Abstract
Single spore isolates of Plasmodiophora brassicae e4 and e9 obtained from diseased Chinese cabbage were identified as race 4 and race 9, respectively, by the Williams' differential variety set. To confirm the possibility of variation in same generation and progeny of a single spore isolate of P. brassicae, random amplified polymorphic DNA (RAPD) analysis was conducted using the URP 3, 6 and OPA 7 primers. There was no difference in band type at each part of the gall of Chinese cabbage obtained by inoculation of e4 and e9 and amplification using the URP 3 and 6 primers when the same generation was analyzed. In addition, the progeny analysis, which was expanded to the third generation and conducted using the URP 3 and OPA 7 primers, revealed no differences in the band type of the e4 isolate. Based on these results, the single spore isolate of P. brassicae was genetically stable.
Keywords: Genetic variation, Plasmodiophora brassicae, Progeny analysis, RAPD, Single spore isolate
Plasmodiophora brassicae, which generates gall in the Cruciferae, belongs to a group of obligate plasmodiophorids and causes serious economic damage in cabbage, Chinese cabbage, and radish crops worldwide. In an effort to reduce the damage caused by this disease, various cultivation control methods have been attempted, including soil pH control, crop rotation and calcium and boron trials; however, no sufficient outcomes have been obtained as a result of these trials. Additionally, the agrochemicals known to be effective for the treatment of Plasmodiophora brassicae are either expensive or restricted by environmental law. Thus, there is a need to develop a resistant cultivar that might fundamentally reduce the damage associated with this disease. To achieve this, a great deal of mycological and etiological research has been conducted on Plasmodiophora brassicae (Voorrips, 1995; Some et al., 1996).
P. brassicae only propagates in living host plants, and the acquisition of a pure isolate is quite difficult because it is too small to isolate as a single spore (3.8 µm). Furthermore, successful infection of the plant root via inoculation using a single spore is difficult. Moreover, with the exception of pathogenicity, almost no forms of expression can be distinguished among single spore isolates (SSIs). Thus, only basic research such as pathogen tests conducted using differential varieties has been conducted (Klewer et al., 2001). Indeed, even though many single spore isolation methods have been designed by researchers (Buczacki, 1977; Tinggal and Webster, 1981; Kageyama, 1995), the pure isolation ratio and inoculation success ratio remain very low, and are both time- and labor-intensive. Accordingly, the development of an effective method for single spore isolation has been the target of a variety of researchers in many countries, including Korea.
Research into race using molecular biological methods has also met with mixed results due to the limitations of the single spore isolation method, as well as the difficulty of acquisition of pure DNA from pathogens alone. The single spore isolation method has not generated consistent accurate DNA extraction results due to host DNA contamination and low DNA quantities in the collected pathogen (Möller and Harling, 1996; Klewer et al., 2001; Graf et al., 2004). As a result, research on P. brassicae is in a relatively nascent stage when compared to that of general plant pathogens.
Among the many doubts and questions regarding sexual reproduction, the existence of mating type for the fusion of zoospores has yet to be identified (Voorrips, 1995), and there is currently no clear evidence of chromosomal recombination as a result of sexual reproduction (Fähling et al., 2004). Additionally, no research has been conducted regarding the manner in which genetically identical asexual spores propagate stably in host plants. Moreover, there are currently no research results available regarding the possibility that physiological differentiation could cause miscarriages in breeding-resistant cultivars as a disease control strategy. Thus, in this study, we extracted DNA from Plasmodiophora brassicae isolates that were propagated from single spores and conducted RAPD (Random Amplified Polymorphic DNA) analysis to determine if its characteristics were maintained genetically over multiple generations. The results constitute important data for the breeding of clubroot disease-resistant Chinese cabbage.
Materials and Methods
Pathogen isolates
Plasmodiophora brassicae SSI e4 and e9 collected as isolates from diseased hosts were identified as race 4 and race 9, respectively, via inoculation into Williams' differential varieties. These SSIs were mass-propagated in the sensitive Chinese cabbage cultivar (Black Pearl, Dongbu Hannong Co.) and subsequently used in this study.
