Abstract
Pituitary cell types arise in a temporally and spatially specific fashion, in response to combinatorial actions of transcription factors induced by transient signaling gradients. The critical transcriptional determinants of the two pituitary cell types that express the pro-opiomelanocortin (POMC) gene, the anterior lobe corticotropes, producing adrenocorticotropin, and the intermediate lobe melanotropes, producing melanocyte-stimulating hormone (MSHα), have remained unknown. Here, we report that a member of the T-box gene family, Tbx19, which is expressed only in the rostral ventral diencephalon and pituitary gland, commencing on e11.5, marks pituitary cells that will subsequently express the POMC gene and is capable of altering progression of ventral cell types and inducing adrenocorticotropin in rostral tip cells. It is suggested that Tbx19, depending on the presence of synergizing transcription factors, can activate POMC gene expression and repress the α glycoprotein subunit and thyroid-stimulating hormone β promoters.
The anterior pituitary gland has provided an instructive model in which to investigate the origin of specific cell types from a common primordium. Initial aspects of development of the gland, from invagination of oral ectoderm to intense proliferation and patterning events, reflect the actions of opposing fibroblast growth factor-, bone morphogenetic protein-, and Shh-signaling gradients (1, 2), commencing with induced expression of the LIM homeodomain factor Lhx3/P-Lim (1–6). After embryonic day (e)10.5, a ventral → dorsal BMP2 gradient appears to be critical for four anterior lobe cell types, but not corticotropes, modulating positional commitment to cell-type determination (6) by inducing overlapping patterns of specific transcription factors. Induction of specific factors correlates with appearance of specific cell types; for example, GATA2 can reprogram cells of the Pit-1 lineage to a gonadotrope fate and Pit-1 conversely, can convert gonadotrope to thyrotrope (7). One of the initially appearing cell types is the corticotropes, which produce adrenocorticotropin (ACTH) from a pro-opiomelanocortin (POMC) precursor. Melanotropes, which also express the POMC gene, initially appear on ≈e13.5–e14.5 in the presumptive intermediate lobe (8). Rostral tip cells also arise on e12.5 and produce α glycoprotein subunit (αGSU) and, at low levels, thyroid-stimulating hormone β (TSHβ).
The 480-bp POMC promoter is sufficient to target expression in developing pituitary (9, 10), whereas additional 5′ information (−2 to −13 kb) is required for expression in ventral diencephalon (11). By using AtT20 murine “corticotropes” as a model, putatively important regions have been defined, uncovering response elements that include sites for β helix–loop–helix function (e.g., Neuro D1), corticotropin-releasing hormone (CRH) and nuclear receptors (12–14), and Pitx factors (15, 16). Two bicoid-related factors, Pitx1 and Pitx2, are expressed in anterior pituitary, and Pitx1 was initially suggested to be corticotrope-specific (16). It has been recently shown in vivo as well as in vitro, that cytokines, such as leukemia inhibitory factor, may play a role in POMC-expressing cell development (17–21) and that the POMC promoter contains signal transducer and activator of transcription 3-binding sites within the −399/−379 region (20). Misexpression of leukemia inhibitory factor in transgenic mice under control of the growth hormone or the αGSU promoter results in expansion of pituitary POMC cells and decrease in the other cell populations (22, 23).
T-box proteins are sequence-specific DNA-binding transcription factors that contain a characteristic ≈180-aa DNA-binding domain and often serve as developmental regulators (24–32). X-ray crystallography indicates that the T-box domain interacts as a dimer with a palindromic DNA duplex (33) but also can bind half-palindromic sequences as a monomer (34). Although most of the T-box proteins act as transcriptional activators, several can repress their target genes (35, 36). Functional interactions between Pitx1 and Tbx4 in organogenesis and the role of T-box factors (37, 38) in mammalian embryogenesis suggested the possibility that a T-box factor might have a role in pituitary development.
In this manuscript, we describe analysis of a T-box factor, Tbx19, that appears to be selectively expressed in precursors of the corticoptrope/melanotrope lineages and in the rostral ventral diencephalon, which may act as a component of the POMC gene activation program. Tbx19 alone does not appear to be epistatic to the program of corticotrope/melanotrope determination. A key aspect of Tbx19 function may be to, directly or indirectly, act as a repressor of αGSU and TSHβ gene expression, perhaps serving as a component of alternative commitment to αGSU or corticotrope lineages. These findings complement those recently reported for T-pit (39) while this manuscript was in preparation.
