Abstract
Many aspergilli that belongs to ascomycetes have sexuality. In a homothallic or self-fertile fungus, a number of fruiting bodies or cleistothecia are formed in a thallus grown from a single haploid conidia or ascospores. Genome-sequencing project revealed that two mating genes (MAT) encoding the regulatory proteins that are necessary for controlling partner recognition in heterothallic fungi were conserved in most aspergilli. The MAT gene products in some self-fertile species were not required for recognition of mating partner at pheromone-signaling stage but required at later stages of sexual development. Various environmental factors such as nutritional status, culture conditions and several stresses, influence the decision or progression of sexual reproduction. A large number of genes are expected to be involved in sexual development of Emericella nidulans (anamorph: Aspergillus nidulans), a genetic and biological model organism in aspergilli. The sexual development process can be grouped into several development stages, including the decision of sexual reproductive cycle, mating process, growth of fruiting body, karyogamy followed by meiosis, and sporulation process. Complicated regulatory networks, such as signal transduction pathways and gene expression controls, may work in each stage and stage-to-stage linkages. In this review, the components joining in the regulatory pathways of sexual development, although they constitute only a small part of the whole regulatory networks, are briefly mentioned. Some of them control sexual development positively and some do negatively. Regarding the difficulties for studying sexual differentiation compare to asexual one, recent progresses in molecular genetics of E. nidulans enlarge the boundaries of understanding sexual development in the non-fertile species as well as in fertile fungi.
Keywords: Aspergillus, Emericella nidulans, Molecular genetics, Sexual development
Originally Aspergillus originated from the shape of conidiophore, an asexual reproductive organ, which resembles aspergillum, the instrument to disperse holy water. Many species belong to genus Aspergillus produce ascospores (Samson, 1994). Emericella nidulans (anamorph: Aspergillus nidulans) having the 'nest-like' fruiting body called cleistothecium, is a representative of the perfect aspergilli. E. nidulans is a homothallic fungus which means a thallus grown from a single haploid conidia can produce a number of cleistothecia. Asci are developed within a cleistothecium and eight ascospores are produced in an ascus as a result of meiosis followed by an additional mitosis.
The ratio of asexual to sexual sporulation varies according to cultural conditions and/or environmental stresses. The process of sexual development takes longer than that of asexual sporulation. More than 1,300 genes are likely involved in the process including genetically programmed acquisition of competence, various signal transduction systems, genetic regulation system for decision of development mode, plasmogamy, karyogamy, meiosis and various types of cellular differentiation (Pontecorvo et al., 1954; Axelrod et al., 1973; Braus et al., 2002). A number of those genes have been screened and identified using a variety of experimental methods, such as mutant isolation, ortholog screening, differentiation-specific ESTs and microarray analysis.
Here, the properties of genes involved in sexual development of E. nidulans are reviewed in point of view of molecular genetic approaches for screening, identifying, and characterizing the genes.
Sexuality of Aspergilli
About half of Aspergilli species are known to have sexuality and some of them are self-fertile, in other word, homothallic. Genome-sequencing of E. nidulans, revealed that two mating type genes (MAT) encoding the regulatory proteins that are necessary for controlling different mating partner recognition in out-cross species of filamentous fungi (Miller et al., 2005; Paolleti et al., 2007). Those are the abox domain protein and high mobility group (HMG) domain protein. Usually, the two genes locate on same genomic position named MAT locus. Each mating partner carries either the a or HMG gene, the protein of which specifies sexual identity and control the mating process.
Many homothallic fungi contain functional mating type genes which are important for maintaining homothallic characteristics. In Cocchliobulous and Fusarium species, deletion of either the mat1 (HMG) or mat2 (adomain) gene converted the strains heterothallic, producing fruiting bodies exclusively by out crosses (Yun et al., 1999; Lee et al., 2003). In E. nidulans, two mating type loci namely MAT-1 (matB) and MAT-2 (matA) which encode an a box protein and a HMG box protein, respectively, were reported (Miller et al., 2005; Paoletti et al., 2007). Unlike other fungal mating type loci, these two mating type genes are located on the different chromosome, MAT-1on chromsome VI and MAT-2 on chromosome III. This finding suggests that the homothallism of E. nidulans might not be caused by fusion of mating type genes from its heterothallic ancestor. The separation of two mating type genes may be due to a chromosome translocation (Scazzocchio, 2006; Paoletti et al., 2007). Galagan et al. (2005) compared the genomic organization of the MAT loci in various aspergilli and suggested that homothallism was ancestral. A imperfect fungus, Aspergillus oryzae and a heterothallic fungus Aspergillus fumigatus (teleomorph: Neosartorya fumigata) carry either the a domain or HMG gene on MAT locus, indicating that the two species could be out-crossing. It was proposed that the heterothallism of the two species resulted from the loss of either of two genes on MAT locus. Indeed, mating experiment between different mating type strains of A. fumigatus revealed that they successfully produced matured cleistothecia and ascospores (O'Gorman et al., 2008). More analytic data concerning the MAT sequences organization in more various aspergilli are necessary to conclude whether self-fertility or out-crossing was ancestral mating property.
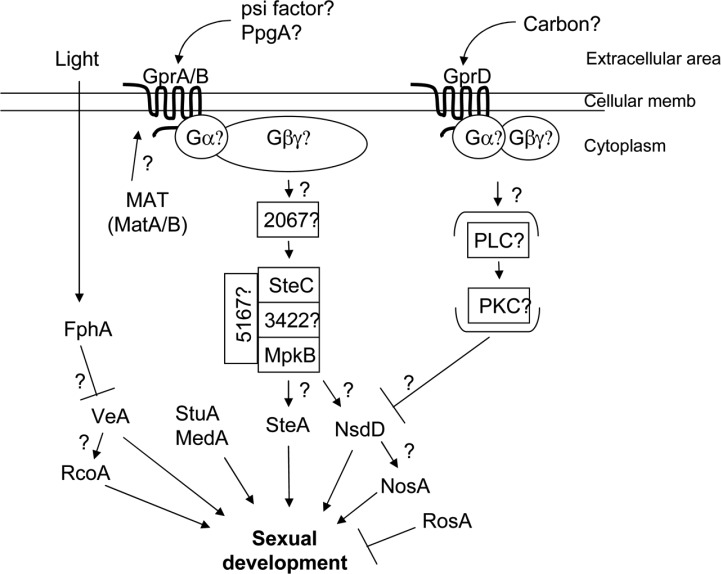
The products of mating type genes in out-crossing fungi such as Neurospora crassa or Saccharomyces cerevisae are known to control the expression of signaling genes necessary for partner recognition or for initiation of sexual development. The MAT gene products are needed for the expression of components of the pheromone-signaling pathway. The mating pheromone signaling pathway is one of the well-known heterotrimeric G-protein signaling processes in budding yeast (Slessareva and Dohlman, 2006). In S. cerevisiae, pheromone such as α- or a-factor binds to its receptor, Ste2p or Ste3p, of the opposite mating type. The pheromone-bound G-protein coupled receptor (GPCR) stimulates a subsequent Gpa1p (Gα) subunit and Ste4p/Ste18p (Gβ/γ) subunits to activate the Fus3p/Kss1p MAPK cascade and up-regulates the expression of Ste12p transcription factor regulating the gene expression of sexual development (Fig. 1).
Fig. 1.
Hypothesized genetic model for sexual development in E. nidulans. Adapted and modified from Seo et al., 2004.
