Abstract
The search for expression quantitative trait loci (eQTL) has traditionally centered entirely on the process of transcription, whereas variants with effects on mRNA translation have not been systematically studied. Here we present a high throughput approach for measuring translational cis-regulation in the human genome. Using ribosomal association as proxy for translational efficiency of polymorphic mRNAs, we test the ratio of polysomal/nonpolysomal mRNA level as a quantitative trait for association with single-nucleotide polymorphisms on the same mRNA transcript. We identify one important ribosomal-distribution effect, from rs1131017 in the 5’UTR of RPS26 , that is in high linkage disequilibrium (LD) with the 12q13 locus for susceptibility to type 1 diabetes. The effect on translation is confirmed at the protein level by quantitative Western blots, both ex vivo and after in vitro translation. Our results are a proof-of-principle that allelic effects on translation can be detected at a transcriptome-wide scale.
Introduction
Genetic effects on gene expression, or expression quantitative trait loci (eQTLs), are common in the human genome 1-7 and their elucidation is critical to understanding human genetic diseases 8-10, especially those with complex-trait inheritance.
Previous genome-wide eQTL searches have focused exclusively on levels of steady-state mRNA, for the discovery of variants modulating transcription or RNA stability. However, as the biological effect of altering expression levels of most genes depends on allelic protein levels, the potential effect of polymorphic RNA on translation may be equally important and is required to complete the picture 11,12. DNA sequence variants can affect translational efficiency through different mechanisms 12: (1) At the 5’UTR, a variant can abolish or create a upstream translation-initiation codon or open reading frame 13-15, or change an internal ribosome entry site (IRES) by loss-of-function16 or gain-of-function17; (2) At the translation start site, a mutation can change the highly conserved Kozak sequence gccRCCAUGG 18,19; (3) At a splice-site of the exon-intron junction, a mutation can cause the production of an mRNA isoform with missing 5’UTR functional elements20; (4) At the 3'UTR, a mutation changing the upstream core polyadenylation signal sequence (UCPAS) consensus sequence, the hexamer AAUAAA, may lead to alternative mRNA isoforms with different 3’ UTR structure and poly(A) tail and therefore differential translational properties21-23. Most of these examples have been demonstrated with mutations causing rare monogenic diseases. For common DNA variations, much fewer examples exist. Perhaps the best-known example is the case of a haplotype of synonymous SNPs (sSNPs) at the COMT gene encoding the enzyme Catechol-OMethyltransferase, which alter mRNA secondary structure and efficiency of protein expression, a likely explanation of their effect on pain sensitivity 24. In addition, a sSNP may change translational efficiency based on differential tRNA abundance for different codons that shape codon usage25,26. Evidence of selective pressure27 indeed suggests a functional role for some sSNPs.
To date, the role of common DNA variation on mRNA translation has received little attention, likely because of the obvious methodological limitations of high-throughput quantitative proteomics as compared to oligonucleotide expression microarrays or RNAseq. As a potential remedy for this gap, we undertook this study to examine the feasibility of systematically screening human genetic variants that affect ribosomal distribution of mRNA, taken as a proxy for translational efficiency. This is based on the principle that efficiently translated mRNAs associate with multiple ribosomes, while less active ones with fewer or none. We leveraged the extensively validated ultracentrifugal method for estimating translational activity based on the sedimentation velocity of mRNA-ribosomal complexes 28-30, using immortalized lymphoblastoid cell lines (LCLs), a cell type extensively used in functional-genomics studies 31.
Results
Genome-wide screening for SNPs with translational effect
We studied human immortalized lymphoblastoid cell lines (LCL) of 38 parents of the HapMap European families of Utah residents with Northern and Western European ancestry (CEU) from the Centre de l'Étude du Polymorphisme Humain (CEPH). A total of 52,737 exonic SNPs in 15,265 autosomal genes with minor allele count (MAC) ≥5 (i.e. MAF≥6.6%) were evaluated (60,425 exonic SNP-gene pairs, Supplementary Data 1), setting a Bonferroni threshold of 8.3 × 10−7. Because of the common existence of overlapping genes, one SNP may map to two genes at the same time. Among these SNPs, 35,620 (67.5%) were genotyped in HapMap and 17,117 (32.5%) SNPs were detected by the 1000 Genomes Project in 29 of the 38 subjects. The distribution of imputation quality scores of SNPs in the 9 samples without the 1000 genomes genotyping is shown in Supplementary Fig. S1. Eleven SNPs in 6 genes met the Bonferroni threshold, while 74 SNPs in 29 genes met the false discovery rate Q-value<0.1 (Supplementary Table S1). Among SNPs with Q<0.1, 5’-UTR SNPs were significantly enriched (20.3% vs. 9.9%, P=2.78×10−3) (Table 1). Among the 74 SNPs with FDR<0.1, 4 SNPs are in linkage disequilibrium (LD, r2>0.5) with SNPs identified as associated with disease susceptibility (Table 2).