Methods
Cloning of the Mouse Tbx19 Gene.
The murine Tbx19-coding sequence was amplified by PCR from cDNA from AtT20 cells by using Pwo DNA polymerase (Roche Molecular Biochemicals) with degenerate primers based on the human TBX19 sequence, and directly cloned into PCR-Blunt II-TOPO vector by using the Zero Blunt TOPO PCR cloning kit (Invitrogen). Two independent clones were sequenced and a full-length cDNA clone was obtained by screening a pituitary library.
Generation of Transgenic Mice and Transfection Assays.
Transgenic constructs were made as described (1) by inserting the N-terminally hemagglutinin (HA)-tagged Tbx19 cDNA between a rabbit 0.65-kb β-globin intron fragment at the 5′ end and a 0.63-kb poly(A) fragment of the human growth hormone (GH) gene at the 3′ end. The transgene was driven by a 14-kb murine αGSU promoter or a 8-kb fragment of 5′-flanking Pitx1 gene information inserted 5′ of the cassette. Appropriate Southern blot and PCR analysis were used for genotyping.
HeLa cells were plated out and grown for 24 h in 12-well dishes. FuGene (Roche Molecular Biochemicals)-mediated transfection was carried out with pGL2 (Promega)-derived rat POMC (−480 ≈+64) or αGSU luciferase reporter and pcDNA3 (Invitrogen)-based expression vectors for cytomegalovirus-driven expression of mouse Pitx1, Pitx2, Lhx3, and Tbx19 for 48 h.
Immunohistochemistry.
Heads from e14.5–e17.5 mouse embryos were fixed in ethanol-formalin mixture and embedded in paraffin as described (40). Serial 6-μm sections were stained with mouse mAbs against HA tag (Babco, Richmond, CA, 1:1,000), rabbit polyclonal antibodies against ACTH (Sigma, 1:1,000), αGSU, TSHβ (National Hormone and Pituitary Program, National Institute of Diabetes and Digestive and Kidney Diseases, rabbit, 1:240), and Pit-1 (1:1,000). Secondary peroxidase-, FITC-, and tetramethylrhodamine B isothiocyanate-conjugated antibodies were from Jackson ImmunoResearch.
Results
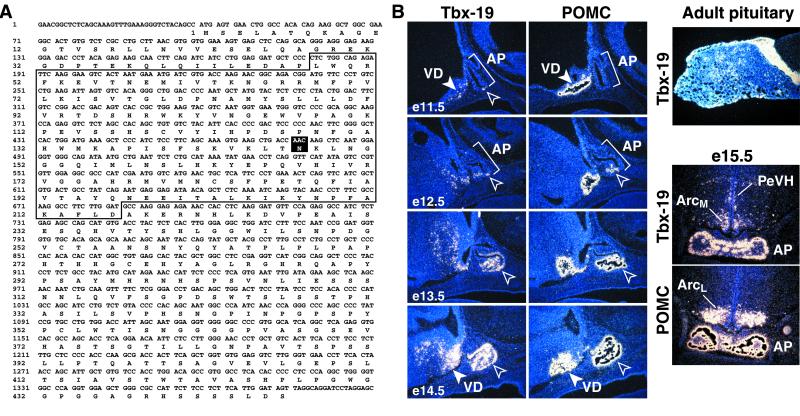
Given the important developmental roles of Tbx factors, we systematically examined new family members in the database to determine whether any might exhibit pituitary expression. We isolated and determined the DNA sequence of the murine Tbx19 cDNA, which has >90% homology to the human Tbx19, including all known critical residues in the T-box DNA-binding domain (41) (Fig. 1A), which was recently reported as T-pit (39). Tbx19 expression is not observed in the invaginating Rathke's pouch, but beginning on e11.5 Tbx19 expression appears in the most caudoventral aspect of the developing gland, with subsequent extension of Tbx19-expressing cells both rostrally and caudally (Fig. 1B). By e12.5, Tbx19 expression is observed in the most peripheral, ventral aspects of the anterior pituitary, as well as adjacent to the region of mesenchymal invagination dorsal to the rostral tip (Fig. 1B). Tbx19 is not expressed in the rostral tip, and its expression is spatially and temporally linked with the subsequent initial appearance of POMC gene expression (Fig. 1B). Immunohistochemistry/in situ hybridization double labeling confirmed coexpression of Tbx19 in pituitary POMC-producing cells (data not shown).