In E. nidulans, deletion of either MAT-1 (matB) or MAT-2 (matA) affects sexual development but not vegetative growth or asexual development. The fruiting body production was severely delayed and diminished in a matA deletion strain. Deletion of matB blocked the meiosis, which eventually resulted in cleistothecium lysis, although cleistothecia were normally formed (Miller et al., 2005; Paoletti et al., 2007). These results indicate that mating type genes are required for sexual differentiation in self-fertile fungi. Two putative pheromone receptor genes gprA and gprB which are homologs of yeast STE2 and STE3, respectively, were identified in E. ndulans (Seo et al., 2004). Deletion of either of two or both genes affected self-fertilized fruiting body formation but not out-crossing sexual development, indicating that GprA and GprB proteins play an important role in self-fertilization. Unlike the out-crossing species, the MAT genes are not necessary for the expression of those components in pheromone-signaling pathway (Paoletti et al., 2007). Thus, it was suggested that the MAT gene products were not required for recognition of mating partner at pheromone-signaling stage but required at later stages of sexual development in self-fertile species.
Sexual Structures of E. nidulans
In most of fertile aspergilli, eight ascospores are produced as a result of sexual reproduction. Meiosis takes place within a sac called ascus and is followed by an additional mitosis. Asci are formed within an ascocarp, the fruiting body of ascomycetes, which develops from ascogenous hyphae. Most ascocarps of Aspergillus species look like a globe named cleistothecium which means a closed container. In some Aspergillus species, the appearance of ascogonial coils which fuse each other is looked upon as the initiation of sexual development. However, the first morphological indication for sexual development in E. nidulans is the appearance of thick-walled globose Hülle cells which are observed around 24 h after germination. A number of Hülle cells aggregate like a bunch of grapes and form a nest-like structure, within which primodia are formed from the ascogenous hyphae. The Hülle cells are not directly related to sexuality and regarded as nurse cells (Braus et al., 2002; Hermann et al., 1983; Scherer and Fischer, 1998). A primordium matures into a cleistothecium where ascogeneous hyphae grow and develop into croziers. The nuclei are sorted in crozier to form dikaryotic cells in which two nuclei are fused forming a zygote and meiosis takes place (Sohn and Yoon, 2002). After one round of meiosis and mitosis, eight ascospores are developed and an additional mitotic division occurs in the mature ascospore resulting in binucleate ascospores.
Environmental Factors Affecting Sexual Development
Environmental conditions are important for most fungi to survive in nature and inadequate conditions. In E. nidulans, development mode as well as growth rate is largely affected by the surrounding environment. There are various environmental factors such as nutritional status, culture conditions and many stresses, that affect not only growth of mycelia, but also the developmental decision between sexual and asexual reproduction (Han et al., 2003b). Generally, well-nourished conditions without any environmental stress favor sexual development. Stress such as starvation of a carbon or nitrogen source, oxidative stress, high osmolarity or intense visible light inhibits cleistothecium formation and promotes asexual development exclusively (Champe et al., 1994; Han et al., 1994a, b). The effects of various cultural or environmental conditions on development are summarized in Table 1.
Table 1.
Effect of environmental factors on development of Emericella nidulansa
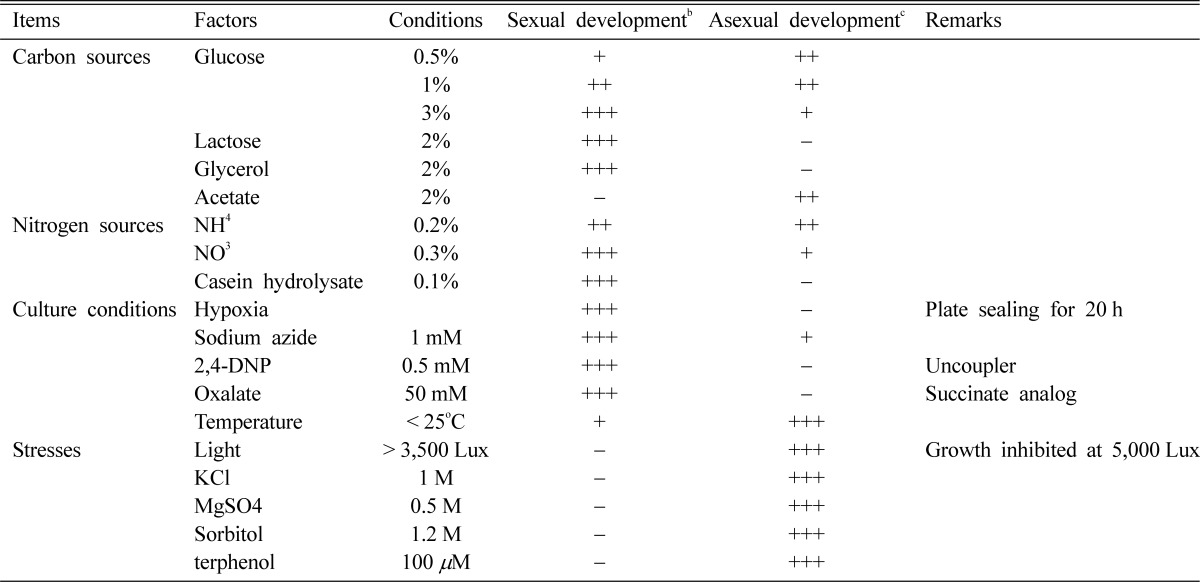
aAdapted and modified from Han et al., 2003b.
bSexual development. The amount of cleistothecia within cm2 area: -, < 1; +, 1~10; ++, 1~50; +++, 50~100.
cAsexual development. The amount of conidia within a circled area of 1 cm diameter: -, < 104; +, 104~105; ++, 105~5 × 106; +++, > 5 × 106.
Nutrients
Since the time or the amount of fruiting body formation is affected by trivial factors such as inoculums size or medium volume, Han et al. (1990) established a standard culture condition of medium volume (30 ml), inoculum size (105/plate), incubation temperature (37℃) and medium type (minimal medium), for examining the effects of various environmental factors. In most out-crossing ascomycetes such as N. crassa and S. cerevisiae nitrogen limitation is a key induction condition for mating or sexual sporulation (Glass and Loremer, 1991). On the contrary, sufficient and favorable nitrogen sources are better condition for sexual development than asexual development in E. nidulans (Han et al., 1990, 1994a, 2003). Sexual development of E. nidulans is also greatly affected by the amount and types of carbon sources. The cleistothecia development was restricted at concentrations lower than 0.5% glucose indicating that a certain level of carbon was required for the induction or completion of sexual development. The ratio of sex/asex sporulation increases as the glucose concentration increases and reaches maximum at 3%. At the concentration higher than 6%, few cleistothecia are produced. However, the inhibitory effect of a high concentration of glucose was relieved by increasing nitrate concentration or the addition of organic nitrogen (N) sources such as casein hydrolysate. The result supports the idea that a balanced C : N ratio is important for the preferential development of fruiting body (Zonneveld, 1977). In addition to sufficient amount of glucose, several carbon sources including lactose, galactose and glycerol also favor sexual development. On acetate medium, however, no cleistothecia or Hülle cells are ever formed. The contrasting effects of these C sources raised the possibility that the energy metabolic pathway may be a significant factor that affects the determination of reproductive cycles. Acetate could be utilized only via aerobic respiration (Hondmann and Visser, 1994) and it is likely that asexual sporulation is favored when cellular energy is provided mainly by aerobic respiration. There have been many reports presenting the essential role of the oxidative metabolism for normal conidiation of ascomycetes (Ng et al., 1973; Galbraith and Smith 1969; Urey, 1971). The low level of aerobic respiration induced by plate-sealing or culture in an hypoxic chamber inhibits asexual sporulation but favors sexual development, which also supports the idea that asexual sporulation is favored by aerobic condition (Han et al., 1990; 2003). The regulatory mechanism of sexual development determination in response to hypoxic condition or the carbon sources including lactose and glycerol has not been investigated.
