Abstract
The specification of tissue size during development involves the coordinated action of many signalling pathways responding to organ-intrinsic signals, such as morphogen gradients, and systemic cues, such as nutrient status. The conserved Hippo (Hpo) pathway, which promotes both cell cycle exit and apoptosis, is a major determinant of size control.The pathway core is a kinase cassette, comprising the kinases Hpo and Warts (Wts) and the scaffold proteins Salvador (Sav) and Mats, which inactivates the pro-growth transcriptional co-activator Yorkie (Yki). We performed a split TEV-based genome-wide RNAi screen for modulators of Hpo signalling. We characterise the Drosophila salt-inducible kinases (Sik2 and Sik3) as negative regulators of Hpo signalling. Activated Sik kinases increase Yki target expression and promote tissue overgrowth through phosphorylation of Sav at Ser413. Since Sik kinases have been implicated in nutrient sensing, this suggests a link between the Hippo pathway and systemic growth control.
Keywords: Hpo signalling, proliferation, Sik2, Sik3, split TEV, RNAi screening
Introduction
The determination of organ size is a central unresolved aspect of development. During the last decade, the Hippo (Hpo) pathway, which is highly conserved from Drosophila to humans, was shown to regulate final organ size by promoting both apoptosis and cell cycle exit 1,2. The Hpo pathway was also reported to be de-regulated in various cancer types.
The core of the Hpo pathway in Drosophila is a kinase cassette in which the Ste20-like kinase Hpo phosphorylates the downstream Dbf2-related kinase Wts3–8. Hpo and Wts kinase activity is facilitated by binding to the scaffold proteins Salvador (Sav) and Mats, resulting in the Wts-mediated phosphorylation of the pro-growth transcriptional co-activator Yki9–12. 14.3.3 proteins sequester phosphorylated Yki in the cytosol, thereby preventing the activation of Yki targets13,14. Nuclear Yki binds to a number of transcription factor partners such as the TEA domain (TEAD) containing protein Scalloped (Sd), and induces the expression of growth-promoting genes15–19. In humans, the Hpo orthologues MST1 and −2 phosphorylate the Wts orthologues LATS1 and −2 leading to the phosphorylation of the Yki orthologues YAP and TAZ14,20,21.
The core kinase complex is activated by the apically localised KEM complex, which comprises Kibra (Kib) and the two FERM domain proteins Expanded (Ex) and Merlin (Mer), probably acting via the Tao-1 kinase22–29. Interestingly, Ex also binds to Yki directly30,31, suggesting a kinase-independent regulation of Yki activity by membrane tethering, with AMOT proteins and α-catenin performing a similar function in mammals32–35. The atypical cadherin Fat also promotes Hpo pathway activity, both through Ex and by repressing the atypical myosin Dachs36–40.
The upstream signals that modulate Hpo pathway activity remain poorly understood. Through the KEM and AMOT complexes, which associate with the apical polarity protein Crumbs (Crb), and junctional components such as α-catenin, Hpo signalling is believed to respond to cell density41–48. Fat signalling has been proposed to mediate the effects of morphogens on Hpo signalling49. Finally, mechanical forces, sensed through the actin cytoskeleton, have also been proposed to modulate Yki/YAP activity, though the molecular mechanism remains unclear50–54. Thus, the Hpo pathway is believed to respond to a variety of signals such as local tissue architecture (cell crowding, tissue mechanics) as well as long-range patterning cues (morphogen gradients), but the details of upstream signalling remain to be elucidated.
Here, we combine the split TEV protein-protein interaction detection method recently described in mammalian cells55 with high-throughput RNAi screening to identify modulators of Hpo signalling. Split TEV is based on the functional complementation of two TEV protease fragments fused to potential interaction partners56. Using a Hpo pathway-specific readout for a genome-wide RNAi screen, we identified the Salt-inducible kinases (Sik2 and Sik3) as negative modulators of Hpo signalling. In vivo overexpression of active Sik2 induces tissue overgrowth and upregulation of the Yki transcriptional target ex. We show that the Hpo partner Sav is the target of Sik2 and Sik3 in the Hpo pathway. Sik2 and Sik3 associate with and phosphorylate Sav, reducing its activity.
Results
The split TEV technique as a tool to monitor protein-protein interactions in Drosophila
Cell-based RNAi screens based on transcriptional readouts of Yki activity have already proved a powerful means of identifying Hpo pathway members 53,57. To increase the sensitivity and specificity of Hpo pathway readouts, we sought to adapt the split TEV protein-protein interaction technique to Drosophila cell culture55.
We used Hpo dimerisation through the C-terminal SARAH (Salvador RASSF Hippo) domain to test the split TEV technique in Drosophila cells. We fused the N-terminal part (amino acids 1-118) and the C-terminal part (amino acids 119-221) of an improved TEV protease mutant (TEV-S219P) to Hpo. In S2R+ cells, dimerisation of Hpo led to the functional reconstitution of TEV protease activity, which in turn strongly activated a specific cytosolic TEV reporter (Supplementary Information, Fig.S1). A Hpo truncation lacking the SARAH domain, or the L619P mutation, which inactivates the SARAH domain60, proved the specificity of the split TEV technique. We obtained similar results for MST1, an orthologue of Hpo in HEK293 cells. Thus, the split TEV method can be used in Drosophila cell culture to monitor protein-protein interactions.