Table 1.
SNP functional classifications
| Function | SNPs with Q<0.1 | SNPs with Q≥0.1 | total | Enrichment/diminishment χ2 test |
|---|---|---|---|---|
| coding-notMod3* | 0 | 335 | 335 | - |
| coding-synonymous | 10 | 13906 | 13916 | χ2=3.79, p=0.052 |
| missense | 15 | 11539 | 11554 | χ2=0.06, p=0.801 |
| nonsense | 1 | 115 | 116 | χ2=0.90, p=0.342 |
| splice-3 | 0 | 21 | 21 | - |
| splice-5 | 0 | 36 | 36 | - |
| utr-3 | 33 | 28435 | 28468 | χ2=0.19, p=0.664 |
| utr-5 | 15 | 5964 | 5979 | χ2=8.95, p=2.78×10−3 |
| Total | 74 | 60351 | 60425 | - |
coding-notMod3: SNP within an exon and translated, while number of coding bases is not a multiple of 3.
Table 2.
SNPs with possible translational cis-regulatory effects and disease association
| chr | SNP with translational effect | Q-value | SNP with disease association* | r2 between two SNPs | Gene | Disease/Trait |
|---|---|---|---|---|---|---|
| 5 | rs177252 | 0.047895 | rs3119854 | 0.737685 | CATSPER3 | Height |
| 22 | rs139562 | 1.29E-05 | rs482202445 | 0.537935 | MEI1 | Vitiligo |
| 22 | rs139561 | 1.92E-06 | rs482202445 | 0.537935 | MEI1 | Vitiligo |
| 12 | rs1131017 | 2.08E-08 | rs170170437,38,42,43 | 0.717906 | RPS26 | Type 1 diabetes autoantibodies/Asthma/Alopecia areata/ Type 1 diabetes |
| 12 | rs1131017 | 2.08E-08 | rs245697345 | 0.717906 | RPS26 | Vitiligo |
| 12 | rs1131017 | 2.08E-08 | rs1117173939 | 0.859427 | RPS26 | Type 1 diabetes |
| 12 | rs1131017 | 2.08E-08 | rs229223936,38,40,41 | 0.587861 | RPS26 | Type 1 diabetes autoantibodies/Type 1 diabetes |
Reported by Genome-Wide Association Studies (http://www.genome.gov/gwastudies/).
To look for alternative splicing as the mechanism of such effects, we intersected the set of our SNPs with a list of 187 splicing quantitative trait loci (sQTLs), discovered by Pickrell et al. using RNA sequencing 32. We found two overlaps, interestingly both in highly polymorphic genes of the major histocompatibility complex on 6p21. The first is SNP rs241451, associated with the splicing of the last exon of TAP2. As our group has previously shown 33, it creates two transcripts (GenBank Nucleotide Accession codes NM_000544 and NM_018833) with completely distinct carboxy terminals and 3' UTRs, which explains the translational effect. The second one is at HLA-C, where rs7767581 is associated with the splicing of an exon encoding the highly conserved functional α3 domain34. Such overlap is not surprising. If both isoforms encode the same protein, splicing is just one of the mechanisms of polymorphic translational control. Even if the proteins are different, the effect on translation should be factored into the mechanisms by which the locus exerts phenotypic effects.
We also found that two of our hits overlapped with eQTLs for steady state mRNA in LCLs 6 (http://www.sph.umich.edu/csg/liang/asthma/). They involve TAP2 (as shown in our previous report 33), and ZNF584. In both cases, the haplotype associated with higher translational efficiency has also higher mRNA level, which may be related to the ribosome protection of mRNA from endonucleolytic attacks for the actively translated allele35.
Translational effects in RPS26 at the protein level
One of the most significant effects on ribosomal distribution was found in the RPS26 gene (Fig. 1), by a 5’ UTR SNP that is in tight LD with the most significant SNP at the type 1 diabetes risk locus on 12q13 36,37. It was chosen for validation at the protein level.
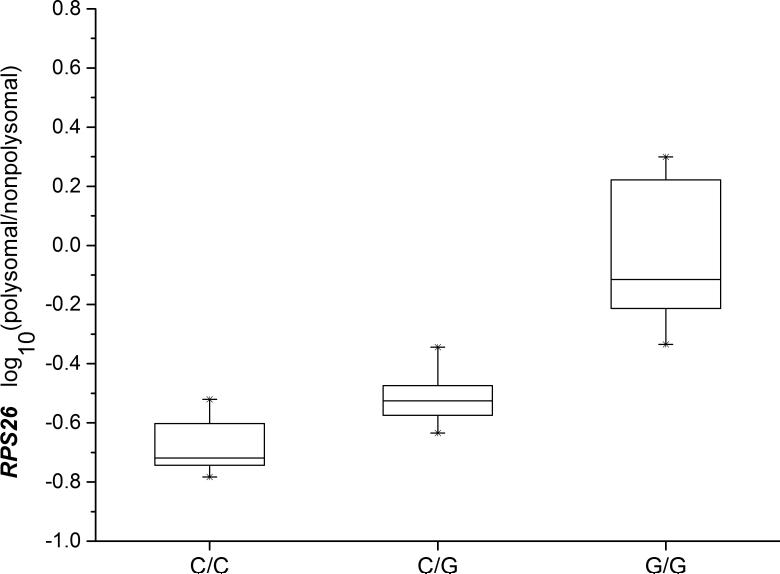
Fig.1. RPS26 mRNA polysomal/nonpolysomal ratio and rs1131017 genotypes.