Figure 1.
Expression of Tbx19 in pituitary development. (A) Nucleotide sequence of murine Tbx19 cDNA ORF predicts a 446-aa gene product containing a T-box (boxed). Reverse lettering on amino acid 147 indicates the site of mutation for Fig. 5 (N → T). (B) In situ hybridization using Tbx19 or POMC probes demonstrates expression of Tbx19 on e11.5 (open arrowhead) in the caudoventral aspect of developing anterior pituitary, followed by expression of POMC in Tbx19-expressing cells by e12–e12.5. Tbx19 expression is apparently restricted to POMC lineages. In ventral diencephalon, Tbx19 (closed arrowhead) is expressed from ≈e10.5, late in the arcuate and paraventricular nucleus, but is not expressed in the lateral regions of the arcuate nucleus where POMC is expressed (closed arrowhead).
Tbx19 also is expressed during development of rostral regions of the ventral diencephalon. Examination of its ontogeny revealed that its expression in the medial aspect of the developing arcuate nucleus, an area of dopaminergic neurons expressing neuropeptide Y and AGRP; however, Tbx19 is not expressed in cells expressing POMC or growth hormone-releasing hormone (Fig. 1B). The domain of Tbx19 expression later expands to include other areas, including the somatostatin-expressing neurons of the periventricular nucleus of hypothalamus (PeVH), another area in which the POMC gene is not expressed (Fig. 1B). We were therefore particularly interested in exploring the potential role, if any, of Tbx19 in initiating corticotrope cell type determination and/or POMC gene expression.
Role of BETA2/Neuro D1 in Corticotrope Development.
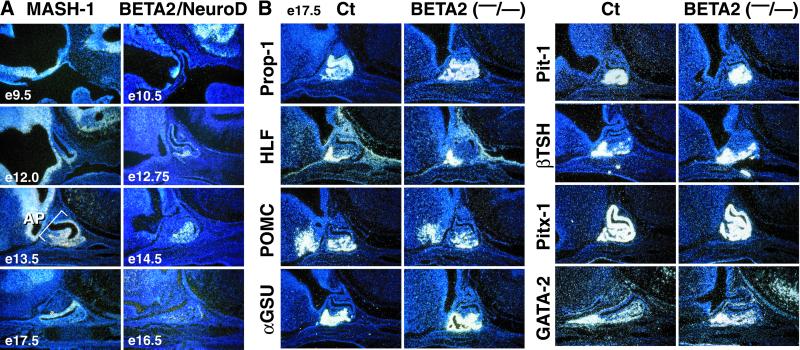
A series of basic helix–loop–helix factors are expressed in the anterior pituitary gland, including BETA2/Neuro D1, which have been noted to have a transient expression in ventral regions of the anterior pituitary gland and which have been claimed, by immunohistochemistry, to be coexpressed with POMC. BETA2/Neuro D1 transcripts, also initially observed at e11.5 in a rostroventral fashion (42, 43), become scarcely detectable by e14.5–e15.5, and never appears in the presumptive intermediate lobe, a temporal and spatial pattern that is not strongly supportive of a role in POMC-expressing cells. In contrast, MASH-1 expression is more intensively sustained in the intermediate lobe (Fig. 2A) in a pattern fully corresponding to POMC cells (Fig. 3B). Deletion of BETA2/Neuro D1 (44, 45) permits evaluation of potential roles of BETA2/Neuro D1 in pituitary development. As illustrated in Fig. 2B, all cell types in BETA2/Neuro D1(−/−) mice appear with only a minor alteration in POMC, αGSU, and TSHβ transcripts. Although BETA2/Neuro D1 may not be required for anterior lobe corticotropes, redundancy with other members of the basic helix–loop–helix family remains possible.
Figure 2.
BETA2/NeuroD1 is not alone required for pituitary development. (A) Expression patterns of Mash-1 and BETA2/Neuro D1 showing initial ventral expression, and continued expression of MASH-1 on POMC lineages, but with extinction of BETA2 by e16.5. (B) In situ hybridization analysis of e17.5 pituitary, using the indicated cDNA probes, showing normal development of BETA2 (−/−) mice.