A split TEV assay for Yki/14.3.3 binding and its use in an RNAi screen
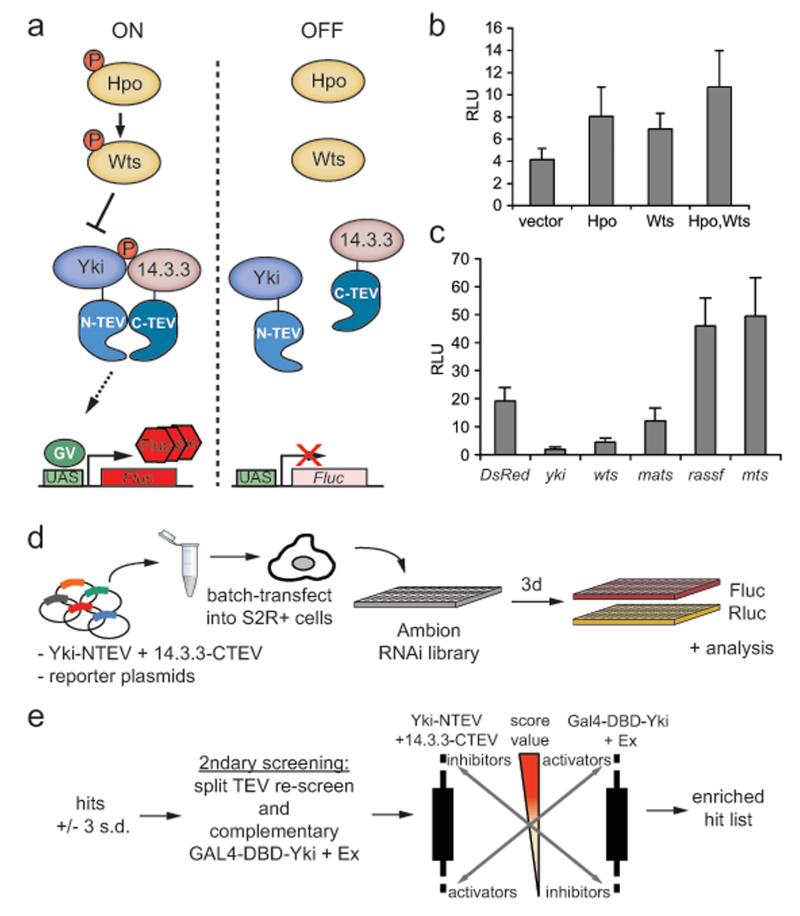
To design a split TEV assay for Hpo signalling activity, we chose the interaction between Yki and 14.3.3, which is dependent on phosphorylation of Yki at serine 168 by Wts 13,14. Using an interaction module consisting of Yki-NTEV and 14.3.3-CTEV (Fig.1a), we could detect the kinase cassette-induced Yki/14.3.3 association. Hpo and Wts expression, either alone or in combination, increased the readout (Fig.1b). Likewise, using RNAi to deplete known pathway inhibitors, such as Rassf and the PP2A catalytic subunit mts57, increased the signal (Fig.1c). Conversely, pathway inactivation by RNAi-mediated depletion of Wts and Mats decreased the luciferase signal (Fig.1c). These data suggest that the Yki/14.3.3 split TEV pair is a faithful reporter of Hpo pathway activity in cell culture.
Figure 1. A split TEV screen for modulators of the Yki/14.3.3 interaction.
(a) Schematic representation of the Yki-NTEV/14.3.3-CTEV split TEV interaction readout to monitor Hpo pathway activity.
(b, c) Overexpression (b) or RNAi-mediated depletion (c) of known Hpo pathway components results in the modulation of the split TEV-based Yki/14.3.3 readout. S2R+ cells were transfected with Yki-NTEV, 14.3.3-CTEV, GV-2ER, UAS-Fluc and a Renilla luciferase under the control of the piZ promoter (pIZ-RL). Cells were lysed 72h later and assayed. RLU denote average of relative luciferase units, all error bars represent s.d., n=6.
(d) Schematic overview of the genome-wide RNAi screen in Drosophila S2R+ cells using the Yki-NTEV/14.3.3-CTEV split TEV assay. Yki-NTEV and 14.3.3-CTEV plasmids along with the reporter plasmids GV-2ER (cytosolic TEV reporter), UAS-Fluc and pIZ-RL were transfected into S2R+ cells, and seeded onto a genome-wide dsRNA library in 384 well plates. After three days of incubation, cells were lysed and subjected to a Dual Luciferase Reporter Assay (Promega)
(e) Schematic overview of the secondary screening involving the Yki-NTEV/14.3.3-CTEV split TEV assay, the complementary GAL4-DBD-Yki/Ex assay and the cross-correlation of potential candidates to generate an enriched hit list. See main text for details. To further eliminate cell-intrinsic variations from the hit list, the pIZ-Rluc used in the genome-wide screen was replaced with an Actin5C promoter-driven Renilla luciferase (pAc-Rluc) in the re-screening process. Furthermore, candidates displaying Rluc values less than 25% of the mean reading per plate were removed as these tended to have distorted Fluc/Rluc ratios
Next, we performed a genome-wide RNAi screen in S2R+ cells using the Yki/14.3.3 split TEV readout and the Ambion Silencer Drosophila RNAi Library, which comprises 13,059 unique dsRNAs targeting 92% of the Drosophila genome (Fig.1d). From this screen, we selected a primary hit list of candidates that were more than 3 standard deviations away from the mean (Fig1e, Supplementary Information, Fig.S2a). These candidates were then subjected to re-screening using a set of non-overlapping dsRNA amplicons to eliminate off-target effects.
For secondary screening, we used a Yki transcriptional reporter based on a GAL4 DNA-binding domain-Yki fusion protein (GAL4-DBD-Yki)11. In order to improve the sensitivity and reproducibility of the assay, we co-transfected a plasmid encoding the upstream Hpo pathway member Ex 53(Supplementary Information, Fig.S2b). In this GAL4-DBD-Yki/Ex assay, known Hpo pathway members behaved as expected when depleted using RNAi (Supplementary Information, Fig.S2c).