Light
Illumination of light is an important environmental factor, which controls the growth and development of various fungal species (Tan, 1978). In E. nidulans, hyphal growth is affected by high dose of visible light above approximately 25 W/m2, suggesting that intense light can be regarded as a general stress (Han et al., 2003b). It has been also suggested that asexual spores are predominantly produced in the light and sexual spores in the dark (Raper and Fennell, 1965; Zonneveld, 1977; Mooney and Yager, 1990; Han et al., 1990, 2003). A plenty of conidiophores but few cleistothecia developed at the dose higher than 10W/m2. Mooney and Yager (1990) reported that far-red light is responsible for the predominant development of asexual spores. Fischer and his colleagues found that a fungal phytochrome of E. nidulans is a photoreceptor which can recognize red light and control sexual development.
Phytochrome is a photoreceptor originally found in photosynthetic organisms such as plants and cyanobacteria. Recently, however, it has been found that heterotrophic bacteria and fungi possess phytochrome (Kehoe and Grossman, 1996; Yeh et al., 1997). A gene encoding E. nidulans phytochrome FphA (fungal phytochrome A) was identified (Blumenstein et al., 2005). The FphA was suggested to bind a biliverdin chromophore and repress sexual development under red light condition. Deletion of the fphA gene overcame the inhibitory effect of red light on cleistothecia development. This derepression was only detectable in veA+ wild type strain but not in veA1 mutant, suggesting that VeA is also necessary in red light repression of sexual development and acts downstream of FphA (Blumenstein et al., 2005). The light responsiveness of development implies that the existence of delicate regulation process including reception and translocation of light signaling and determination of development. Recently, several mutants that can develop normal fruiting bodies in the presence of intensive visible light were isolated (Min et al., 2007). However, neither the structure nor the function of the genes was illustrated yet.
High osmolarity
One of the environmental factors that induce asexual sporulation but repress the fruiting body formation is a high concentration of salt such as sodium chloride or potassium chloride (Song et al., 2001; Han et al., 2003b). The high osmolarity is thought to be responsible for the preferential development of asexual spores (Lee and Adams, 1994). Not only the salts but also high concentration of sorbitol or glycerol can induce the conidiation only (Han et al., 2003b). Sexual cycle is completely inhibited by addition of 1M KCl, 1M NaCl or 0.5M MgCl2. The effect of salts on developmental balance was dose-dependent below these concentrations. As the concentration increased higher than those levels, the amount of conidia is gradually reduced and the growth of aerial mycelia was inhibited at higher concentrations than 2M. Like other stresses such as starvation and exposure to high dose of light, the high osmolarity below the inhibitory dose promotes asexual development but represses sexual development. It can be argued that that the effect of salts is not only due to the osmolarity but also to physiological effects of individual cations because of the fact that the concentrations of KCl and MgCl2 affecting the growth or development pattern were different from each other. In A. oryzae which has no sexual cycle asexual sporulation is also promoted by high concentration of salts, suggesting that the preferential development of asexual spores is not simply due to the balance shift by the inhibition of sexual cycle but due to the induction of asexual development by high osmolarity (Song et al., 2001).
Classical Genetic Approaches for Studying Sexual Development
Screening of sterile mutants
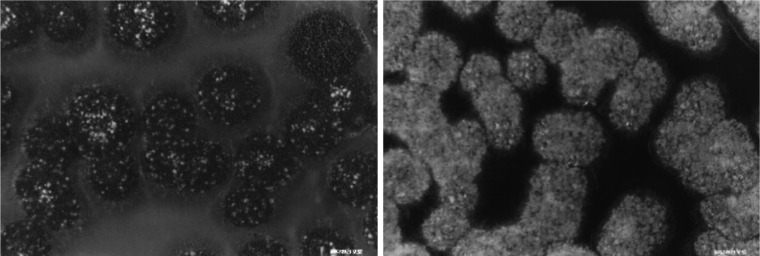
In classical era of genetics, the most successful way of identification of genes with a certain function is screening of mutants showing defective phenotypes related to the function. Since E. nidulans was selected and studied as a genetic model organism by Pontecorovo (1953), a great number of mutants with various functions have been isolated. Mutants deficient in important stages of asexual sporulation have been extensively isolated and characterized by Clutterbuck (1969) and Martinelli and Clutterbuck (1971). However, any massive screening of mutant related to sexual reproduction has not been ever attempted until Han's group applied the plate-sealing or hypoxic condition under which sexual development preferentially took place to distinguish the mutant colonies clearly (Han et al., 1990). Conidia are usually developed prior to cleistothecia and the amount or the time of cleistothecia formation in a colony is different according to the position of the colony in a plate. That is the one of reasons why the phenotypes of the sexual development-defective mutants could not be easily discernable and it makes difficult to isolate mutants deficient in sexual reproduction. Solving the problem, protection of the culture plate from the aeration by sealing with parafilm inhibited the asexual sporulation of the veA+ strains but sexual differentiation process could be easily observed in all colonies (Fig. 2; Han et al., 1990).
Fig. 2.
Shape of colonies arisen on the normoxia or hypoxia plate. In most of the colonies grown normally both asexual (dark part of colony) and sexual (bright spots) organs are formed (A), while in those on hypoxia plate only sexual organs develop exclusively (Han et al., 1990).
Mutants defective in the initiation of sexual cycle
Hundreds of mutants that were defective in sexual development were isolated using a veA+ strain (Han et al., 1990), and they were classified into three groups: (1) mutants that were unable to form any sexual structures (NSD: never in sexual development), (2) mutants that produced immature sexual organs (BSD: block in sexual development), and (3) mutants that produced fully matured sexual organs but showed differences in amount or timing compared to wild-type (ASD: abnormal in sexual development). Since NSD mutants form neither Hülle cells nor any primordia-like structures, the corresponding genes could be expected to act in determinative stage of sexual development. More than twenty NSD mutants were analyzed with their phenotypic and genetic properties and at least four complementation groups (nsdA, nsdB, nsdC and nsdD) were identified (Han et al., 1994b, 1998, 2001; Kim et al., 2009). In addition to the defect in sexual development, the NSD mutants shared several common phenotypes such as apical growth retardation, earlier development of asexual spores, and production of dark pigments at the bottom of the colonies. These mutations were all recessive (possibly loss-of-function mutations).
Among the four genes, nsdC and nsdD were isolated and characterized (Han et al., 2001; Kim et al., 2009). The two genes encode putative transcription factors that regulate sexual development positively. The nsdC gene product contains C2H2-C2H2-C2HC zinc finger DNA binding domain in the middle of the polypeptide with 673 amino acids. The zinc finger domain consists of 80 amino acids whose sequence is highly conserved in homologous proteins of various filamentous fungi, especially in ascomycetes, including A. fumigates, A. oryzae and N. crassa. The NsdC type zinc finger domain is not found in plant or animal. In nsdC6 allele, a single T insertion occurred between 407~408 bp leading to the frameshift mutation and early termination of translation producing the truncated protein, which had only 139 amino acids. The nsdD gene encodes a putative GATA type transcription factor that has a type IVb C2/C2 zinc finger DNA binding domain at C terminus. Four allelic mutations were identified in the nsdD locus. All of them were predicted to produce truncated polypeptides lacking the zinc finger DNA binding domain, which caused the complete loss of function. The nsdD homologs are well conserved in A. fumigatus and A. oryzae although the functions of the genes are not clearly illustrated yet. Since the most NSD mutants in E. nidulans are affected in not only decision of development but also vegetative growth, the mutants of AfnsdD may show phenotypes related to the growth. However, it is not clear yet what stage or system is blocked or what kind of the genes are crucially responsible for the loss of fertility of those aspergilli.