The log-transformed mRNA ratio of normalized signals of polysomal/nonpolysomal is associated with the rs1131017 genotypes with high statistical significance (n=38, Spearman rank correlation P=3.79×10−13). Error bars represent 25 and 75% percentiles. The G allele of rs1131017 is correlated with higher polysomal/nonpolysomal ratio.
To confirm that ribosomal distribution does reflect a translational effect at the protein level, we tested the assumption that the mRNA allele found at a higher proportion in the polyribosomal fraction than in the soluble RNA will produce more protein product under the same translational environment. We tested this expectation in the case of allelic effects of rs1131017 on RPS26 protein translation by two independent approaches, i.e. protein quantification ex vivo and following in vitro translation of allelic constructs. In ex vivo translation quantification, the RPS26 RNA and the RPS26 protein from 36 LCL samples were quantified and compared based on their genotypes. We compared the mRNA-normalized protein levels of RPS26 between the three genotypes. Statistically significant correlation (Student's t-test P = 0.005) was found with the rs1131017genotype (Fig.2a). The ex vivo results show that mRNA with the G allele of rs1131017 produces significantly more protein, which is concordant with the microarray result. Protein also correlates with the ribosomal distribution ratio, independently of genotype: r= 0.449, correlated samples t-test P=0.028). In the in vitro translation quantification, paired t-test was applied to compare the constructs containing each allele of rs1131017 in seven distinct in vitro translation experiments with each Western blot membrane containing at least 3 replicates of each genotype. We found again that the G allele of rs1131017 showed higher translational efficiency by producing more protein (paired t-test P=0.020) (Fig.2b,c). This observation confirms the ex vivo results, both results showing that the G allele of rs1131017 that was found to be most strongly associated with polysomal distribution is also producing more protein.
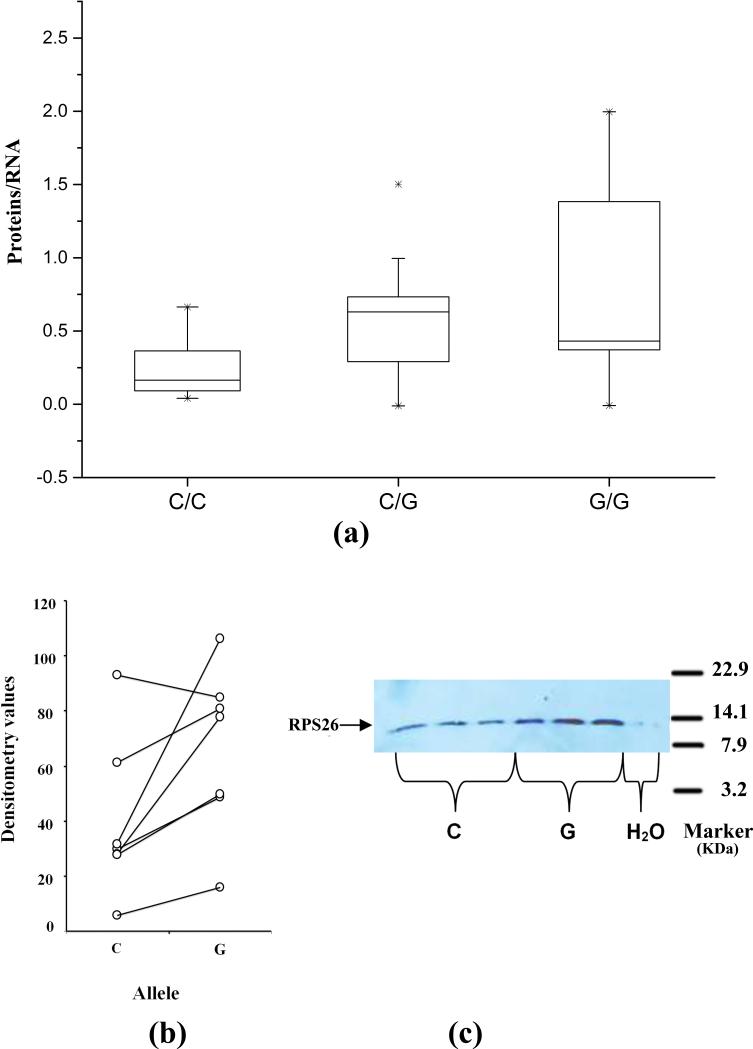
Fig.2. Confirmation of translational effects in RPS26 at the protein level.