Using the complementary GAL4-DBD-Yki/Ex assay for re-screening we assembled a refined hit list by efficiently removing false-positives (Fig.1e). In this cross-correlation analysis, potential pathway activators displayed a high score in the GAL4-DBD-Yki/Ex assay and a low score in the Yki-NTEV/14.3.3-CTEV assay, and vice versa for inhibitors. The candidate list was then refined using an anti-phospho-Ser168-Yki (pS168-Yki) antibody, which detects Wts-dependent Yki phosphorylation14,28. S2R+ cells were individually depleted for candidate genes and western blots of the lysates were probed for pS168-Yki levels (summarised in Supplementary Information, Supplementary Table 1).
Identification of Salt-Inducible Kinase as a Hpo Pathway Regulator
From this screening approach we recovered the gene CG42856, a serine/threonine kinase, the Drosophila orthologue of human Salt-Inducible Kinase 3, Sik3 (Supplementary Information, Fig.S2a). Sik3 and the closely related Sik2 (CG4290) have been shown to regulate gluconeogenesis both in flies and humans58–60. Humans have three salt-inducible kinases, SIK1-3. In flies, Sik2 and Sik3 represent the only two members of the salt-inducible kinase family, Sik2 being the orthologue of human SIK1 and SIK261,62.
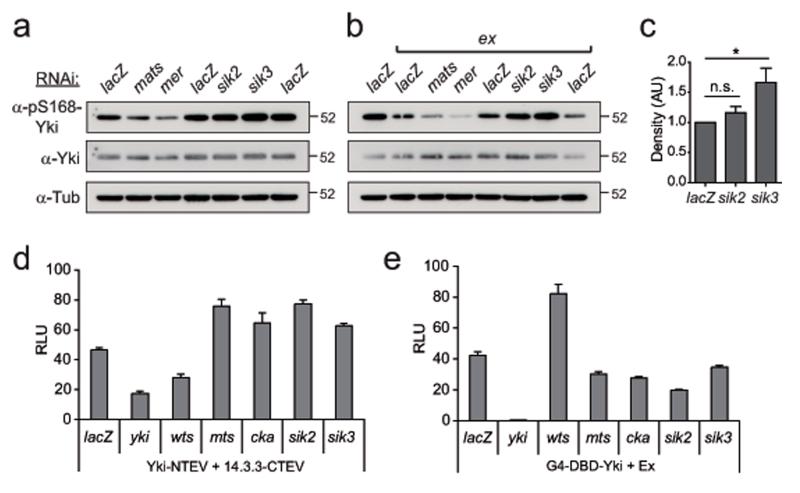
As Siks were identified as potential negative regulators of the Hpo pathway in our screen, we wanted to confirm the effect of Sik2 and Sik3 on Hpo signalling using the pS168-Yki antibody. S2R+ cells depleted for Sik3 show a moderate increase in Yki phosphorylation levels (Fig.2a). Moreover, upon depletion of Ex, where baseline pS168-Yki levels are reduced, we observed a marked increase in Yki phosphorylation levels for Sik3-depleted cells and a modest increase for Sik2-depleted cells, supporting the notion that Siks negatively regulate Yki activity (Fig.2b,c). Upon re-testing using the Yki/14.3.3 split TEV and GAL4-DBD-Yki/Ex assays, we confirmed the inhibitory effect for both Sik2 and Sik3 (Fig.2d,e).
Figure 2. Salt-inducible kinases inhibit the Hpo pathway.
(a, b) Sik2 and Sik3 inhibit Yki phosphorylation. S2R+ cells were treated for 3d with individual dsRNA as indicated (a) or in combination with dsRNA targeting ex (b). Endogenous pS168-Yki and pan-Yki protein levels were determined by western blot analysis, and Tubulin levels served as a loading control
(c) Quantification of 3 independent experiments shown in (b) for lacZ/ex, Sik2/ex, and Sik3/ex-depleted S2R+ cells. Western blot data was processed using ImageJ. Asterisk indicates p<0.05; n.s., not significant. Error bars represent s.e.m., n=3.
(d) Sik2- and Sik3-depleted-cells show reduced Hpo pathway activity in the Yki-NTEV/14.3.3-CTEV split TEV assay. Yki-NTEV, 14.3.3-CTEV, GV-2ER, UAS-Fluc and pAFW-hRL plasmids were transfected into S2R+ cells, expressed for 72h, and assayed. RLU denote average of relative luciferase units, all error bars represent s.d., n=6
(e) Sik2 and Sik3 depleted-cells show an enhanced readout in the G4-Yki/Ex assay. GAL4-DBD-Yki, Ex-HA, UAS-Fluc and pAFW-hRL plasmids were transfected into S2R+ cells. Same experimental setup as in (c)
Sik2 regulates wing tissue size by restricting Hpo signalling
To examine the effect of Sik activity on the Hpo pathway in vivo, we generated transgenic fly lines expressing Sik2 and Sik3-PA, both wild type and activated forms mutant for a regulatory PKA site (Sik2-S1032A and Sik3-PA-S563A61), under the control of the GAL4/UAS system. Ex is both an upstream Hpo pathway regulator and a transcriptional target of Yki, acting in a negative feedback loop63. Thus, we can use an enhancer trap fly line bearing a lacZ reporter inserted in the ex locus (ex-lacZ) as a readout of Yki transcriptional activity.