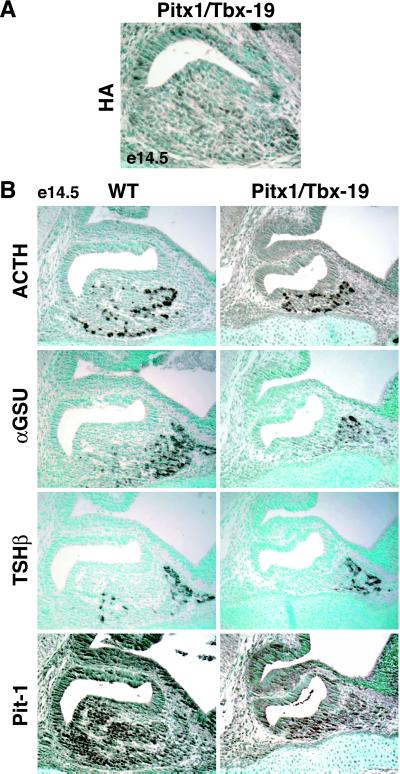
Figure 3.
Cell type development in the hypoplastic pituitary of Pitx1/Tbx19 transgenic mice. (A) Representative section (e14.5) of a pituitary expressing HA-tagged Tbx19, under control of the 8-kb Pitx1 promoter. (B) The pituitary exhibits no alteration of ACTH-expression but decreased numbers of αGSU+ and Pit-1+ cells.
Tbx19 Roles in Vivo.
To explore whether Tbx19 might exert a determining role in development of the corticotrope/melanotrope cell lineages in the developing pituitary gland, we took advantage of the availability of targeting vectors that could effectively express Tbx19 in specific temporal and spatial fashion during pituitary development. They have successfully permitted documentation of the ability of GATA2 and Pit-1 to alter Pit-1 and gonadotrope lineages, respectively (7). Tbx19 was first placed into transcription units under control of 8-kb Pitx1 regulatory sequences, which have been documented to target transgene expression from initial appearance of Rathke's pouch (6), and remain strongly expressed in all cells until ≈e11.5–e12.5, when its expression becomes progressively restricted. Analysis of five founders with effective expression of the Pitx1/Tbx19 transgene, harvested at e14.5, revealed a decrease in gland size and broad expression of HA/Tbx19. All cell lineages were proportionately represented (Fig. 3A). By using specific anti-Pit-1, anti-αGSU, and anti-POMC antibodies, no induced expression of POMC was detected in any other lineages; however, there was a marked decrease in Pit-1 expression, which might account for the decreased TSHβ level (Fig. 3B). Thus, although the transgene was effectively expressed and caused no POMC gene activation, it did inhibit proliferation of both Pit-1 and αGSU lineages. Therefore, early high levels of Tbx19 expression were not sufficient to alter lineage commitments but could alter programs of cell proliferation of all cell types.
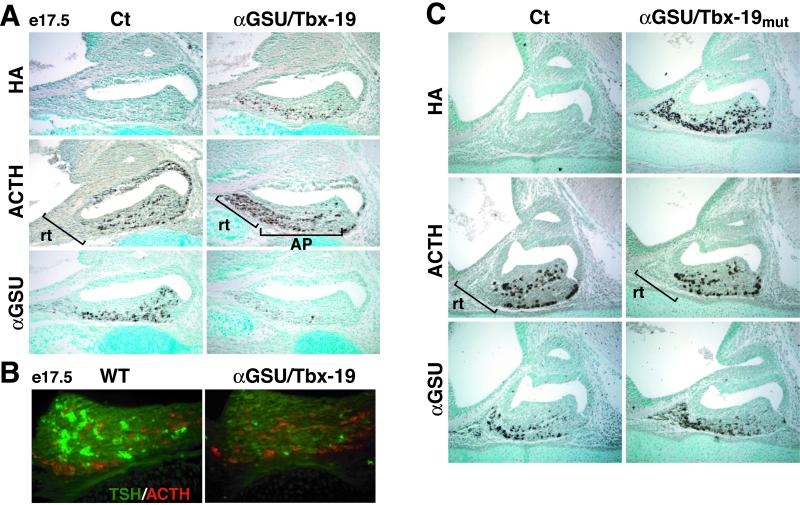
We next used the 14.5-kb αGSU regulatory sequences to sustain high levels of Tbx19 expression across the rostral tip and the ventromedial anterior pituitary in ventral cell types on e17.5. ACTH was found to be expressed in virtually every cell of the rostral tip; however, no clear increase in ACTH was evident in cells that would normally constitute the gonadotrope or caudomedial Pit-1-dependent thyrotrope population (Fig. 4A). Double-label immunohistochemistry revealed that there was a striking decrease in αGSU expression in Tbx19-expressing cells, including caudomedial thyrotrope, gonadotrope, as well as rostral tip populations. Further, caudomedial expression of TSHβ was decreased in Tbx19-expressing cells (Fig. 4B). Based on these data, we suggest that Tbx19 is capable of modifying patterns of gene expression in cell types in which it is not normally expressed. In light of the Pitx1/Tbx19 transgene phenotype, most of these effects appear to occur later and require the sustained, high level of Tbx19 expression. Conversely, although the rostral tip cells could represent “conversion” to a POMC cell type, this is clearly not the case for the ventrally arising gonadotropes or the more dorsally arising thyrotropes. These data raise the question of whether Tbx19 might be functioning in a fashion analogous to that of Pit-1, with respect to its inhibitory actions on the GATA2-dependent developmental program. In this case, Pit-1 acts in a fashion independent of binding to its cognate response elements, but via protein–protein interaction, to transrepress GATA2 on gonadotrope-specific genes (7). To evaluate whether a similar mechanism might operate in the case of Tbx19, we took advantage of the structural definition of the T-box DNA-binding domain by mutating a single residue (N147 → T), which caused a loss of high affinity, specific DNA binding (data not shown). When this mutant form of Tbx19 was similarly targeted under control of the αGSU regulatory sequence, in vivo, there was no expression of ACTH in rostral tip nor any decrease in αGSU expression in anterior pituitary (Fig. 4C). Thus, Tbx19 appears to exert its actions in a DNA-binding-dependent fashion.
Figure 4.
Phenotype of the αGSU/Tbx19 mouse pituitary gland. (A) The expression of HA-tagged Tbx19 follows the normal pattern of αGSU-expression, but levels of αGSU are markedly decreased, whereas ACTH is now expressed in virtually all rostral tip cells, no change in expression of ventral anterior pituitary. (B) Double label immunohistochemistry, showing a decreased TSHβ expression in αGSU/Tbx19-expressing cells of the Pit-1 lineage, but with no coexpression of POMC and TSHβ. (C) A Tbx19 mutant incapable of specific binding to cognate DNA elements no longer induced rostral tip expression of POMC or decreased αGSU gene expression in ventral pituitary.
Tbx19 Can Regulate the POMC Gene Promoter.
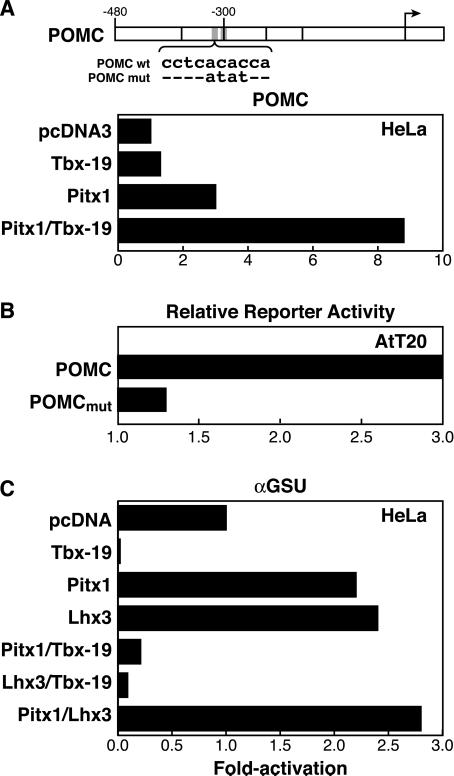
Based on extensive transient transfection analyses by using different cell types, a quantitative role has been suggested for a number of DNA-binding sites on the POMC gene promoter, including putative effects of Pitx1 and BETA2/Neuro D1/Pan (39). There is a putative T-box-binding element, a “half site” at −310 bp in the POMC regulatory sequence (Fig. 5A). Tbx19 can bind with lower affinity to this site, as evidenced by protein-dependent gel shift analysis (data not shown). In transient transfection assays, using a 480-bp POMC promoter, Tbx19 can cause a very weak activation in HeLa cells but not in AtT20 cells nor in 293 cells, in which it generally causes repression. Tbx19 appears to exert synergistic activation effects with either Pitx1 or Pitx2 factors (Fig. 5A, and data not shown). Mutation of the Tbx19-binding site decreased expression of POMC in AtT20 cells (Fig. 5B). Tbx19 was actually inhibitory to the expression of αGSU promoter in transient transfection assays (Fig. 5C). In light of its observed repressive effects on αGSU expression in vivo, a major role of Tbx19 might be to repress αGSU lineage progression in cells signaled to assume a POMC lineage fate, as well as to serve as a potential activator of POMC gene expression.