(a) RPS26 protein levels by ex vivo translation quantification. The mRNA-normalized protein levels of RPS26 between the three genotypes showed statistically significant correlation (n=36, Student's t-test P = 0.005), and the G allele of rs1131017 is correlated with higher protein levels. Error bars represent 25 and 75% percentiles. (b) RPS26 protein levels by in vitro translation and immunoblot quantification. Comparison of RPS26 constructs with rs1131017 C/G alleles. Each western blot contained at least 3 replicates for each allele (C or G). Statistical significance favoring the construct with the G allele was found (paired t-test P =0.0204). (c) The RPS26 constructs containing different alleles of rs1131017 produced different levels of RPS26 protein. The G allele of rs1131017 produced higher level of RPS26 protein. A representative of seven translation runs is shown.
RPS26 and type 1 diabetes susceptibility
RPS26 maps to a region of Chr12q13.2, known to confer type 1 diabetes susceptibility in the GWASs by the Wellcome Trust Case-Control Consortium (WTCCC), our group, and the Type 1 Diabetes Genetics Consortium (T1DGC) 36-41, as well as other autoimmune diseases42,43. The SNP identified to have cis-translational effect is in high LD with SNPs tagging T1D genetic susceptibility, i.e. rs170170437, rs1117173939, and rs229223936,40,41 (Supplementary Fig. S2). The allele with lower translational activity is in LD with the T1D predisposing effect. The G allele interrupts a oligopyrimidine tract, which plays a role in translational repression44.
Discussion
Because of the availability of microarray technologies for high throughput transcriptome profiling, numerous eQTL studies on gene transcription have successfully acquired knowledge of genetic regulation of gene transcription in human diseases. However, no such high throughput method exists to study genetic regulation of translation genome-wide. Despite the rapid progress of proteomics technologies, quantitative assays of proteomics are much more complicated and expensive, but also lack the precision and dynamic range of transcriptome assays. In this study, we created a novel high throughput approach to discover the genetic effect on mRNA translation using ribosomal association as proxy, to drastically narrow down the range of targets for proteomic validation.
Because ribosomal fractionation is costly and labor intensive, we felt that a small pilot study was justified prior to investing in more powerful sample sizes. Despite the very small sample size and consequent reduced statistical power, we were able to detect 29 genes subject to genetic modulation in cis at the translational level. Nevertheless, 29 represents a much smaller number of translational eQTLs than found at the transcriptional level. We propose that this is only a matter of statistical power and that these 29 genes represent the “tip of the iceberg”, as indicated by the QQ plot (Supplementary Fig. S3). The same approach, applied to hundreds of samples will likely reveal a much larger number of loci, conceivably comparable to that of genetic effects on transcription. Regardless of their actual numbers, the utility of discovering genetic loci affecting translation is enhanced by the ability to easily identify the causal variant as it is constrained to map on the mRNA, while transcriptional variants must be sought over genomic regions orders of magnitude wider
Aligning our results with GWAS findings, we found three of the 29 loci mapping to loci of susceptibility to human diseases. The most interesting example is the ribosomal protein S26 gene (RPS26) at a risk locus for type 1 diabetes and other autoimmune diseases36-41,43,45. The RPS26 SNP rs1131017 identified to have cis-translational effect is in high LD with genetic markers of T1D and Vitiligo susceptibility. The SNP rs1131017 maps to a 5' terminal oligopyrimidine tract (TOP) identified in the mRNAs encoding the ribosomal proteins in mammalia, which plays the role of translational repression44. Interruption of the TOP by rs1131017 may thus alleviate the translational repression. The same SNP has been reported to also have a transcriptional effect on the gene which, however, goes in the opposite direction 6,46. The two opposing effects probably cancel each other, at least partially, which may be consistent with statistical analysis indicating that the transcriptional effect is probably not causal in this association47. However, many transcriptional effects (and, in all likelihood translational ones) are tissue-specific. It is quite possible that, in some T1D-relevant tissues only one of the two effects modulates expression, in which case this conclusion will need to be re-evaluated. Translational eQTL studies with larger sample sizes will be needed to address this question.
In conclusion, the approach developed in this study covers an aspect of functional genomics that has not received much attention so far, and adds an important tool in the evaluation of genetic loci associated with complex disorders. This study screened human genome systematically for cis-regulation effect on gene translation in LCLs. It is readily expandable to study cell- or tissue-specific genetic effect of gene translation by investigating different cell lines or tissues. Larger sample sizes are likely to reveal a number of effects that rivals that of transcriptional effects. Ideally such questions should be answered by direct quantification of proteins. However, hypothesis-free methodologies for proteome-wide quantification have very significant drawbacks48 while targeted proteomics with selected reaction monitoring (SRM) now allows sensitive quantification with no need for affinity reagents49. Our methodology will be most useful in identifying candidates for confirmation by SRM. The latter can also be used to detect post-translational modifications50 thus covering all processes by which genetic variation affects gene expression.