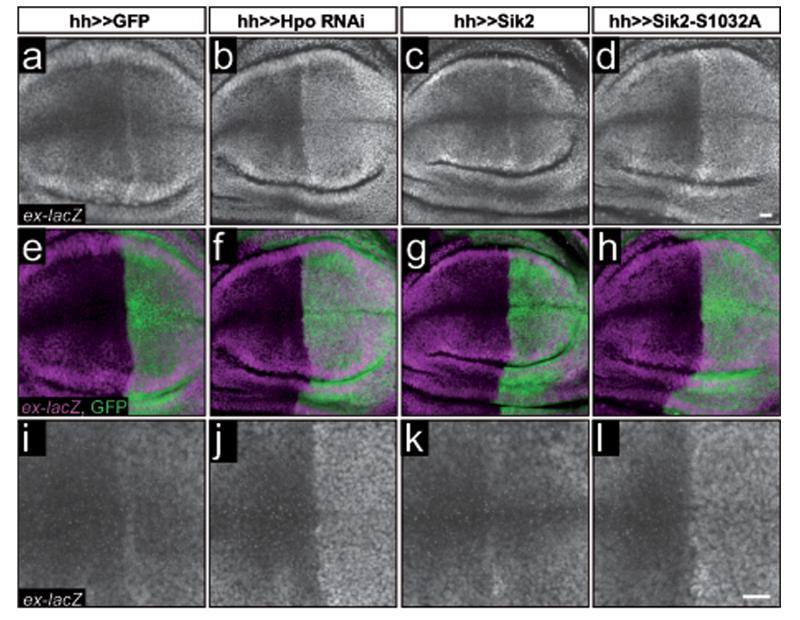
Hpo depletion using RNAi under the hedgehog-GAL4 driver line (hh-GAL4) leads to ex-lacZ upregulation in the wing disc posterior half (Fig.3a,e,i,b,f,j). Interestingly, overexpression of Sik2-S1032A, but not wild type Sik2, induced a robust increase in ex-lacZ expression (Fig.3c,g,k,d,h,l). We observed the same effect on ex-lacZ expression upon either Hpo depletion or Sik2 overexpression in flip-out clones (Supplementary Information, Fig.S3a-j). Apically localised Ex protein levels were also increased in both Hpo-depleted (Fig.4a,d,g,j) and Sik2-S1032A-overexpressing clones (Fig.4c,f,i,l) but not in Sik2 wt clones (Fig.4b,e,h,k). In contrast, staining for the septate junction component Discs-large was unaffected, indicating that cellular architecture was not altered (Fig.4m-r). Expression of Sik3-PA led to severe defects and cell delamination both under the hh-GAL4 driver and in clones. However, when cells were rescued by expression of the caspase inhibitor p35, we obtained small flip-out clones of Sik3-PA-S563A-expressing cells, which showed increased Ex protein staining (Supplementary Information, Fig.S3k,l). Thus, the Sik kinases can promote Yki activity in vivo, in accordance with our cell-based data.
Figure 3. Sik2 promotes Yki target gene expression.
(a-l) Overexpression of Sik2-S1032A leads to an increase in Yki/SD-driven ex transcription. Hpo pathway activity was monitored in imaginal wing discs from 3rd instar larvae using the ex697-lacZ (ex-lacZ) enhancer trap fly line. GFP (a,e,i), Hpo RNAi (b,f,j), Sik2 wild type (c,g,k), and Sik2-S1032A (d,h,l) were expressed under the control of hh-GAL4. Planar sections depict anti-βGal nuclear staining in grey; e-h are merge images of βGal staining (magenta) and GFP (green); i-l are magnifications from a-d. Scale bars, 10 μm
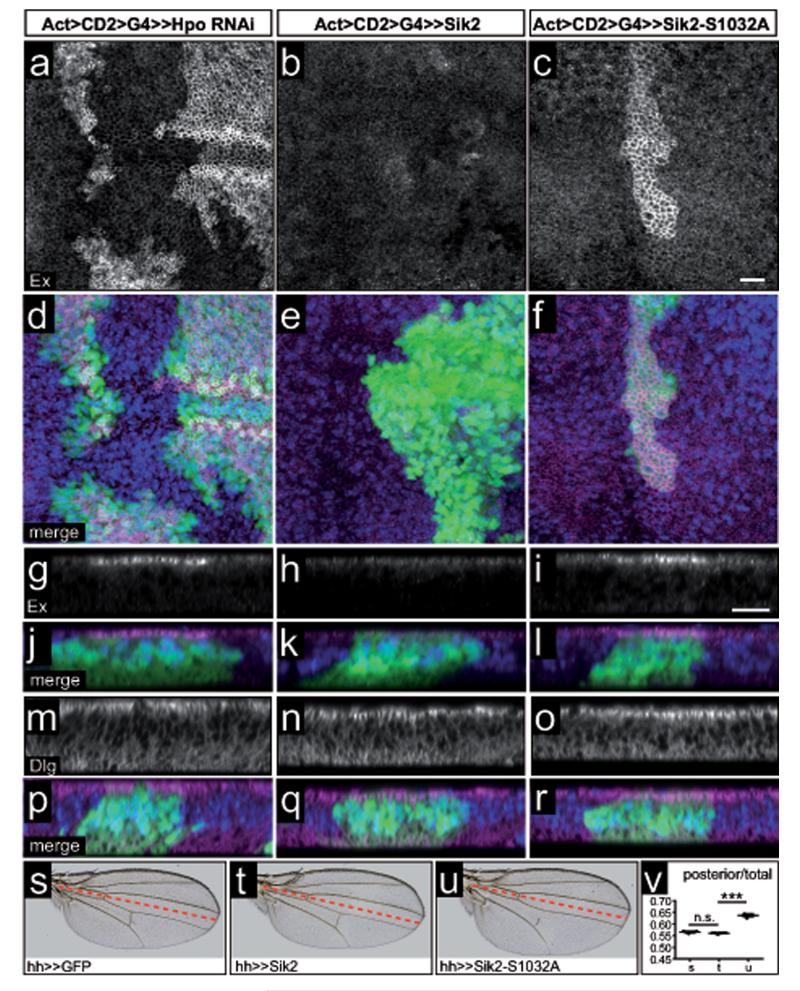
Figure 4. Activated Sik2 induces wing growth through the Hpo pathway.
(a-l) Expression of Hpo RNAi (a, d,g,j) and Sik2-S1032A (c,f,i,l), but not Sik2 (b,e,h,k) in flip-out clones leads to the upregulation of Ex protein levels at the apical membrane of imaginal wing discs.