Single spore isolation
Single spores were isolated via the coverglass method designed by Kageyama et al. (1995). The sucrose method used by Castlebyry et al. (1994) was conducted in a side-by-side fashion to eliminate the impurities, except for resting spores originating from plant tissue. First, the root galls were washed several times with running water to eliminate soil particles and foreign materials, after which the resting spores of P. brassicae were separated after grinding the samples in sterilized water with a homogenizer (13,500 rpm/5minutes). Plant tissues and impurities of Chinese cabbage were eliminated from a suspension prepared from root galls via filtration through sterilized gauze folded eight times followed by 15 minutes of centrifugation at 3,000 rpm, after which the pellet was recovered and subjected to the sucrose method.
The concentration of the refined spore suspension was adjusted to 1 × 103 spores/ml using a haemacytometer, and 0.5 µl of the suspension was then dropped onto a cover slip using a micro-pipette. The resting spores were then examined microscopically to identify single spores. Once the single resting spores were confirmed, two- or three-day-grown seedlings were placed on the cover glass containing the spores and then covered lightly with sterilized fine soil, after which they were planted in pots and covered with vinyl for one day to maintain humidity. The plants were subsequently cultivated for seven to eight weeks at 20~25℃ in a greenhouse (Kageyama et al., 1995). The soil pH was adjusted to 6.0~6.5 to induce optimal disease outbreak.
DNA extraction. Genomic DNA of P. brassicae was extracted via the CTAB method (Doyle and Doyle, 1990). By performing the single spore isolation mentioned above in conjunction with the sucrose method, foreign materials originating from a host plant were removed. The sample was then centrifuged at 3,000 rpm for 15 minutes, after which the pellet was removed and treated with 50 µl DNase I(Sigma, D-4263) at 50 unit/ml. Next, 300 ppm streptomycin and 100 ppm rifampicin were applied and the sample was incubated at 37℃ for one hour. To inactivate the DNase I, 5 µl of proteinase K (25 µg/µl) were added to the sample, which was subsequently incubated at 37℃ for one hour. Next, 300 µl of 5mM EDTA was added and the sample was then treated at 80℃ in a heating block for 5 minutes. The conidial suspension with inactivated DNase I was then centrifuged again at 4,000 rpm, after which the supernatant was removed and the pellet was freeze-dried and stored until subsequent use.
For DNA extraction, the pellets were thoroughly ground with 0.2 g glass beads (0.09~0.15 mm diameter) using a pellet pestle (Sigma), after which 400 µl of extraction buffer [200 mM Tris-Cl (pH 8.0), 200 mM EDTA, 1.4M NaCl, 1% PVP] were added. The mixture was then extracted with 600 µl of chloroform: isoamyl alcohol (24 : 1), after which DNA extraction and purification was conducted using a G-spinTMGenomic DNA extraction kit (Intron Biotechnology, Korea). The purified DNA was then eluted in 200 µl of H2O in the final step, and the samples were maintained at -20℃ until subsequent analysis.
RAPD analysis
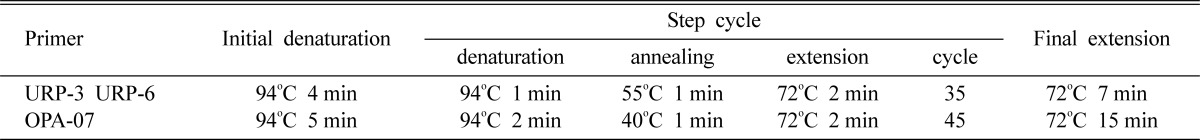
The primers tested in this study were the 12 URP primers provided in a UniPrimerTM kit (Seoulin Bioscience Institute, Korea), Operon 10-base primer for DNA polymorphism analysis (Möller and Harling, 1996) and the RFM8-1, RFM8-2 primer set for race determination (Klewer et al., 2001). The effective primers, UPR-3 (5'-CCCAGCAACTGATCGCACAC-3'), URP-6 (5'-ATGTGTGCGATCAGTTGCTG-3') and OPA-07 (5'-AGTCAGCCAC-3'), were employed in this study.
