:5425–5430. doi: 10.1073/pnas.0608030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moolenaar WH. Lysophosphatidic acid, a multifunctional phospholipid messenger. J Biol Chem. 1995;270:12949–12952. doi: 10.1074/jbc.270.22.12949. [DOI] [PubMed] [Google Scholar]
- 55.Kolodney MS, Elson EL. Correlation of myosin light chain phosphorylation with isometric contraction of fibroblasts. J Biol Chem. 1993;268:23850–23855. [PubMed] [Google Scholar]
- 56.Zhang Q, Checovich WJ, Peters DM, Albrecht RM, Mosher DF. Modulation of cell surface fibronectin assembly sites by lysophosphatidic acid. J Cell Biol. 1994;127:1447–1459. doi: 10.1083/jcb.127.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.