(m-r) Dlg protein levels at the apical membrane are not affected by Hpo RNAi (m, p), Sik2 (n,q) and Sik2-S1032A (o,r) expression.
Planar (a-f) and transverse (g-r) sections showing Ex (a-f) or Dlg (g-i) protein levels (grey/magenta) in hsFlp, Act>>Hpo RNAi (a,d,g,j), hsFlp, Act>>Sik2 (b,e,h,k), and hsFlp, Act>>Sik2-S1032A (c,f,I,l) 3rd instar wing discs. j,k,l,p,q,r are merge images with Ex/Dlg in magenta, GFP in green, and DAPI in blue. Scale bars, 10 μm
(s-v) Overexpression of GFP (s) and Sik2 wild type (t) do not affect wing size, but activated Sik2-S1032A (u) increases growth. Sik2 transgenes were driven by hh-GAL4. The red line denotes the border between anterior (above) and posterior compartment (below) of the wing
(v) Quantification of wing phenotypes shown from (s) to (u). The values represent the ratio of the posterior compartment divided by the total wing area. 3 asterisks indicate p<0.001; n.s., not significant. Error bars represent s.d., n=6.
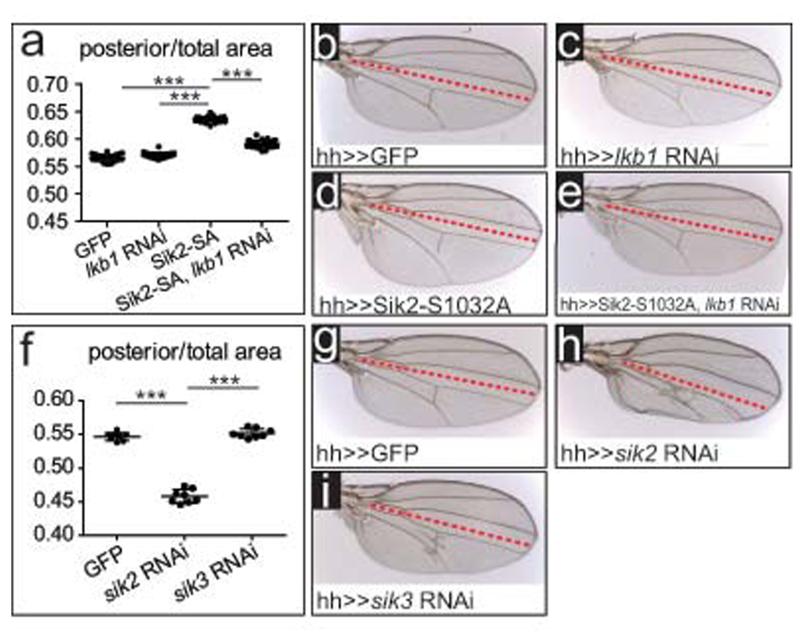
To examine the effects of Sik2 and Sik3 on growth, we measured the wings of adult flies expressing our transgenes. Interestingly, overexpression of activated Sik2-S1032A, but not wt Sik2, leads to an increase in posterior compartment size (Fig.4s-v), consistent with the Ex stainings. Overexpression of Sik3-PA and activated Sik3-PA-S563A caused collapse of the posterior part of the wing (Supplementary Information, Fig.S4i,l). Like other AMPK family members, Sik kinases can be activated by the upstream master kinase Lkb164. Depletion of Lkb1 using RNAi had no effect on wing growth (Fig.5a-c), however co-expression of lkb1 RNAi reverted the overgrowth caused by activated Sik2 (Fig.5a,d,e). Thus, activated Sik2 can drive wing growth in an Lkb1-dependent manner.
Figure 5. Depletion of the Sik kinases reduces wing growth.
(a-e) Expression of lkb1 RNAi rescues Sik2-S1032A-induced overgrowth (e) in comparison with expression of Sik2-S1032A alone (d). Expression of lkb1 RNAi alone does not decrease wing growth (c). (a) Quantification of the wing phenotypes shown in (b) to (e).
(f-i) Expression of sik2 RNAi (h) reduces wing growth, but sik3 RNAi does not affect wing size (i). (f) Quantification of the wing phenotypes shown from (g) to (i).
Transgenes were driven using hh-GAL4. The values in (a) and (f) represent the ratio of the posterior compartment divided by the total wing area. 3 asterisks indicate p<0.001. Error bars represent s.d. In (a), n=25 (GFP and lkb1 RNAi), n=29 (Sik2-S1032A), n=35 (Sik2-S1032A, lkb1 RNAi). In (f), n=8 for each genotype
Depletion of Sik2 and Sik3 reduces wing size
To overcome the potential redundancy between Sik2 and Sik3 in loss-of-function studies, we took advantage of the VDRC sik2 RNAi line KK103739, which is predicted to target both Sik2 and Sik3. We verified this by co-expressing this RNAi line, or a sik3 RNAi line, with either activated Sik2 (Supplementary Information, Fig.S4a-e) or wt Sik3 (Supplementary Information, Fig.S4f-k). While the sik3 RNAi line could revert the Sik3 but not the Sik2 overexpression phenotype, the sik2 RNAi line could suppress both the Sik2 and Sik3 phenotypes, confirming it can be used to silence both kinases. Depleting Sik2 and Sik3 with this RNAi line in the posterior part of the wing significantly reduced posterior compartment size, while Sik3 depletion alone had no effect on size (Fig.5f-i). The Sik kinases therefore promote growth in the wing, consistent with their antagonistic effect on Hpo pathway activity.