URP-PCR reactions were conducted in a reaction mixture with a final volume of 30 µl that contained 5 ng of genomic DNA, 100 ng URP primers, 800 µM dNTPs, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 and 2.5 units of Taq polymerase (Solgent Co., Ltd. Korea). PCR reactions using OPA-07 were conducted in a reaction mixture with a final volume 50 µl that contained 10 ng of genomic DNA, 1 µM Operon 10-base primer, 2 mM dNTPs, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 and 2.5 units of Taq polymerase (Solgent Co., Ltd. Korea) (Table 1).
Table 1.
PCR conditions for DNA polymorphic analysis
Variation analysis in the same generation
For variation analysis in the same generation of P. brassicae, root galls that were obtained from the Black Pearl cultivar after inoculation with the e4 and e9 isolates were divided into five pieces and the DNA was extracted from each piece, after which RAPD analysis was conducted using the URP 3 and URP 6 primers to reveal the active polymorphisms between the pathogen isolates.
Variation analysis through progeny generations
To investigate patterns of longitudinal variation of the pathogen, second and third generations were made through regrowth after single spore inoculation with e4 as an original generation. To determine the genetic stability among generations, RAPD analysis was conducted using the URP 3 and OPA-7 primers. The isolate, e4, was used for this study because it showed the strongest pathogenicity (Jang, 2006) and is easy to form the gall by inoculation of single resting spore.
Results and Discussion
Because the resting spores of P. brassicae are haploid, single spore isolates propagated from these spores should constitute a genetically homothallic genotype (Voorrips, 1995). However, evidence of this issue has not been clearly demonstrated in most molecular biological studies, including those conducted to analyze genetic polymorphisms of this pathogen due to difficulties in the mass extraction of DNA and contamination of foreign materials originating from the host plant. This pathogen can grow only in a host plant; therefore, it is difficult to obtain a large number of pathogens when compared to other pathogens that can be propagated through pure cultivation in media. DNA extraction of P. brassicae includes two complex steps, isolation of resting spores of the pathogen from a host plant and a second step in which DNA is extracted from the isolated resting spores. Indeed, all procedures involved in the extraction of DNA from P. brassicae involve long and complex processes to remove the large amounts of foreign materials that originate from host cells and contamination of the host DNA. Nevertheless, these processes still result in the loss of target DNA. Additionally, if the amount of DNA recovered using these procedures was small or contaminated with host DNA, the RAPD and RFLP bands will be unclear or indistinct, even though the experiments were properly conducted. Thus, the development of an effective DNA extraction method, as well as a mass pathogen propagation method within the host plants remains acutely necessary.
The mass propagation of an experimental material from single spore isolates might be resolved to some degree by using a susceptible host plant, but isolates demonstrating very little pathogenicity are still being propagated. Accordingly, the development of a more effective propagation method independent of the degree of the pathogenicity of the single spore isolate should be resolved in future studies.
This study used race 4 (e4) and race 9 (e9), which are both known as prevalent pathogens in Korea, to secure sufficiently propagated single spore isolates as the experimental material (Heo, 2004; Jang, 2006). Additionally, during the DNA extraction stage, the extraction efficiency was maximized by a physical grinding method using a glass bead and a pellet pestle. Resting spores have a spherical shape, are only 3.8 µm in diameter and are composed of hard chitin compounds; therefore, they are not damaged when they are examined using a CCD phase-contrast microscope (Iponacology, Japan) (× 2,161) after treating them with EDTA or SDS. Conversely, the method of grinding them with very small glass beads (0.09~0.15 mm diameter) can damage the resting spores, leading to over 80% of the spores being destroyed (Jang, 2005, 2006).