Other known mutations
veA1
The original E. nidulans laboratory strain carried the veA1 mutant allele. The veA1 mutation causes delayed and reduced production of fruiting bodies and eventually results in the preferential development of asexual spores, which is the one of the reasons that made the genetic studies of sexual development difficult (Käfer, 1965; Champe et al., 1981). The asexual development of veA1 mutant is much less affected by various environmental factors, including nutrients, light and temperature (Käfer, 1965; Han et al., 1994a). The veA1 mutant can form cleistothecia at 30℃ though less amount than wild type, but can not at 42℃. A transversion from G in the translation initiation codon of the wild type veA to T was occurred in the veA1 mutation, which results in the use of ATG at the 37th codon as a translation initiation codon. The lack of the 36 amino acids at N-terminus of the VeA protein is responsible for the temperature-sensitivity of the veA1 mutation (Kim et al., 2002). A bipartite nuclear localization signal (NLS) locates in N terminus of VeA polypeptide (Stinnett et al., 2007). The VeA1 mutant protein carries the truncated bipartite NLS and is found predominantly in the cytoplasm. This indicates that VeA with the truncated bipartite NLS cannot migrate to nucleus and fails to regulate the genes required for sexual development.
stuA1
Several mutations that were originally identified as for other phenotype show an additional phenotype related sexual development. Stunted (stuA) mutant that shows defect in conidiophore development also blocks fruiting body formation (Martinelli, 1979; Yager, 1992). They produce diminutive conidiophores bearing apparently normal conidia without distinctive metulae and phialides (Clutterbuck, 1969). StuA is a member of APSES (Asm1, Phd1, Sok2, Efg1, and StuA) family fungal protein, which is required for the correct spatial distribution of BrlA and AbaA (Miller et al., 1992). The stuA mutant develops neither cleistothecia nor Hülle cells (Dutton et al., 1997; Wu and Miller, 1997). StuA regulates the expression of some genes, whose products are found in sexual organ, such as cpeA which encodes a Hülle cell specific catalase-peroxidase (Scherer et al., 2002).
medA1
As with stuA1 mutant, medusa (medA) mutants that form multilayered metulae with normal conidiophore vesicle also are sterile (Clutterbuck, 1969; Martinelli, 1979). The MedA is necessary for proper temporal expression of brlA transcripts and also functions as co-activator of abaA expression (Busby et al., 1996). The medA1 mutant is not able to develop cleistothecia but able to produce Hülle cells (Vallim et al., 2000).
dopA1 (aco586)
One of aconidial mutant genes, aco586, was recently identified as dopey gene (dopA) which regulates not only asexual sporulation but also the initiation of sexual development (Axelrod et al., 1976; Pascon and Miller, 2000). The dopA gene encodes an 1858 amino acids polypeptide which has wide range of similarity with yeast Dop1. The DopA protein is suggested to be a transcription factor which carries three leucine zipper domains in both N and C termini and a transcription activation domain of C/EBP (CAAT/enhancer binding protein) family at its C-terminus (Pascon and Miller, 2000). The DopA affects the expression of some important transcription factor involved in asexual or sexual development such as brlA or steA, respectively. The expression of the brlA gene is delayed and diminished, while the expression of the steA gene is upregulated (Pascon and Miller, 2000).
argB2 and trpC801
Some auxotrophs for certain amino acids are sterile. The mutations in trpC or argB gene cause a specific block during fruiting body formation (Kafer, 1977; Selupi-Crescenzi et al., 1983; Eckert et al., 1999). Hoffmann et al. (2000) suggested that the arrest of sexual development by the mutations could be caused by activation of cross-pathway control via amino acid starvation signals. The 'cross-pathway control' is a complex transcriptional network by which biosynthesis of amino acid or level of charged tRNA is controlled in filamentous fungi. In E. nidulans, amino acid starvation does not significantly affect growth or asexual development. However, maturation of primordia to cleistothecia is blocked by amino acid starvation. When amino acid starvation was induced by adding the histidine analogue 3-amino-1,2,4-triazole (3-AT), sexual development was blocked at the primordial stage. The defect was restored and sexual sporulation proceeds further if the primordial, which were grown under amino acid starvation, were transferred into the normal conditions (Hoffmann et al., 2000), indicating that the activation of the cross-pathway control is responsible for the blockage of sexual development. The activation of the cross-pathway control induced by amino acid starvation or auxotrophy of trpC or argB results in a specific blockage of sexual development in E. nidulans.
Important sexual development-specific genes in E. nidulans
veA
As mentioned above, the mutation of the veA gene (veA1) is responsible for the velvet-like phenotype of colony. The veA gene encodes a polypeptide of 573 amino acids without any similarity with other known proteins. However, genome sequencing of various fungal species revealed that the veA gene is highly conserved in filamentous fungi. The mRNA of veA was detected in all stages of life cycle suggesting that it acts not only during vegetative growth but also developmental stages. The deletion mutants of the veA gene never undergo sexual development even under the conditions where fruiting bodies were preferentially produced in wild type. Forced expression of the gene resulted in the formation of larger numbers of sexual structures even under conditions where wild type strains form a few sexual structures but form conidiophores very well. These phenotypes indicate that the veA gene plays a key role in positive regulator of sexual development.
VeA is also necessary in red-light repression of sexual development and acts downstream of FphA (Blumenstein et al., 2005). VeA carries a putative bipartite nuclear localization signal (NLS) motif in N terminus. Interestingly, VeA migration to the nucleus is light-dependent (Stinnett et al., 2007). In the dark VeA is located mainly in the nuclei, while under light VeA is found abundantly in the cytoplasm. The VeA1 mutant protein lacking the first 36 amino acids at the N-terminus is found predominantly in the cytoplasm, which indicates that the truncated bipartite NLS in VeA1 is not functional and fails to regulate the genes required for sexual development.
The veA gene negatively regulates the expression of various genes involved in secondary metabolism including the aflR gene for the transcription factor AflR which activates the gene cluster involved in the production of ST and the acvA gene, the key gene in the first step of penicillin biosynthesis (Kato et al., 2003).
The veA gene represses the transcription of the rosA gene which represses sexual development (Vienken et al., 2005). All of these results suggest that the veA gene regulates the expression of wide range genes involved in growth, sexual development as well as asexual development and secondary metabolism.
The veA homologs are identified in the aflatoxin-producing aspergilli such as A. flavus and A. parasiticus. They form structures called sclerotia that allow for survival under adverse conditions. Deletion of the veA gene in A. flavus or A. parasiticus blocks production of aflatoxin as well as sclerotial formation (Calvo et al., 2004; Cary et al., 2007). VeA of A. parasiticus is required for the expression of aflR and aflJ, which regulate the activation of the aflatoxin biosynthesis. The veA homolog in A. flavus also regulates the synthesis of the mycotoxins cyclopiazonic acid and aflatrem (Duran et al., 2007).
nsdD
The nsdD gene was isolated and identified as a positive regulator of sexual development (Han et al., 2001). The nsdD gene encodes a GATA-type transcription factor carrying the type IVb zinc finger DNA binding domain at its C-terminus. The nsdD deletion mutants produce no cleistothecia, even under the conditions that promote sexual development, indicating that the nsdD gene is necessary for sexual development. On the other hand, when the nsdD gene is over-expressed by niiA promoter, not only the number of cleistothecia increases dramatically on a solid medium but also they develop in the presence of stresses. Furthermore, Hülle cells, a sexual-specific organ are formed even in a submerged culture where sexual development is completely blocked in wild types. These results suggest that the nsdD gene functions as an activator of sexual development (Han et al., 2001). The nsdD gene was expressed in almost constant level during the vegetative growth, but the expression level increased during sexual development, implying that NsdD may act not only in decision stage but also during sexual development. When the nsdD gene was overexpressed, cleistothecia were formed even in the presence of 0.6M KCl that inhibits sexual development in a wild type. A Northern blot analysis revealed that the expression of the nsdD gene was repressed by 0.6M KCl. These results strongly suggest that the inhibition of sexual development by salts was carried out via the nsdD gene-mediated regulatory network (Han et al., 2003a).