Sik2 and Sik3 bind to Sav
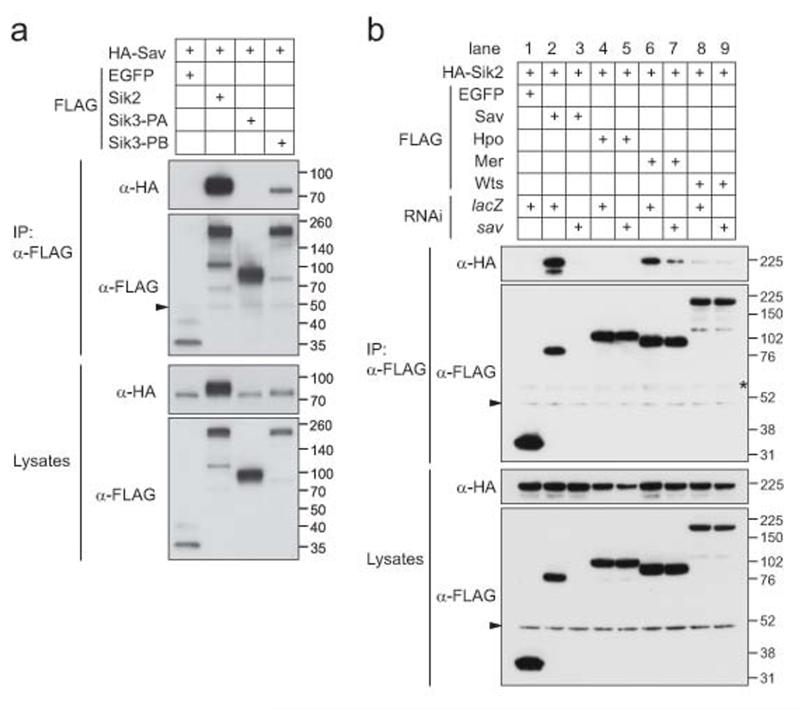
To clarify the molecular mechanism of the Siks’ input into Hpo signalling, we tested if Siks associate with Hpo pathway components, using co-immunoprecipitation (co-IP) experiments in S2 cells between Sik2 and core Hpo pathway members. We tested a number of pathway components (such as Hpo, Sav, Wts, and Mer, Fig.6a,b). Sik2 associates robustly with Sav, but not with the other tested proteins (Fig.6b). Interestingly, we also observed a modest co-IP between Sik2 and Mer, which was largely abolished upon Sav depletion (Fig.6b, lanes 6 and 7). Sav contains an N-terminal FERM-binding motif (FBM, Fig.7a), which binds to the FERM-domain of Mer29, supporting the notion that the Sik2/Mer interaction is bridged by Sav.
Figure 6. Sik kinases bind to and phosphorylate the Hpo pathway core member Sav.
(a) Sik2 and Sik3-PB, but not Sik3-PA, interact with Sav.
(b) Sik2 interacts with Mer in a Sav-dependent manner
Co-IP experiments were performed in S2 cells transfected with the indicated plasmids, and cells were lysed 48h later. Lysates and FLAG-purified immunoprecipitates were subjected to western blot analysis using the indicated antibodies. For samples treated with RNAi, cells were pre-incubated with indicated dsRNA before transfection. The asterisk denotes an unspecific band, and the arrowheads indicate the IgG heavy chain
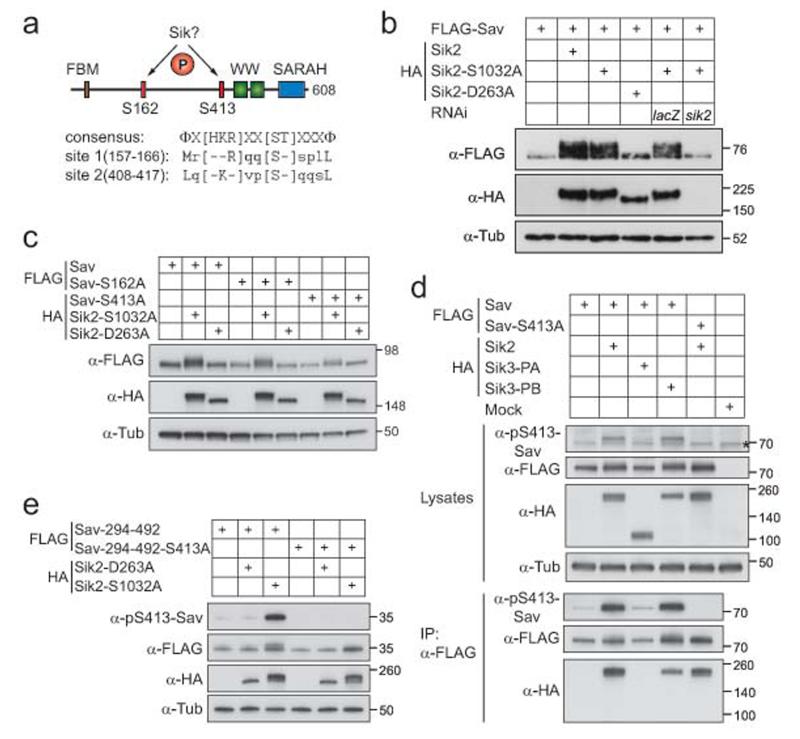
Figure 7. Sik kinases phosphorylate the Hpo pathway core member Sav.
(a) Structure of Sav indicating potential Sik2 phosphorylation sites. Below, the Sik consensus phosphorylation sequence59 and the corresponding sites in Sav are shown. FBM, FERM-binding motif; W, WW domain; SARAH, SARAH domain; Φ, hydrophobic residue
(b) Sav phosphorylation is dependent on Sik2 and its kinase activit
(c) Sik2 phosphorylates Sav on Ser-413 but not Ser-162.