Materials & Methods
Cell lines
The LCLs were acquired from the Coriell Institute for Medical Research (http://ccr.coriell.org/Sections/Collections/NHGRI/hapmap.aspx?PgId=266&coll=HG, New Jersey, USA). These LCLs have been genotyped for ~9 millions SNPs on autosomes by the HapMap project (http://www.hapmap.org/) and the 1000 Genomes Project (the March 2010 release, http://www.1000genomes.org). The cells were cultured using RPMI-1640 containing 15% FBS, 1% L-glutamine, 1% penicillin and 1% streptomycin, in 37°C 5%CO2 incubator. The Research Ethics Board of the Montreal Children's Hospital approved the study.
Polyribosome fractionation
RNA molecules were fractionated on the basis of the number of ribosomes they were associated with, by ultracentrifugal sedimentation velocity in viscous media (sucrose gradients). The larger aggregates of the more actively translated RNA sediment faster than smaller aggregates, while RNA not associated with ribosomes remains at the top layer28,51. From each LCL, 1×108 cells were incubated with 100μg/μl cycloheximide for 5 mins at 37°C to freeze translational activity and then washed with phosphate buffered saline (PBS) containing 100μg/ml cycloheximide. Cell pellets, collected by centrifugation at 1,000 rpm for 10 min at 4°C, were lysed with hypotonic polysome lysis buffer [5 mmol/L Tris-HCL (pH 7.5), 2.5 mmol/L MgCl2, 1.5 mmol/L KCL, 100μg/mL cycloheximide , 2 mmol/L dithiothreitol (DTT), 10% Triton X-100, and 10% sodium deoxycholate]. After 2 min of incubation on ice in a prechilled Eppendorf tube, with occasional vortexing, the extracts were centrifuged for 2 mins at 13,000 rpm at 4°C to remove cellular debris. The supernatant was directly loaded on a 10%–50% linear sucrose gradient [20 mmol/L HEPES-KOH (pH 7.6), 100 mmol/L KCl, 5 mmol/L MgCl2] and then centrifuged in a Beckman SW41 rotor for 120 minutes at 35,000 rpm at 4°C. As stated earlier, mRNA directing enhanced protein synthesis at many ribosomes simultaneously, is expected to be more abundant in the heavier polysome fractions, whereas mRNA subjected to repressed translation should be more abundant in fractions spanning low-number polysomes, monosomes, and free ribosomal subunits. Polyribosomal fractions were collected using the Brandel fraction collector (Gaithersburg, Maryland) with real-time monitoring of UV optical density at 254 nm using an ISCO type 11 optical unit connected with a recorder. There were 24 fractions/cell line.
Sample preparation and microarray analysis
After the fractions were collected, RNA from each fraction was extracted using TRIzol (Invitrogen, California) followed by the phenol–chloroform purification. The mRNA concentration of each fraction was quantified by slot-blot hybridization using DIG-labeled oligodT to quantify poly-A RNA (Supplementary Fig. S4, S5). Equal amounts of poly-A RNA (5ng/μl) from unfractionated RNA, nonpolysomal RNA (4 fractions before the 80S polysome peak), and polysomal RNA (4 fractions after the polysomal fraction peak) of each individual were evaluated for genome-wide expression profiling on the Illumina HumanRef-8 v3.0 Expression BeadChip (San Diego, California). The selection of polysome fractions of each individual was based on the realtime OD detection in the fractionation process. The selection of nonpolysomal fraction and the polysomal fraction is illustrated in Fig.3. If the profile of a cell line did not fit the exact pattern, the extraction was repeated. All decisions to repeat were made prior to the availability of any microarray results. Our quality control (QC) process showed the fractionation introduced relatively little random noise (Supplementary Fig. S6). The Principal Component Analysis (PCA) showed distinctive genome-wide expression profiles in different fractions with the fractionation as the first principal component (PC1, Supplementary Fig. S7).
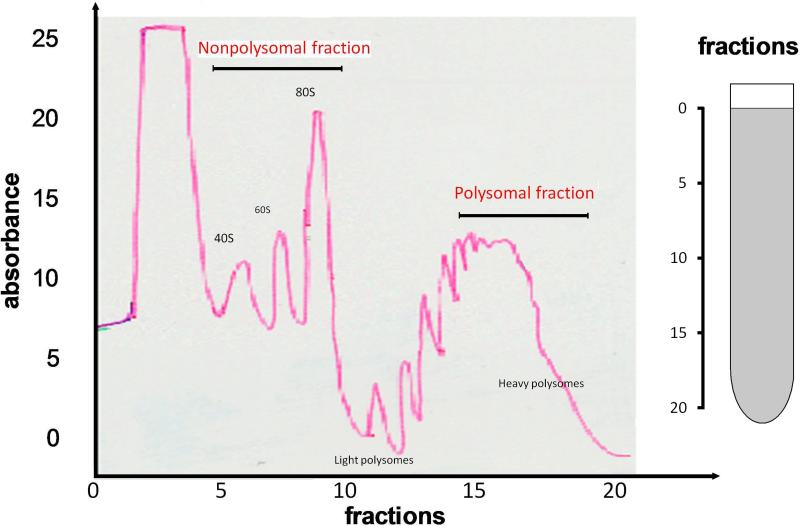
Fig.3. Realtime OD detection of the polysome fractions.