Identification of Sexual Development Genes by Reverse Genetics
Many genes having specific functions can be identified by searching homologs whose functions are known in other organisms. Some components of signal transduction pathways or several transcription factors involved in mating or sexual sporulation in yeast are well-conserved in filamentous fungi including E. nidulans.
Genes in signal transduction pathways
In out-cross species, mating partner is recognized via the reception of pheromone secreted by opposite mating type partner. After the pheromone binds to the specific receptor, the signal is transferred into nucleus through complicated signal transduction pathway(s). In yeast, the signaling components including receptors, G proteins and MAPK cascade proteins are well characterized (Bidaut et al., 2006). Orthologs are identified in self-fertile E. nidulans and some of them are revealed to play important roles in sexual development.
steC (encoding a MAPKK kinase)
The MAPK (mitogen-activated protein kinase) cascade is one of the central signal transduction pathways conserved in almost every eukaryotic cell. There are several protein kinases in the MAPK cascade, which are sequentially activated by induction of extracellular mitogens. Ste11p is a yeast MAP kinase kinase kinase (MAPKKK or MAPKK kinase) which is involved in mating, pseudohyphal growth and osmoregulation processes. It phosphorylates a downstream MAP kinase kinase (MAPKK or MAPK kinase). E. nidulans homolog of yeast Ste11p, SteC, was identified by Wei et al. (2003). In deletion mutants of steC, curled and branched hyphae with reduced growth are formed (Wei et al., 2003). As was expected according to the function of yeast homolog STE11, sexual development of the deletion mutants was affected. The steC deletion mutant develops no cleistothecia. And it is not crossed with wild type by the conventional hyphal fusion method. Rather, heterokaryotic mycelia can be constructed by the protoplast fusion method. The heterokaryon constructed with homozygous steC deletion mutants was sterile, indicating that the steC gene is required for self-fertile cross and cleistothecia development (Wei et al., 2003). Not only sexual development but also conidiophore development of the deletion mutant was altered. Very large conidia and secondary conidiophores which come out from the vesicle of primary conidiophores were observed with the low frequency (2%) in the ΔsteC mutant. Furthermore, the steC transcript was more abundant during asexual sporulation than fruiting body formation. A western blot analysis with antiphosphoantibodies of p44/42, SAPK/JNK and p38 showed that SteC can activate at least two downstream MAPKs, a p44/42 homolog and a SAPK/JNK homolog, in E. nidulans. Although evidences that the downstream MAPK(s) are regulated by SteC are not enough yet, it is quite clear that the MAPK cascade is involved in sexual development as well as normal conidiophore formation.
hogA/sakA (encoding a MAP kinase)
The hogA gene, together with an identical gene, sakA, which is identified as a homolog of yeast HOG1in E. nidulans, encodes a member of the stress MAPK family. Han and Prade (2002) reported that the hogA gene in E. nidulans plays a crucial role for regulating the osmotic stress response. Later, the role of the hogA/sakA gene in the response to an oxidative stress and in sexual development was illustrated by Kawasaki et al. (2002). The sakA gene encodes a 379 amino acid protein the sequence of which has similarity to those of the stress-activated MAPK family (SAPK), containing the conserved TGY phosphorylation site. SakA is phosphorylated immediately after the fungus is exposed to an oxidative stress as well as an osmotic stress, indicating that SakA is a functional SAPK activated by various external stresses. The conidia of a sakA mutant lost their viability faster than a wild type, indicating that the sakA gene plays an important role in the spore viability as well as the stress resistance (Kawasaki et al., 2002). And also the deletion mutant produced more cleistothecia than wild type suggesting that SakA represses sexual development or that other signaling pathways are derepressed by inactivation of the sakA gene. In this process the steA gene might be necessary because the ΔsakA ΔsteA double mutant were not able to produce cleistothecia.
Transcription factors that control sexual development
Several transcription factors, which are known to regulate sexual development, are characterized as homologs of other fungi. An important transcription factor SteA which was identified as a homolog of S. cerevisiae Ste12p plays a positive regulator of sexual development (Vallim et al., 2000). The RosA and the NosA which were isolated by ortholog screening of Pro1 of Sodaria macrospora contain a fungal specific Zn(II)2Cys6 binuclear cluster and play as a negative regulator of sexual development (Vienken et al., 2005; Vienken and Fischer, 2006).
SteA
In S. cerevisiae, Ste12p plays an important role in the regulating cellular morphogenesis and the mating process, especially in the pseudohyphal growth and the karyogamy. A homolog of yeast STE12, steA, was identified by the degenerate PCR method in E. nidulans (Vallim et al., 2000). The steA gene encodes a 692 amino acid polypeptide carrying a conserved homeodomain in its Nterminus N-terminus with an additional two tandem C2H2 Zn-finger domains on its C-terminus, which is not found in yeast Ste12p. Deletion of the steA gene did not affect vegetative hyphal growth and asexual sporulation. However, the deletion mutants did not develop any cleistothecia and ascospores, even under the condition sexual development is induced by restriction of oxygen supply. Only Hülle cells were observed after 4 days of the induction. Overexpression of the steA gene under the alcA promoter resulted in the delayed formation of conidiophores with an irregular and abnormal morphology similar to the ascogenous tissue. All of these results suggest that SteA plays as a positive regulator of sexual development (Vallim et al., 2000).
RosA
RosA is a homolog of Pro1 of S. macrospora, which is a Zn(II)2Cys6 transcription factor controlling perithecia development (Masloff et al., 1999). The rosA gene (repressor of sexual development) encodes a 713 amino acid polypeptide carrying a Zn(II)2Cys6 domain. Deletion of the rosA gene caused an increment of cleistothecia production in the dark conditions, suggesting that the rosA gene at least partially represses sexual development in E. nidulans. The deletion mutant can form cleistothecia in the low concentration of glucose or the presence of 0.6M KCl under which sexual development is highly repressed, and also produces Hülle cells even in a submerged culture. These phenotypes are very similar to overexpression of the positive sexual regulator, nsdD or veA (Han et al., 2001; Kim et al., 2002). When the rosA gene is overexpressed, profuse aerial hyphae were formed without any sexual or asexual development, which is a quite similar phenotype to that of the fadA-dominant activating mutant, but a genetic analysis revealed that the FadA-signaling is independent of the RosA. The nsdD, veA, and even stuA genes in a submerged culture are upregulated in rosA deletion mutant (Vienken et al., 2005).