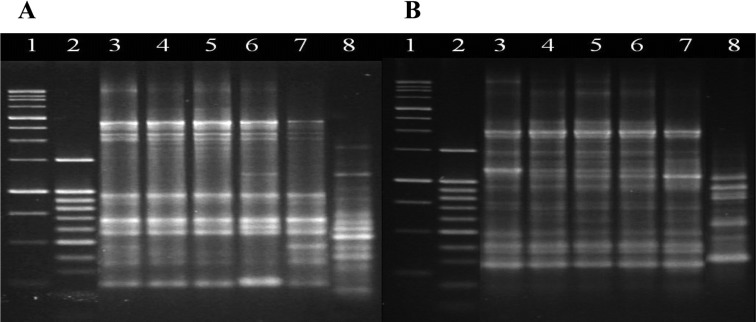
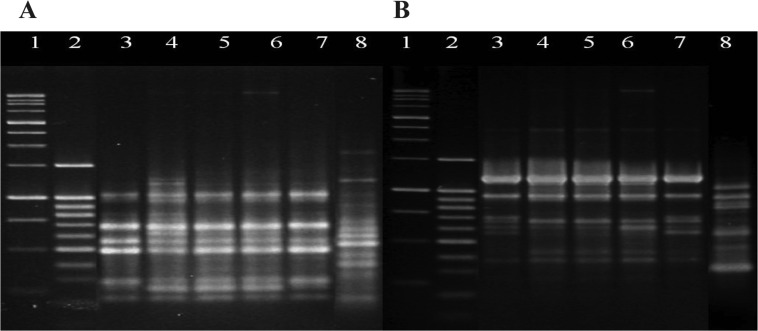
Variation analysis within a same generation performed using URP 3 and URP 6 revealed nearly no variation in the patterns of bands of e4 and e9 (Fig. 1, 2). Additionally, RAPD analysis conducted to investigate the occurrence of longitudinal variation through progeny generations revealed that only the band patterns produced using the URP 3 and 6 primers and OPA-07 primer produced varying patterns. In a previous study, only the three primers out of a group of primers evaluated for their effectiveness for variation analysis and race differentiation using P. brassicae showed a high enough number of bands and differences in band patterns to separate organisms according to race (Heo, 2004). Since Williams' differential system (Williams, 1966) and the European clubroot differential system (Buczacki et al., 1975) were suggested, an extremely small number of single spore isolates (SSI) including only a few races have been reported, and the development of genetic markers has not be progressed because the diversity of SSIs has not been fully elucidated.
Fig. 1.
Comparisons of PCR amplification of Plasmodiophora brassicae DNA using URP 3 (A) and URP 6 (B) from five pieces that were divided from the same gall infected by e4 isolate to investigate genetic variation within a same generation. Lane 1; 1,000 bp ladder, 2; 100 bp ladder, 3~7; DNAs from each of five pieces, respectively, 8; B. rapa.
Fig. 2.
Genetic variation analysis of PCR amplification of Plasmodiophora brassicae DNA by URP 3 (A) and URP 6 (B) from five pieces that were divided from the same gall infected by e4 isolate to investigate genetic variation within a same generation. Lane 1; 1,000 bp ladder, 2; 100 bp ladder, 3~7; DNA from each of five pieces, respectively, 8; B. rapa.
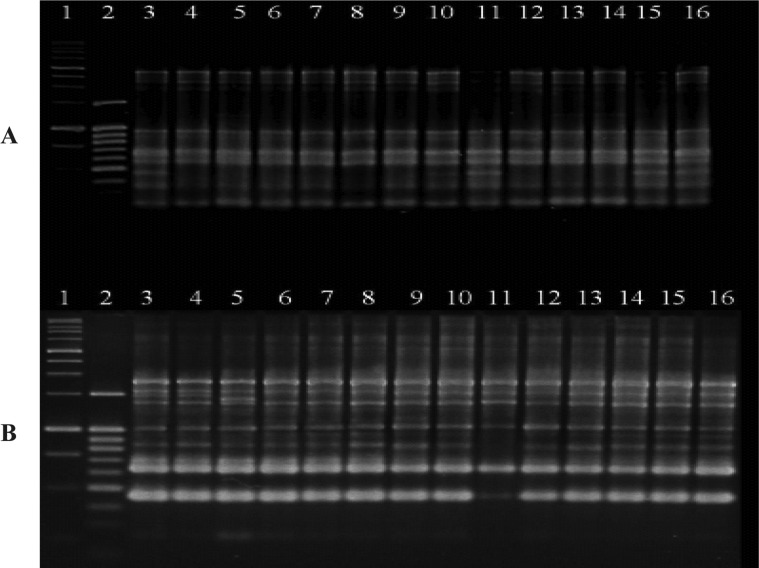
We detected no differences in band types in our variation analysis of the progeny generation using URP 3 and OPA 7 primers among generations from the original e4 to the third generation (Fig. 3). Thus, the results of this study showed that the SSI of P. brassicae are stably genetically inherited following infection and residence within the host independently, and this occurs via an asexual process. However, to make more accurate conclusions regarding the genetic stability following the growth of this pathogen, the possibility of genetic variation due to combined inoculation of isolates of different races should also be examined.