Figure 5.
Tbx19 can repress the αGSU promoter in transient transfection assays. (A) Effects of cytomegalovirus/Tbx19 and cytomegalovirus/Pitx1 on expression of a POMC (−480 to +64) luciferase fusion gene in HeLa cells. (B) Effects of mutation of the consensus T-box half site in the POMC promoter on expression in AtT20 corticotropes. (C) Effects of cytomegalovirus/Tbx19 on αGSU expression, and effects of Tbx19, on Pitx1- or Lhx3-induced expression of the αGSU promoter (−400 to +34) in HeLa cells; results are an average of duplicates differing by <5%.
Discussion
Although it is well established that pituitary development is controlled by actions of diverse classes of transcriptional activators, many of which have proved to be homeodomain proteins (8), achieving a clear understanding of physiological roles of the numerous factors capable of binding the regulatory regions of the POMC promoter has proved to be a difficult problem. Here, we have described a T-box protein, Tbx19, recently reported as T-pit (39), that limited its expression to pituitary and rostral regions of the ventral diencephalon. Tbx19 exhibits a spatial and temporal pattern of pituitary expression that is predictive of the subsequent appearance of POMC gene expression and does not overlap with POMC expression in the arcuate nucleus. Based on the inability of early misexpression of Tbx19 in Rathke's pouch to convert other lineages to a POMC-producing cell fate, although limiting proliferation, it would not appear that Tbx19/T-pit alone is sufficient to commit primordial pituitary ectoderm to a POMC lineage developmental program. Sustained ventral expression under αGSU regulatory sequence is sufficient to activate POMC gene expression in the rostral tip, which normally does not differentiate into any definitive cell types, as shown in studies with T-pit at e14.5 (39). Tbx19, however, fails to activate POMC gene expression in the anterior pituitary gonadotropes or thyrotropes, although dramatically inhibiting both αGSU expression in rostral tip, and anterior lobe gonadotropes and thyrotropes, and TSHβ expression in the Pit-1-dependent caudomedial thyrotropes. Together, these data indicate that although Tbx19/T-pit alone is not sufficient to activate a corticotrope program, it is capable of, directly or indirectly, imposing repression on aspects of the developmental program of cells that would generate the αGSU lineage. Given the ability of the cytokine, leukemia inhibitory factor, to induce POMC expression and inhibit proliferation of other lineages, and its linkage with signal transducer and activator of transcription 3 activation (18), it is reasonable to suggest that signals from the cytokine pathway may be critical to aspects of corticoptrope development. Indeed, one suggestion is that the spatial pattern of corticotrope appearance is consistent with its proximity to mesenchyme and microvasculature, including the mesenchyme invaginating dorsally to the rostral tip on e12.5, which could be the critical source of cytokine signals.
A second issue is whether Tbx19 acts as an activator or a repressor. Tbx19/T-pit (39) exerts only modest activating effects on the POMC gene expression in transient cotransfection assays, but is capable of synergizing with Pitx factors on this promoter in heterologous cell types. Mutation of the potential Tbx19-binding site on the POMC promoter actually causes decrease of the POMC promoter activity in AtT20 cells. Paradoxically, however, increased expression of Tbx19 in AtT20 cells is inhibitory to POMC expression. As the effects of Tbx19/T-pit seem to require the DNA-binding function of the T-box, the observed developmental effects in transgenic studies do not appear to reflect DNA-binding-independent transrepression effects, ruling out transrepression mechanisms. The finding that a human kindred with low ACTH levels exhibits a mutation in the T-pit locus provides the initial in vivo suggestion of its physiological positive role in production of ACTH (41). The ability of Tbx19 to inhibit αGSU and TSHβ expression can be interpreted as an effect on αGSU program activity in terminally differentiated cells, perhaps causing an indirect cascade leading to POMC gene expression events. Interestingly, Pitx1 levels are higher on rostral tip at e12.5, perhaps accounting, in part, for the selective ability of Tbx19 to induce POMC only in the rostral tip.