NosA
NosA (number of sexual spores) was also isolated by ortholog screening with the Pro1 from S. macrospora (Masloff et al., 1999; Vienken and Fischer, 2006). The NosA protein was predicted to be composed of 675 amino acids, having 44% of the sequence identity with the Pro1 of S. macrospora. NosA shows about 43% similarity with RosA. The two Pro1 homologs have been found only in aspergilli including A. oryzae and A. fumigatus while other fungi have only one. The expression of the nosA gene is increased at the late stage of asexual sporulation. However, very low steady-state level of the nosA transcript is detected during the early sexual stage (Vienken and Fischer, 2006). Similar to the rosA gene, transcript of nosA accumulates within 3 h after glucose starvation. In deletion mutant of the nosA gene with the veA1 allele, cleistothecia are not produced both in the normal and favored condition, such as at the increased CO2 concentration. In the veA+ background, sexual development of the ΔnosA mutant was blocked at the primodial stage, indicating that the nosA gene is necessary for maturation of fruiting body. The nosA deletion strain in the constitutively induced nsdD background does not complete sexual development and blocked at the primordia stage, suggesting that the nosA gene is in the downstream of the nsdD gene in the same pathway (or parallel to nsdD). Northern blot analysis showed that nosA expression was upregulated in a ÄrosA strain when compared to a wild type, indicating that the rosA gene represses the expression of the nosA gene (Vienken and Fischer, 2006). The Zn(II)2Cys6 binuclear cluster transcription factor is found only in fungi. According to the genomic database of E. nidulans, 123 potential Zn(II)2Cys6 binuclear cluster proteins exist but only a few including RosA and NosA have been characterized (Vienken et al., 2005).
Metabolic regulators
Decision of development in fungi is largely dependent upon energy metabolism or nitrogen starvation. There should be complex regulatory pathways or mechanisms in deciding the reproductive cycle after monitoring the cellular state created by various metabolisms. Unfortunately, little information on the genetic or molecular mechanism for the process is available yet. Reactive oxygen species (ROS) are generated inevitably during aerobic respiration of eukaryotic organisms. There may be a regulatory connection between ROS production and the control of development.
Appropriate protein degradation is very important for regulating cell growth and differentiation. Ubiquitinylation is a well-known mechanism for the targeted degradation of protein and E3 ubiquitin ligase complex is a part of the enzymatic cascade. CSN, the constitutive photomorphogenesis complex 9 (COP9) signalosome, which directly interacts with E3 ubiquitin ligases, has been known to be an important regulator of development.
NoxA
Reactive Oxygen Species (ROS) generation during respiration is inevitable process in aerobic organisms. The active production of ROS, which is usually governed by NADPH oxidase (Nox), is important in response to pathogenic infection. Recently, new roles of Nox-generated ROS in eukaryotes, such as regulation of cell growth, oxygen sensing, growth factor signaling and fertilization, were reported (Lambeth, 2004). In E. nidulans, the noxA gene encoding a novel microbial NADPH oxidase homologous to mammalian gp91phox was identified as a regulator of sexual development (Lara-Ortiz et al., 2003). The noxA gene is predicted to encode a polypeptide containing 550-amino acid residues which is well-conserved in most filamentous fungi but not in S. cerevisiae and Schizosaccharomyces pombe (Lara-Ortiz et al., 2003). A deletion mutant of noxA does not show any change in hyphal growth and asexual development. However, the mutant develops immature primordia and Hülle cells, implying that it can initiate sexual development but is arrested at the primordial stage. The young primordia and Hülle cells of wild type produce superoxide, H2O2, and other ROS which are essential for cleistothecium and ascospore formation. But they are generated in the noxA deletion mutant. This result suggests that ROS yielded by NADPH oxidase (Nox) activity is required for the progression of development from primordia to cleistothecia (Lara-Ortiz et al., 2003).
CSN (COP9 signalosome)
Two components of the COP9 signalosome, which can be a potent regulator of sexual development, have been identified in E. nidulans, (Busch et al., 2003). One of the components is csnD that encodes a PCI domain protein similar to the fourth subunit of CSN. Deletion of csnD arrests sexual development at the primordial stage, indicating that the gene is also required for maturation of cleistothecia. Although the mutant shows additional phenotypes of reduced radial growth and accumulation of red pigment, conidiophore and conidia formation are not affected by the deletion of csnD, indicating that the function of COP9 signalosome may be specific to sexual development. The csnE gene encoding the fifth CSN subunit contains conserved MPN domain. The csnE deletion mutant shows almost identical phenotypes of ΔcsnD, block in sexual development, slow growth and red hyphae formation. The identical phenotypes of deletion of either csnD or csnE indicate that both genes are involved in the same function including several physiological and developmental processes (Busch et al., 2003). The CSN components also are concerned in light response in development, but have no significant relationship with the veA gene.
Conclusion and Perspectives
In contrast to asexual sporulation, the genetic and molecular mechanisms of sexual sporulation process in aspergilli are not much understood yet. Given the much more complexity of the developmental process, it might have been predicted that far more genes, which play critical roles in sexual development, would have been identified. The reason why the mutants defective in sexual sporulation could not be easily isolated in E. nidulans is that the early laboratory strain carried the veA1 mutation which causes delayed and decreased sexual development. The other reason is that asexual sporulation takes place prior to fruiting body formation and, thus, most colonies are covered with conidia, which makes it difficult to observe the process of fruiting body formation. Han et al. (1990) made a success in massive screening of mutants defective in sexual development by mutagenesis of veA+ wild type strain followed by culture in hypoxic condition. Now a variety of mutants showing alternative developmental pattern is easily obtainable. However, there is a technical limitation to isolate the genes by complementation of mutants even though various genomic libraries have been developed. A number of reverse genetic techniques are now available and have been applied for searching for genes involved in sexual development.
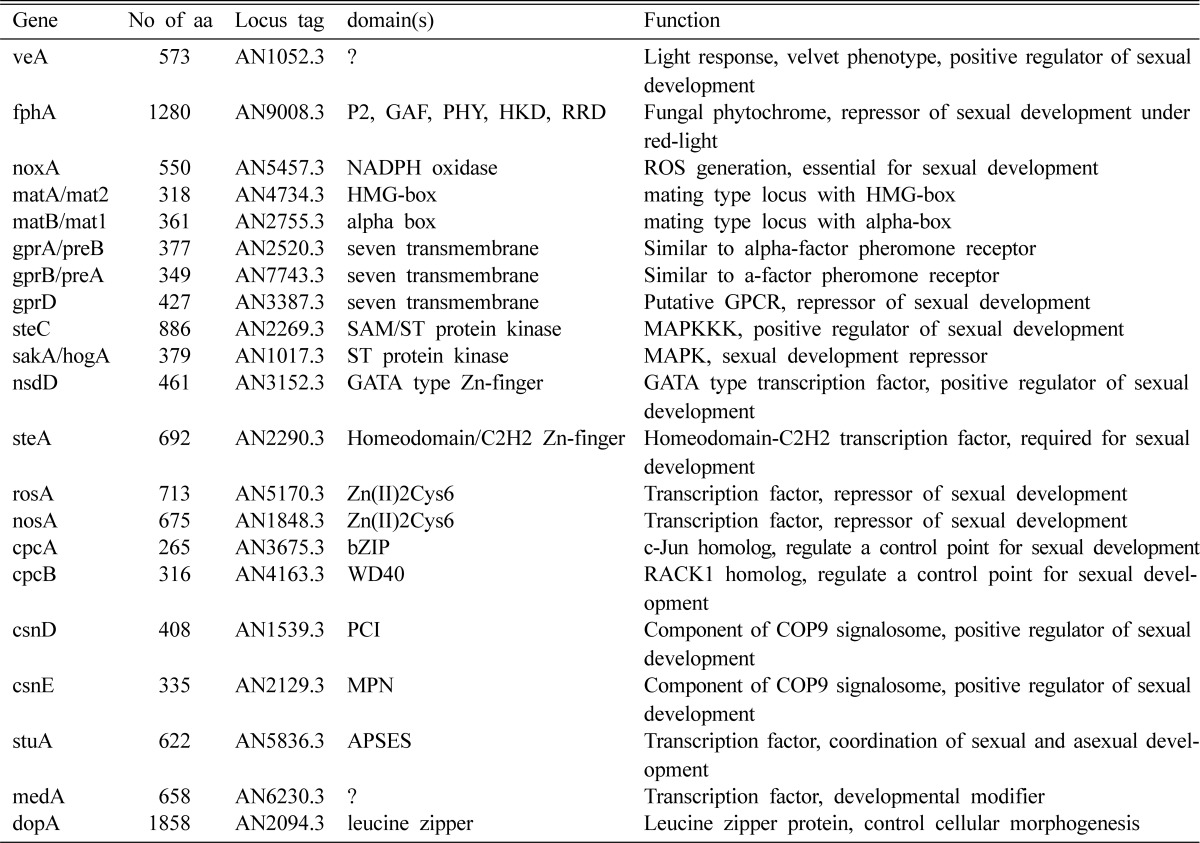
A large number of genes are expected to be involved in sexual development of E. nidulans. The properties of the genes that have been characterized so far are summarized in Table 2. Most of the genes act in the reproduction decision stage or early sexual development. Some of them control sexual development positively (veA, nsdD, steA, etc.) and some negatively (nosA, rosA). Several components in signal transduction pathways and protein kinases play some roles in decision of sexual development or in formation of fruiting bodies, although the information on the signal they response to and on the transcription factors they connect with are very poor.