S2 cells (b, c) were transfected with indicated plasmids, and cells were lysed 48h later and subjected to western blot analysis using a 7.5% SDS-PAGE gel and the antibodies as indicated
(d) Sav is phosphorylated at S413 by Sik2 and Sik3. Indicated plasmids were transfected into S2R+ cells, and lysates (upper panel) and FLAG-purified immunoprecipitates (lower panel) were subjected to western blot analysis using the indicated antibodies. The asterisk indicates an unspecific band
(e) The Sav fragment 294-492 does not shift when harbouring the S413A mutation. S2R+ cells were transfected with the indicated plasmids, and cells were lysed 48h later and subjected to western blot analysis using a 4-15% gradient SDS-PAGE gel and the indicated antibodies
Sik2-S1032A, active Sik2; Sik2-D263A, kinase-dead form. When samples were treated with RNAi, cells were pre-incubated with the indicated dsRNA
For Sik3, there are multiple splice isoforms to consider. Sik3-PA, a short isoform, was initially described65, but two longer isoforms (PB and PC) with extended C-termini were recently annotated in Flybase. By performing RT-PCR on RNA extracted from larval preparations we isolated cDNAs coding for a total of six isoforms (PA to PF, see Supplementary Information, Fig.S5). Interestingly, we compared the short PA isoform with one of the longer isoforms (PB), and found that only the latter could bind Sav, suggesting that the extended C-terminus is required for Sav binding (Fig.6a).
Next, we mapped the domains in Sik2 and Sav that mediate their interaction. The Sik2-binding domain in Sav is located within the C-terminal part, covering the two WW domains and the region located in between the WW domains and the SARAH domain (Supplementary Information, Fig.S6a,b). The Sav-binding region in Sik2 is located near the C-terminus, covering a region from aa 1054 to 1250 (Supplementary Information, Fig.S6c,d).
Sik2 and Sik3 phosphorylate Sav
Sav contains two potential Sik phosphorylation sites (Fig.7a), prompting us to test whether Sik can phosphorylate Sav. Overexpression of either Sik2 or Sik2-S1032A leads to a Sav mobility shift, while a kinase-dead form had no effect, and depletion of Sik2-S1032A by RNAi reverted this band shift (Fig.7b). Both Ser162 and Ser413 of Sav fit the Sik phosphorylation consensus site59. A Sav-S413A mutant showed a decreased mobility shift upon Sik2 expression, whereas Sav-S162A was unaffected (Fig.7c). The Sav-S162/413A double mutant did not further decrease the shift (Supplementary Information, Fig.S7a). These results indicate that S413 is phosphorylated by Sik2.
To validate this finding, we raised a phospho-specific antibody against Sav-S413. This antibody detected a robust signal when wt Sav but not Sav-S413A was co-expressed with Sik2-S1032A (Fig.7d). In addition, lambda phosphatase treatment abolished the phospho-signal and upshift, confirming the antibody’s specificity (Supplementary Information, Fig.S7b). To characterise the S413 site in more detail, we used the truncations we generated to map the Sav/Sik2 interaction (Supplementary Information, Fig.S6a,b). Sav fragment 294-492, which associates with Sik2, was shifted and phosphorylated at S413 upon Sik2 kinase expression, while the S413A mutation reverted the shift (Fig.7e). However, the band shift reversal in the in full-length Sav S413 mutant was incomplete, so there are likely other Sik sites in Sav. The Sav N-terminus (aa 1-300) did not shift upon Sik2 co-expression, presumably because it does not bind to the kinase (Supplementary Information, Fig.S6b, second lane), but may still harbour phosphorylation sites. We tested eight other potential Sik sites in Sav, but no mutant other than the S413A showed a decreased mobility shift (Supplementary Information, Fig.S7c,d), supporting the notion that S413 is the critical Sik site in Sav. Similar to Sik2, Sik3-PB strongly phosphorylated Sav, while Sik3-PA had little effect (Fig.7d). Together, our data suggest that Sik2 and Sik3 associate with and phosphorylate Sav at S413.
Sik2 antagonises Hpo pathway activity by phosphorylating Sav
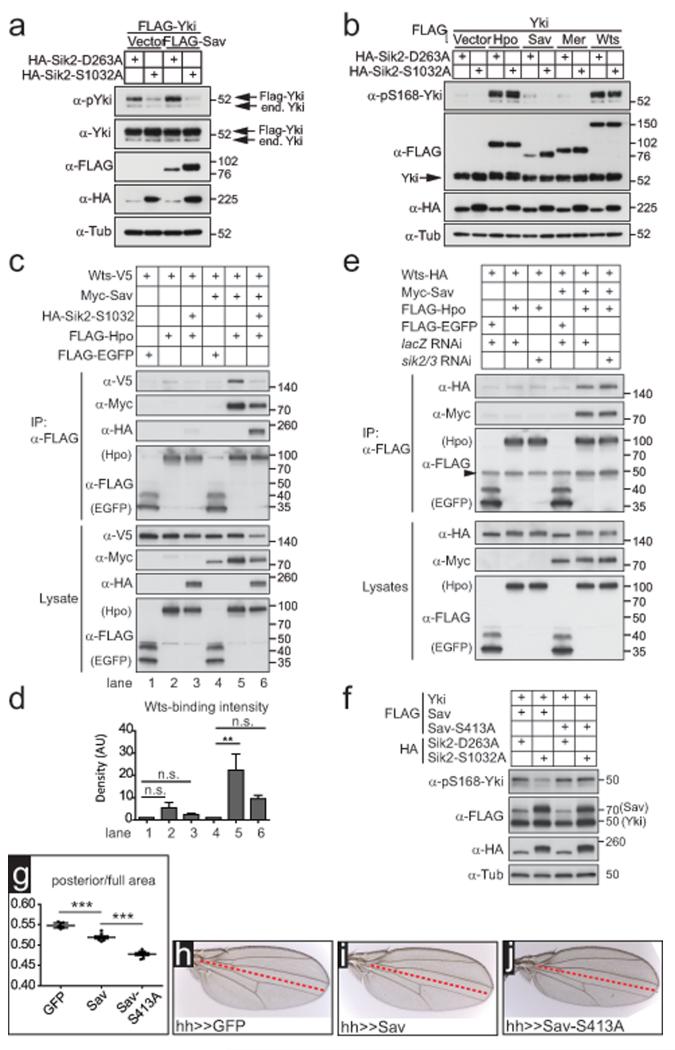
We wished to determine if Sav phosphorylation by Sik accounts for its ability to promote Yki activity. Co-expression of Sav with Yki in S2 cells induced a modest increase in Yki phosphorylation on S168 (Fig.8a). Co-expression of activated Sik2 abolished baseline, Sav-induced and Mer-induced Yki phosphorylation, while kinase-dead Sik2 had no effect (Fig.8a,b). In contrast, activated Sik2 had no effect on Yki phosphorylation induced by Hpo or Wts (Fig.8b), suggesting that Sik2 acts at the level of Sav, downstream of Mer, and upstream of the kinases Hpo and Wts.