Thus, analogous to their actions in development of other organs, a tissue-specific T-box protein may ultimately prove to be an important component of development and function of the POMC lineages in pituitary development. The relationship between Tbx19/T-pit and signal transducer and activator of transcription 3, its role in gene activation/repression, and the question of an indirect or a direct effect on POMC gene activation events remain intriguing issues for future investigation.
Acknowledgments
We thank J. Zhao for help with transgenic mice, P. Meyer for preparation of illustrations, and M. Fisher for assistance in manuscript preparation. M.G.R. is an Investigator with Howard Hughes Medical Institute. This research was supported by National Institutes of Health grants to M.J.T. and M.G.R.
Abbreviations
- POMC
pro-opiomelanocortin
- ACTH
adrenocorticotropin
- TSHβ
thyroid-stimulating hormone β
- αGSU
α glycoprotein subunit
- en
embryonic day
- HA
hemagglutinin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF345520).
References
- 1.Treier M, Gleiberman A S, O'Connell S M, Szeto D P, McMahon J A, McMahon A P, Rosenfeld M G. Genes Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ericson J, Norlin S, Jessell T M, Edlund T. Development (Cambridge, UK) 1998;125:1005–1015. doi: 10.1242/dev.125.6.1005. [DOI] [PubMed] [Google Scholar]
- 3.Sheng H Z, Zhadanov A B, Mosinger B, Jr, Fujii T, Bertuzzi S, Grinberg A, Lee E J, Huang S P, Mahon K A, Westphal H. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- 4.Bach I, Rhodes S J, Pearse R V, 2nd, Heinzel T, Gloss B, Scully K M, Sawchenko P E, Rosenfeld M G. Proc Natl Acad Sci USA. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheng H Z, Moriyama K, Yamashita T, Li H, Potter S S, Mahon K A, Westphal H. Science. 1997;278:1809–1812. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- 6.Treier M, Gleiberman A S, O'Connell S M, Szeto D P, McMahon J A, McMahon A P, Rosenfeld M G. Genes Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasen J S, O'Connell S M, Flynn S E, Treier M, Gleiberman A S, Szeto D P, Hooshmand F, Aggarwal A K, Rosenfeld M G. Cell. 1999;97:587–598. doi: 10.1016/s0092-8674(00)80770-9. [DOI] [PubMed] [Google Scholar]
- 8.Dasen J, Rosenfeld M G. Annu Rev Neurosci. 2001;24:327–355. doi: 10.1146/annurev.neuro.24.1.327. [DOI] [PubMed] [Google Scholar]
- 9.Tremblay Y, Tretjakoff I, Peterson A, Antakly T, Zhang C X, Drouin J. Proc Natl Acad Sci USA. 1988;85:8890–8894. doi: 10.1073/pnas.85.23.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammer G D, Fairchild-Huntress V, Low M J. Mol Endocrinol. 1990;4:1689–1697. doi: 10.1210/mend-4-11-1689. [DOI] [PubMed] [Google Scholar]
- 11.Young J I, Otero V, Cerdan M G, Falzone T L, Chan E C, Low M J, Rubinstein M. J Neurosci. 1998;18:6631–6640. doi: 10.1523/JNEUROSCI.18-17-06631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philips A, Maira M, Mullick A, Chamberland M, Lesage S, Hugo P, Drouin J. Mol Cell Biol. 1997;17:5952–5959. doi: 10.1128/mcb.17.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philips A, Lesage S, Gingras R, Maira M H, Gauthier Y, Hugo P, Drouin J. Mol Cell Biol. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maira M, Martens C, Philips A, Drouin J. Mol Cell Biol. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szeto D P, Ryan A K, O'Connell S M, Rosenfeld M G. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamonerie T, Tremblay J J, Lanctot C, Therrien M, Gauthier Y, Drouin J. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 17.Arzt E, Pereda M P, Castro C P, Pagotto U, Renner U, Stalla G K. Front Neuroendocrinol. 1999;20:71–95. doi: 10.1006/frne.1998.0176. [DOI] [PubMed] [Google Scholar]
- 18.Bousquet C, Melmed S. J Biol Chem. 1999;274:10723–10730. doi: 10.1074/jbc.274.16.10723. [DOI] [PubMed] [Google Scholar]
- 19.Guignat L, Bertherat J. Eur J Endocrinol. 1999;141:214–215. doi: 10.1530/eje.0.1410214. [DOI] [PubMed] [Google Scholar]