Table 2.
Comprehensive genetic and genomic information of the genes involved in sexual development
Regarding the importance of sexual development in filamentous fungi, recent progresses in molecular genetics and genomics enlarge the boundaries of biological understanding of sexual development not only in the fertile species but also in the sterile species.
Acknowledgements
This work was supported by Korea Science and Engineering Foundation (KOSEF) grant (R01-2006-000-11204-0) and Korea Research Foundation (KRF) grant (1998-516) funded by the Korean government (MEST), and in part of Woosuk University (2009).
References
- 1.Axelrod DE, Gealt M, Pastushok M. Gene control of developmental competence in Aspergillus nidulans. Dev Biol. 1973;34:9–15. doi: 10.1016/0012-1606(73)90335-7. [DOI] [PubMed] [Google Scholar]
- 2.Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, Frankenbergdinkel N, Fischer R. The phytochrome FphA represses sexual development in red light. Curr Biol. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 3.Braus GH, Krappmann S, Eckert SE. Sexual development in ascomycetes: fruit body formation of Aspergillus nidulans. In: Osiewacz HD, editor. Molecular biology of fungal development. Boca Raton, FL: CRC Press; 2002. pp. 215–244. [Google Scholar]
- 4.Busby TM, Miller KY, Miller BL. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics. 1996;143:155–163. doi: 10.1093/genetics/143.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busch S, Eckert SE, Krappmann S, Braus GH. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 2003;49:717–730. doi: 10.1046/j.1365-2958.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- 6.Calvo AM, Bok J, Brooks W, Keller NP. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl Environ Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cary JW, GR OB, Nielsen DM, Nierman W, Harris-Coward P, Yu J, Bhatnagar D, Cleveland TE, Payne GA, Calvo AM. Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics. Appl Microbiol Biotechnol. 2007;76:1107–1118. doi: 10.1007/s00253-007-1081-y. [DOI] [PubMed] [Google Scholar]
- 8.Champe SP, Kurtz MB, Yager LN, Butnick NJ, Axelrod DE. Spore formation in Aspergillus nidulans: competence and other developmental processes. In: Turian G, Hohl HR, editors. The fungal spore: morphogenetic controls. New York, NY: Academic Press; 1981. pp. 63–91. [Google Scholar]
- 9.Champe SP, Nagle DL, Yager LN. Sexual sporulation. In: Martinelli SD, Kinghorn JR, editors. Aspergillus: 50 Years On, Progress in Industrial Microbiology. Amsterdam: Kinghorn, Elsevier; 1994. pp. 429–454. [Google Scholar]
- 10.Clutterbuck AJ. A mutational analysis of conidial development in Aspergillus nidulans. Genetics. 1969;63:317–327. doi: 10.1093/genetics/63.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran RM, Cary JW, Calvo AM. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl Microbiol Biotechnol. 2007;73:1158–1168. doi: 10.1007/s00253-006-0581-5. [DOI] [PubMed] [Google Scholar]
- 12.Dutton JR, Johns S, Miller BL. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 1997;16:5710–5721. doi: 10.1093/emboj/16.18.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert SE, Hoffmann B, Wanke C, Braus GH. Sexual development of Aspergillus nidulans in tryptophan auxotrophic strains. Arch Microbiol. 1999;172:157–166. doi: 10.1007/s002030050755. [DOI] [PubMed] [Google Scholar]
- 14.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 15.Galbraith JC, Smith JE. Sporulation of Aspergillus niger in submerged liquid culture. J Gen Microbiol. 1969;59:31–45. doi: 10.1099/00221287-59-1-31. [DOI] [PubMed] [Google Scholar]
- 16.Glass NL, Lorimer I. Ascomycete mating types. In: Bennett JW, Lasure LS, editors. More Gene Manipulations in Fungi. San Diego, CA: Academic Press; 1991. pp. 193–216. [Google Scholar]
- 17.Han DM, Han YJ, Chae KS, Jahng KY, Lee YH. Effects of various carbon sources on the development of Aspergillus nidulans with velA+ or velA1 allele. Korean J Mycol. 1994a;22:332–337. [Google Scholar]
- 18.Han DM, Han YJ, Kim JH, Jahng KY, Chung YS, Chung JH, Chae KS. Isolation and characterization of NSD mutants in Aspergillus nidulans. Korean J Mycol. 1994b;22:1–7. [Google Scholar]
- 19.Han DM, Han YJ, Lee YH, Jahng KY, Jahng SH, Chae KS. Inhibitory conditions of asexual development and their application for the screening of mutants defective in sexual development. Korean J Mycol. 1990;18:225–232. [Google Scholar]
- 20.Han KH, Cheong SS, Hoe HS, Han DM. Characterization of several NSD mutants of Aspergillus nidulans that never undergo sexual development. Korean J Genet. 1998;20:257–264. [Google Scholar]
- 21.Han KH, Han KY, Kim MS, Lee DB, Kim JH, Chae SK, Chae KS, Han DM. Regulation of nsdD expression in Aspergillus nidulans. J Microbiol. 2003a;41:259–261. [Google Scholar]
- 22.Han KH, Han KY, Yu JH, Chae KS, Jahng KY, Han DM. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol Microbiol. 2001;41:299–309. doi: 10.1046/j.1365-2958.2001.02472.x. [DOI] [PubMed] [Google Scholar]
- 23.Han KH, Lee DB, Kim JH, Kim MS, Han KY, Kim WS, Park YS, Kim HB, Han DM. Environmental factors affecting development of Aspergillus nidulans. J Microbiol. 2003b;41:34–40. [Google Scholar]
- 24.Han KH, Prade RA. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol Microbiol. 2002;43:1065–1078. doi: 10.1046/j.1365-2958.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- 25.Hermann TE, Kurtz MB, Champe SP. Laccase localized in hulle cells and cleistothecial primordia of Aspergillus nidulans. J Bacteriol. 1983;154:955–964. doi: 10.1128/jb.154.2.955-964.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann B, Wanke C, Lapaglia SK, Braus GH. c-Jun and RACK1 homologues regulate a control point for sexual development in Aspergillus nidulans. Mol Microbiol. 2000;37:28–41. doi: 10.1046/j.1365-2958.2000.01954.x. [DOI] [PubMed] [Google Scholar]
- 27.Hondmann DH, Visser J. Carbon metabolism. Prog Ind Microbiol. 1994;29:61–139. [PubMed] [Google Scholar]
- 28.Kafer E. Origins of translocations in Aspergillus nidulans. Genet. 1965;52:217–232. doi: 10.1093/genetics/52.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 30.Kato N, Brooks W, Calvo AM. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot Cell. 2003;2:1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki L, Sanchez O, Shiozaki K, Aguirre J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol Microbiol. 2002;45:1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x. [DOI] [PubMed] [Google Scholar]