Fractions were monitored using an ISCO UA-6 UV detector. The positions of 80S ribosomes, light polysomes, and heavy polysomes, in the gradients are labeled. Two fractions, i.e. nonpolysomal fraction (corresponds to Fraction 7-10) and polysomal fraction (corresponds to Fraction 15-18), used in this study were indicated with arrows. X-axis: the fraction number (from Fraction 1 to Fraction 24); Y-axis: the UV absorbance. Increases in polysome size by a single ribosome are indicated by secondary peaks in the up-slope of the broad polysome peak.
Data analysis
The gene expression data were normalized by robust spline normalization using the Lumi Bioconductor package52, implemented in the FlexArray software (genomequebec.mcgill.ca/FlexArray). Probe signals for 17,255 autosomal genes were assayed on the HumanRef-8 v3.0 Expression Beachip, and the expressions of 15,265 genes were detectable in the LCLs. The log10 transformed ratio of normalized polysomal/nonpolysomal signals was taken as the quantitative trait for the examination of the effect of exonic SNPs.
The HapMap Phase 2 and Phase 3 genotyping data of these individuals were bulk-downloaded from the HapMap website ((http://www.hapmap.org/). The 1000 genomes genotyping data were acquired from Dr. Yun Li (http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-06.html). Among the 38 individuals in our study, 9 individuals had no data from the 1000 Genomes Project. The genotypes of the missing 1000 genomes SNPs of these 9 individuals were imputed based on the HapMap data using the MACH 1.0 software (http://www.sph.umich.edu/csg/abecasis/MACH/index.html). The 11.5% of SNPs with imputation quality score <0.90 were discarded. Exonic SNPs were identified according to the SeattleSeq SNP Annotation (http://snp.gs.washington.edu/SeattleSeqAnnotation/) based on dbSNP build 134. The cis-regulation of ribosomal distribution of gene mRNAs on the 22 autosomes by exonic SNPs was investigated. Quantitative trait association between the expression data and SNP genotype were tested using the Spearman rank correlation (SRC), as previously described for transcription data53. The workflow of this project is summarized in Fig.4.
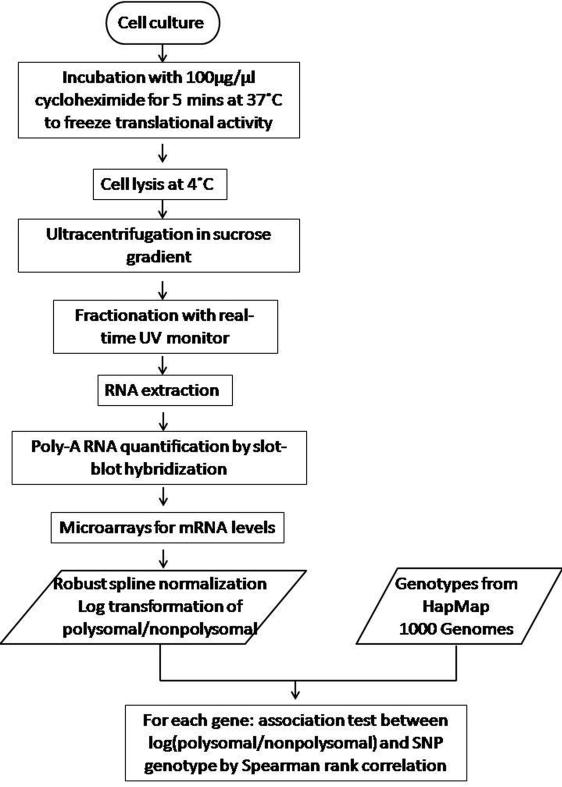
Fig.4. Flowchart of the experimental approach for translational cis-regulation in the human genome.
The sedimentation velocity of mRNA-ribosomal complexes is taken as a proxy for mRNA translational efficiency.