Fig. 3.
Comparison of PCR amplification of the original generation of e4 and its progeny by URP 3 (A) and OPA 7 (B) to investigate the occurrence of genetic variation through progeny generations. Lane 1; 1,000 bp ladder, 2; 100bp ladder, 3~4; original generations, 5~8; second generations, 9~16; third generations.
Acknowledgements
This work was supported in part from the Agricultural Research and Development Promotion Center (ARPC), Ministry for Food Agriculture Forestry and Fisheries (MIFAFF), Republic of Korea.
References
- 1.Buczacki ST, Toxopeus H, Mattush P, Johnston TD, Dixon GR, Hobolth LA. Studies of physiologic specialization in Plasmodiophora brassicae: proposals for attempted rationalization through an international approach. Trans Br Mycol Soc. 1975;65:295–303. [Google Scholar]
- 2.Buczacki ST. Root infections from single resting spores of Plasmodiophora brassicae. Trans Br Mycol Soc. 1977;69:328–329. [Google Scholar]
- 3.Castlebyry LA, Maddox JV, Glawe DA. A technique for the extraction and purification of viable Plasmodiophora brassicae resting spores from host root tissue. Mycologia. 1994;86:458–460. [Google Scholar]
- 4.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13. [Google Scholar]
- 5.Fähling M, Graf H, Siemens J. Characterization of single-spore isolate population of Plasmodiophora brassicae resulting from a single club. J Phytopathology. 2004;152:438–444. [Google Scholar]
- 6.Graf F, Fähling M, Siemens J. Chromosome polymorphism of the obligate biotrophic parasite Plasmodiophora brassicae. J Phytopathology. 2004;152:86–91. [Google Scholar]
- 7.Heo SH. Characteristics analysis of genetic variation in Plasmodiophora brassicae from Korea. Chungnam National University; 2004. Ms. D. Thesis. [Google Scholar]
- 8.Jang CS. Development of diagnostic system of root rot disease of ginseng caused by Cylindrocarpon destructans using nested PCR from soil. Chungnam National University; 2005. Ms. D. Thesis. [Google Scholar]
- 9.Jang SJ. Characteristics of infection and novel single-spore isolation method through two-step inoculation of clubroot pathogen, Plasmodiophora brassicae. Chungnam National University; 2006. Ms. D. Thesis. [Google Scholar]
- 10.Kageyama K, Kamimura Y, Hyakumachi M. A simple inoculation method with a single resting spore of Plasmodiophora brassicae. Ann Phytopathol Soc Jpn. 1995;61:415–418. [Google Scholar]
- 11.Klewer A, Luerben H, Graf H, Siemens J. Restriction fragment length polymorphism markers to characterize Plasmodiophora brassicae single spore isolates with different virulence patterns. J Phytopathology. 2001;149:121–127. [Google Scholar]
- 12.Möller M, Harling R. Randomly amplified polymorphic DNA(RAPD) profiling of Plasmodiophora brassicae. Lett Appl Microbiol. 1996;22:70–75. [Google Scholar]
- 13.Some A, Manzanares MJ, Laurens F, Baron F, Thomas G, Rouxel F. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 1996;45:432–439. [Google Scholar]
- 14.Tinggal SH, Webster J. Technique for single spore infection by Plasmodiophora brassicae. Trans Br Mycol Soc. 1981;76:187–190. [Google Scholar]
- 15.Voorrips RE. Plasmodiophora brassicae: aspects of pathogenesis and resistance in Brassica oleracea. Euphytica. 1995;83:139–146. [Google Scholar]
- 16.Williams PH. A system for the determination of race of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology. 1966;56:624–626. [Google Scholar]