- 32.Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 33.Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 34.Kim H, Han K, Kim K, Han D, Jahng K, Chae K. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet Biol. 2002;37:72–80. doi: 10.1016/s1087-1845(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 35.Kim HR, Chae KS, Han KH, Han DM. The nsdC gene encoding a putative C2H2-type transcription factor is a key activator of sexual development in Aspergillus nidulans. Genetics. 2009;182:771–783. doi: 10.1534/genetics.109.101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 37.Lara-Ortiz T, Riveros-Rosas H, Aguirre J. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol Microbiol. 2003;50:1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Lee T, Lee YW, Yun SH, Turgeon BG. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol. 2003;50:145–152. doi: 10.1046/j.1365-2958.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 39.Martinelli SD. Phenotypes of double conidiation mutants of Aspergillus nidulans. J Gen Microbiol. 1979;114:277–287. doi: 10.1099/00221287-114-2-277. [DOI] [PubMed] [Google Scholar]
- 40.Martinelli SD, Clutterbuck AJ. A quantitative survey of conidiation mutants in Aspergillus nidulans. J Gen Microbiol. 1971;69:261–268. doi: 10.1099/00221287-69-2-261. [DOI] [PubMed] [Google Scholar]
- 41.Masloff S, Poggeler S, Kuck U. The pro1(+) gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics. 1999;152:191–199. doi: 10.1093/genetics/152.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller KY, Nowell A, Miller BL. Differential regulation of fruitbody development and meiosis by the unlinked Aspergillus nidulans mating type loci. Fungal Genet Newslett. 2005;52:184. [Google Scholar]
- 43.Miller KY, Wu J, Miller BL. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- 44.Min JY, Kim HR, Han KH, Han DM. Isolation and characterization of Aspergillus nidulans mutants which undergo sexual development in light exposure. Korean J Microbiol. 2007;43:77–82. [Google Scholar]
- 45.Mooney JL, Yager LN. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 1990;4:1473–1482. doi: 10.1101/gad.4.9.1473. [DOI] [PubMed] [Google Scholar]
- 46.Ng AML, Smith JE, McIntosh AF. Changes in activity of tricarboxic acid cycle and glyoxylate cycle enzymes during synchronous development of Aspergillus niger. Trans Brit Mycol Soc. 1973;61:12–20. [Google Scholar]
- 47.O'Gorman CM, Fuller HT, Dyer P. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2008;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 48.Paoletti M, Seymour F, Alcocer M, Kaur N, Calvo A, Archer D, Dyer P. Mating Type and the Genetic Basis of Self-Fertility in the Model Fungus Aspergillus nidulans. Curr Biol. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Pascon RC, Miller BL. Morphogenesis in Aspergillus nidulans requires Dopey (DopA), a member of a novel family of leucine zipper-like proteins conserved from yeast to humans. Mol Microbiol. 2000;36:1250–1264. doi: 10.1046/j.1365-2958.2000.01950.x. [DOI] [PubMed] [Google Scholar]
- 50.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 51.Raper KB, Fennell DI. The Genus Aspergillus. Baltimore: The Williams and Wilkins Co; 1965. [Google Scholar]
- 52.Samson RA. Current systematics of the genus Aspergillus. In: Powell KA, Renwick A, Peberdy JF, editors. The genus Aspergillus: from taxonomy and genetics to industrial application. London: Plenum Press; 1994. pp. 261–276. [Google Scholar]
- 53.Scazzocchio C. Aspergillus genomes: secret sex and the secrets of sex. Trends in Genetics. 2006;22:521–525. doi: 10.1016/j.tig.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Scherer M, Fischer R. Purification and characterization of laccase II of Aspergillus nidulans. Arch Microbiol. 1998;170:78–84. doi: 10.1007/s002030050617. [DOI] [PubMed] [Google Scholar]
- 55.Scherer M, Wei H, Liese R, Fischer R. Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot Cell. 2002;1:725–735. doi: 10.1128/EC.1.5.725-735.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seo J, Han K, Yu J. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol Microbiol. 2004;53:1611–1623. doi: 10.1111/j.1365-2958.2004.04232.x. [DOI] [PubMed] [Google Scholar]
- 57.Serlupi-Crescenzi O, Kurtz MB, Champe SP. Developmental defects resulting from arginine auxotrophy in Aspergillus nidulans. J Gen Microbiol. 1983;129:3535–3544. doi: 10.1099/00221287-129-11-3535. [DOI] [PubMed] [Google Scholar]
- 58.Sohn KT, Yoon KS. Ultrastructural study on the cleistothecium development in Aspergillus nidulans. Mycobiology. 2002;30:117–127. [Google Scholar]
- 59.Song MH, Nah JY, Han YS, Han DM, Chae KS. Promotion of conidial head formation in Aspergillus oryzae by a salt. Biotechnol Lett. 2001;23:689–691. [Google Scholar]
- 60.Stinnett SM, Espeso EA, Cobeno L, Araujo-Bazan L, Calvo AM. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol Microbiol. 2007;63:242–255. doi: 10.1111/j.1365-2958.2006.05506.x. [DOI] [PubMed] [Google Scholar]
- 61.Tan KK. Light-induced fungal development. In: Smith JE, Berry DR, editors. The Filamentous Fungi. New York, NY: Wiley; 1978. pp. 334–357. [Google Scholar]
- 62.Urey JC. Enzyme patterns and protein synthesis during synchronous conidiation in Neurospora crassa. Dev Biol. 1971;26:17–27. doi: 10.1016/0012-1606(71)90103-5. [DOI] [PubMed] [Google Scholar]
- 63.Vallim MA, Miller KY, Miller BL. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol Microbiol. 2000;36:290–301. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- 64.Vienken K, Fischer R. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol Microbiol. 2006;61:544–554. doi: 10.1111/j.1365-2958.2006.05257.x. [DOI] [PubMed] [Google Scholar]
- 65.Vienken K, Scherer M, Fischer R. The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics. 2005;169:619–630. doi: 10.1534/genetics.104.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei H, Requena N, Fischer R. The MAPKK kinase SteC regulates conidiophore morphology and is essential for heterokaryon formation and sexual development in the homothallic fungus Aspergillus nidulans. Mol Microbiol. 2003;47:1577–1588. doi: 10.1046/j.1365-2958.2003.03405.x. [DOI] [PubMed] [Google Scholar]
- 67.Wu J, Miller BL. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol Cell Biol. 1997;17:6191–6201. doi: 10.1128/mcb.17.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yager LN. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans. Biotechnology. 1992;23:19–41. [PubMed] [Google Scholar]
- 69.Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 70.Yun SH, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci USA. 1999;96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zonneveld BJ. Biochemistry and ultrastructure of sexual development in Aspergillus nidulans. In: Smith JE, Pateman JA, editors. Genetics and Physiology of Aspergillus. London: Academic Press; 1977. pp. 59–80. [Google Scholar]