Ex vivo quantification of RPS26 protein
Protein content in total protein extract was measured in the LCLs of 36 of the CEU subjects and, after adjustment for specific mRNA level, compared among the three genotypes. Cells were washed with PBS and mixed with the appropriate volume of Cell Extraction Buffer (CEB) consisting of 98% TALON buffer, 1% Protease inhibitor and 1% PMSF (phenylmethylsulfonyl fluoride). Samples were subject to vortex every 10 minutes for 10 seconds to ensure complete lysis and after 30 minutes they were centrifuged at 20,000 g for 20 minutes. The supernatant was transferred to a clean tube and stored at −80°C. Total protein concentration was determined using the Pierce BCA (bicinchoninic acid) Protein Assay Kit (Thermo Scientific, USA) and used to dilute samples to the same concentration. Relative content of the target protein was measured by quantitative Western blots. Rabbit RPS26 polyclonal antibody (ProteinTech Group, Illinois, USA) was diluted to 1:2000 and mixed with 5% milk blocking solution (5 g of non-fat milk per 100 ml TBST). Mouse calnexin antibody (BD Biosciences, California, USA) at 1:2000 dilution was used to measure the housekeeping gene control. Bands were visualized with the Western Lightning Chemiluminescence ECL plus detection kit (PerkinElmer, MA, USA). ImageJ software (http://rsb.info.nih.gov/ij/index.html) was used to compare the density of the RPS26 bands of the different samples for relative quantification. In each sample the density of the calnexin band (loading-control band) was used to scale the values for RPS26 protein. To assess translation, protein results was corrected for the amount of mRNA in each sample. In parallel to the protein extraction from the LCLs, RNA was also obtained using the RNeasy Plus Mini Kit (Qiagen, CA, USA). After reverse transcription, Multiplex Ligation-dependent Probe Amplification (MLPA) was used to measure the cDNA levels of RPS26 in all the samples. MLPA peaks were obtained in the ABI 310 Genetic Analyzer (Applied Biosystems, California, USA). The RPS26 signals were normalized using a probe set of the housekeeping microglobulin M2 gene, whose expression was found to be the most invariant in the microarrays.
Quantification of in vitro RPS26 protein translation
Plasmid constructs containing different alleles of rs1131017 were made to study in vitro the effects of rs1131017 on translational efficiency. TA cloning was done using the pGEM vector (Promega, WI, USA). Each allele was constructed twice, each time with a different tag epitope: flag or myc. The linearized DNA plasmids were verified by Sanger sequencing. RNA from each allele construct was generated using an in vitro transcription kit (Ribomax Large Scale RNA Production–T7, Promega, WI, USA). The in vitro produced DNAase-treated RNAs were purified using the RNeasy Plus Mini Kit (Qiagen, CA, USA). The Ribogreen RNA assay (Molecular Probes, OR, USA) was used to quantify the RNA samples. RT-PCR and Sanger sequencing were used to confirm the in vitro produced RNAs. The in vitro synthesis of proteins in cell-free extracts was done in the Flexi rabbit reticulocyte lysate system (Promega, WI, USA). Western Blotting was used to quantify the RPS26 protein produced by the different alleles, as described above.
Supplementary Material
Acknowledgments
This work was funded by Genome Canada, Génome Québec (GRiD project) and the DP3 program of the USA National Institutes of Health (NIDDK). The GWAS meta-analysis data at the RPS26 locus were generated by our collaborators Drs. Jonathan P. Bradfield, Struan F. A. Grant, and Hakon Hakonarson. L.Q. holds a fellowship from Eli Lilly Canada, and the research activities of C.P. are supported by the Sessenwein Award. We are indebted to Dr. Nahum Sonenberg for sharing with us his protocols and expertise in ribosomal fractionation.
Footnotes
Author Contributions: Polychronakos and Qu conceived and designed the study with assistance of Pastinen and Hudson . Data analysis was performed by Qu and Li. Makri and Lu performed the experiments and wrote the Methods section. Li, Polychronakos, and Qu wrote the manuscript. All authors gave input and approved the final version.
Conflict of Interest statement: None declared.
Accession codes
Microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession code GSE46195.