- 20.Bousquet C, Zatelli M C, Melmed S. J Clin Invest. 2000;106:1417–1425. doi: 10.1172/JCI11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chesnokova V, Auernhammer C J, Melmed S. Endocrinology. 1998;139:2201–2208. doi: 10.1210/endo.139.5.6017. [DOI] [PubMed] [Google Scholar]
- 22.Akita S, Readhead C, Stefaneanu L, Fine J, Tampanaru-Sarmesiu A, Kovacs K, Melmed S. J Clin Invest. 1997;99:2462–2469. doi: 10.1172/JCI119430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yano H, Readhead C, Nakashima M, Ren S G, Melmed S. Mol Endocrinol. 1998;12:1708–1720. doi: 10.1210/mend.12.11.0200. [DOI] [PubMed] [Google Scholar]
- 24.Tada M, Smith J C. Dev Growth Differ. 2001;43:1–11. doi: 10.1046/j.1440-169x.2001.00556.x. [DOI] [PubMed] [Google Scholar]
- 25.Papaioannou V E, Silver L M. BioEssays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 26.Smith J. Trends Genet. 1999;15:154–158. doi: 10.1016/s0168-9525(99)01693-5. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi J K, Koshiba-Takeuchi K, Matsumoto K, Vogel-Hopker A, Naitoh-Matsuo M, Ogura K, Takahashi N, Yasuda K, Ogura T. Nature (London) 1999;398:810–814. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua-Belmonte J C. Nature (London) 1999;398:814–818. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- 29.Bamshad M, Lin R C, Law D J, Watkins W C, Krakowiak P A, Moore M E, Franceschini P, Lala R, Holmes L B, Gebuhr T C, et al. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 30.Basson C T, Bachinsky D R, Lin R C, Levi T, Elkins J A, Soults J, Grayzel D, Kroumpouzou E, Traill T A, Leblanc-Straceski J, et al. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 31.Smith J C, Price B M, Green J B, Weigel D, Herrmann B G. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- 32.Warga R M, Nusslein-Volhard C. Dev Biol. 1998;203:116–121. doi: 10.1006/dbio.1998.9022. [DOI] [PubMed] [Google Scholar]
- 33.Muller C W, Herrmann B G. Nature (London) 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 34.Sinha S, Abraham S, Gronostajski R M, Campbell C E. Gene. 2000;258:15–29. doi: 10.1016/s0378-1119(00)00417-0. [DOI] [PubMed] [Google Scholar]
- 35.He M-L, Wen L, Campbell C E, Wu J Y, Rao Y. Proc Natl Acad Sci USA. 1999;96:10212–10217. doi: 10.1073/pnas.96.18.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carreira S, Dexter T J, Yavuzer U, Easty D J, Goding C R. Mol Cell Biol. 1998;18:5099–5108. doi: 10.1128/mcb.18.9.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szeto D P, Rodriguez-Esteban C, Ryan A K, O'Connell S M, Liu F, Kioussi C, Gleiberman A S, Izpisua-Belmonte J C, Rosenfeld M G. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanctot C, Moreau A, Chamberland M, Tremblay M L, Drouin J. Development (Cambridge, UK) 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 39.Lamolet B, Pulichino A M, Lamonerie T, Gauthier Y, Brue T, Enjalbert A, Drouin J. Cell. 2001;104:849–859. doi: 10.1016/s0092-8674(01)00282-3. [DOI] [PubMed] [Google Scholar]
- 40.Gleiberman A S, Fedtsova N G, Rosenfeld M G. Dev Biol. 1999;213:340–353. doi: 10.1006/dbio.1999.9386. [DOI] [PubMed] [Google Scholar]
- 41.Yi C H, Terrett J A, Li Q Y, Ellington K, Packham E A, Armstrong-Buisseret L, McClure P, Slingsby T, Brook J D. Genomics. 1999;55:10–20. doi: 10.1006/geno.1998.5632. [DOI] [PubMed] [Google Scholar]
- 42.Poulin G, Lebel M, Chamberland M, Paradis F W, Drouin J. Mol Cell Biol. 2000;20:4826–4837. doi: 10.1128/mcb.20.13.4826-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naya F J, Stellrech C M, Tsai M K. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 44.Naya F J, Huang H P, Qui Y, Mutoh H, DeMayo F J, Leiter A B, Tsai J J. Genes Dev. 1997;11:2322–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim W Y, Fritzch B, Serls A, Bakel L A, Huang E J, Reichardt L F, Barth D S, Lee J E. Development (Cambridge, UK) 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]