References
- 1.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge B, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–22. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 4.Pastinen T, et al. Mapping common regulatory variants to human haplotypes. Hum Mol Genet. 2005;14:3963–71. doi: 10.1093/hmg/ddi420. [DOI] [PubMed] [Google Scholar]
- 5.Ge B, et al. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584–91. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–40. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadee W. Measuring cis-acting regulatory variants genome-wide: new insights into expression genetics and disease susceptibility. Genome Med. 2009;1:116. doi: 10.1186/gm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson TJ. Wanted: regulatory SNPs. Nat Genet. 2003;33:439–40. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- 11.Cobon GS, Verrills N, Papakostopoulos P, Eastwood H, Linnane AW. The proteomics of ageing. Biogerontology. 2002;3:133–6. doi: 10.1023/a:1015240304287. [DOI] [PubMed] [Google Scholar]
- 12.Polychronakos C. Gene expression as a quantitative trait: what about translation? J Med Genet. 2012;49:554–7. doi: 10.1136/jmedgenet-2012-101199. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi M, et al. Familial thrombocytosis. Br J Haematol. 1995;89:900–2. doi: 10.1111/j.1365-2141.1995.tb08432.x. [DOI] [PubMed] [Google Scholar]
- 14.Ghilardi N, Wiestner A, Kikuchi M, Ohsaka A, Skoda RC. Hereditary thrombocythaemia in a Japanese family is caused by a novel point mutation in the thrombopoietin gene. British Journal of Haematology. 1999;107:310–316. doi: 10.1046/j.1365-2141.1999.01710.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, et al. Mutation of the CDKN2A 5' UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet. 1999;21:128–132. doi: 10.1038/5082. [DOI] [PubMed] [Google Scholar]
- 16.Yoon A, et al. Impaired Control of IRES-Mediated Translation in X-Linked Dyskeratosis Congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 17.Paulin FEM, Chappell SA, Willis AE. A single nucleotide change in the c-myc internal ribosome entry segment leads to enhanced binding of a group of protein factors. Nucl. Acids Res. 1998;26:3097–3103. doi: 10.1093/nar/26.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Angioletti M, Lacerra G, Sabato V, Carestia C. Beta+45 G --> C: a novel silent beta-thalassaemia mutation, the first in the Kozak sequence. Br J Haematol. 2004;124:224–31. doi: 10.1046/j.1365-2141.2003.04754.x. [DOI] [PubMed] [Google Scholar]
- 20.Wiestner A, Schlemper RJ, van der Maas AP, Skoda RC. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nat Genet. 1998;18:49–52. doi: 10.1038/ng0198-49. [DOI] [PubMed] [Google Scholar]
- 21.Chen JM, Ferec C, Cooper DN. A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet. 2006;120:1–21. doi: 10.1007/s00439-006-0180-7. [DOI] [PubMed] [Google Scholar]
- 22.Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH., Jr. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. Embo J. 1985;4:453–6. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rund D, et al. Two mutations in the beta-globin polyadenylylation signal reveal extended transcripts and new RNA polyadenylylation sites. Proc Natl Acad Sci U S A. 1992;89:4324–8. doi: 10.1073/pnas.89.10.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nackley AG, et al. Human Catechol-O-Methyltransferase Haplotypes Modulate Protein Expression by Altering mRNA Secondary Structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 25.Cannarozzi G, et al. A Role for Codon Order in Translation Dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 26.Fredrick K, Ibba M. How the Sequence of a Gene Can Tune Its Translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu HQ, Lawrence SG, Guo F, Majewski J, Polychronakos C. Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: evidence for functional effects of synonymous polymorphisms. BMC Genomics. 2006;7:213. doi: 10.1186/1471-2164-7-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampath P, et al. A Hierarchical Network Controls Protein Translation during Murine Embryonic Stem Cell Self-Renewal and Differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Mills JR, et al. mTORC1 promotes survival through translational control of Mcl-1. Proceedings of the National Academy of Sciences. 2008 doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci Res. 2009;87:1953–9. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- 32.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu HQ, et al. Genetic Control of Alternative Splicing in the TAP2 Gene: Possible Implication in the Genetics of Type 1 Diabetes. Diabetes. 2007;56:270–275. doi: 10.2337/db06-0865. [DOI] [PubMed] [Google Scholar]
- 34.Whitman MC, et al. The isolated major histocompatibility complex class I alpha3 domain binds beta2m and CD8alphaalpha dimers. Mol Immunol. 2000;37:141–9. doi: 10.1016/s0161-5890(00)00034-1. [DOI] [PubMed] [Google Scholar]
- 35.Dreyfus M, Joyce S. The interplay between translation and mRNA decay in procaryotes. Translational mechanisms.[Online.] Landes Bioscience, Georgetown, Tex. 2002 http://www.eurekah.com/abstract.php.
- 36.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakonarson H, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–6. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 38.Plagnol V, et al. Genome-Wide Association Analysis of Autoantibody Positivity in Type 1 Diabetes Cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper JD, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirota T, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. Oligopyrimidine tract at the 5' end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci U S A. 1991;88:3319–23. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin Y, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schadt EE, et al. Mapping the Genetic Architecture of Gene Expression in Human Liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plagnol V, Smyth DJ, Todd JA, Clayton DG. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics. 2009;10:327–34. doi: 10.1093/biostatistics/kxn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghazalpour A, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doerr A. Mass spectrometry-based targeted proteomics. Nat Meth. 2013;10:23–23. doi: 10.1038/nmeth.2286. [DOI] [PubMed] [Google Scholar]
- 50.Altelaar AFM, Munoz J, Heck AJR. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 51.Lin CJ, Robert F, Sukarieh R, Michnick S, Pelletier J. The antidepressant sertraline inhibits translation initiation by curtailing mammalian target of rapamycin signaling. Cancer Res. 2010;70:3199–208. doi: 10.1158/0008-5472.CAN-09-4072. [DOI] [PubMed] [Google Scholar]
- 52.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 53.Dimas AS, et al. Common Regulatory Variation Impacts Gene Expression in a Cell Type–Dependent Manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.