Figure 8. Sik2 inhibits Hpo signalling by phosphorylating Sav.
(a) Activated Sik2 inhibits Hpo signalling by decreasing phospho-Yki levels
Overexpression of activated Sik2 decreases baseline and Sav-induced pS168-Yki levels. Indicated plasmids were transfected into S2 cells, lysed after 48h and subjected to western blot analysis.
(b) Overexpression of activated Sik2 decreases pS168-Yki levels when Sav or Mer are co-expressed, but not when Hpo or Wts are present. Indicated plasmids were transfected into S2 cells, lysed after 48h and subjected to western blot analysis.
(c) Sik2 causes a decreased association of Hpo and Wts in a Sav-dependent manner. Co-IP experiments were performed in S2 cells using the indicated plasmids, and cells were lysed 48h later. FLAG-purified (Hpo, EGFP) immunoprecipitates and lysates were subjected to western blot analysis.
(d) Quantification of 3 independent experiments shown in (c). Band intensities were densitometrically quantified using ImageJ. Wts levels in the IPs were normalised to Wts levels in the lysates to control for different expression levels. Asterisk indicates p<0.05; n.s., not significant. Error bars represent s.e.m., n=3
(e) Depletion of Sik kinases increases Sav-mediated Hpo/Wts association
Co-IP experiments were performed in S2R+ cells treated with indicated RNAi and 6h later transfected with indicated plasmids. Cells were lysed 48h post transfection. Lysates and FLAG-purified immunoprecipitates were subjected to western blot analysis. The arrowhead indicates the IgG heavy chain (at 50kD)
(f) The Sav-S413A mutant prevents the Sik2-induced decrease in pS168-Yki levels. Indicated plasmids were transfected into S2 cells, lysed after 48h and subjected to western blot analysis.
(g)The Sav mutant S413A restricts growth in the fly wing. Quantification of wing phenotypes shown in (h) to (j). Scatter plot representing the ratio of the posterior compartment divided by the total wing area. 3 asterisks indicate p<0.001. Error bars represent s.d., n=8 for each genotype.
(h-j) Overexpression of Sav leads to a decreased posterior compartment size in the adult fly wing (i) compared to GFP (h). This phenotype is enhanced in the Sav-S413A mutant (j). Sav transgenes were driven by hh-GAL4. The red line denotes the border the between anterior (above) and posterior compartments (below) of the wing.
Sav is a scaffold protein known to associate with Hpo through its SARAH domain3–6,8, Mer through an FBM29, and Wts via its WW domains10. To gain further insight into how Sik2 regulates the Hpo pathway core kinase cassette, we characterised the effect of Sik2 upon binding of Sav to its partners. Firstly, Hpo association with Sav was not perturbed by Sik2 expression (Supplementary Information, Fig.S7e), nor did Hpo or kinase-dead Hpo expression prevent the Sav/Sik2 interaction (Supplementary Information, Fig.S7f). The Sav-S413A mutation did not affect Hpo/Sav or Sik2/Sav binding (Supplementary Information, Fig.S8a,b). Secondly, the Sav/Mer association was unchanged upon Sik2 co-expression (Supplementary Information, Fig.S8c). Finally, we tested the ability of Sav to scaffold the Hpo/Wts kinase cassette. In the absence of Sav, the interaction between co-expressed Hpo and Wts was barely detectable (Fig.8c, top panel, lane 2). Upon Sav overexpression, this interaction was markedly increased (Fig8c, lanes 2 and 5). Co-expression of activated Sik2 significantly reduced the Sav-mediated Hpo/Wts association (Fig.8c and quantified in Fig.8d). Furthermore, depletion of Sik2 and Sik3 resulted in a modest increase in baseline Hpo/Wts binding (Fig.8e). Our results suggest that Sik2 inhibits Sav by reducing its ability to bridge the Hpo/Wts association.
Ser 413 in Sav is critical for Hpo signalling
If Sav phosphorylation by Sik2 affects the assembly of the core complex, this would be expected to affect Yki phosphorylation, which is dependent on Hpo’s ability to activate Wts. Indeed, the reduction in pS168-Yki levels observed upon activated Sik2 expression was prevented by co-expression of the non-phosphorylatable Sav-S413A mutant (Fig.8f). Finally, we tested the biological activity of the Sav-S413A mutant in vivo by making transgenic lines expressing wild type Sav or Sav-S413A under the control of the GAL4/UAS system. hh-driven expression of wild type Sav modestly reduced the size of the posterior compartment of the wing, while the Sav-413A mutant had a significantly stronger effect (Fig.8g-j). In fact, while expression of wild type Sav had no effect on viability, the Sav-S413A mutant strongly reduced the number of progeny. Thus, phosphorylation of Sav on serine 413 reduces its biological activity in Hpo signalling and growth control.
To investigate whether the Sik-mediated phosphorylation is conserved in humans, we co-transfected Drosophila Sik2 with human SAV1. We detected a moderate upshift of SAV1, though the phosphorylation site is not conserved, as the pS413-Sav antibody failed to detect SAV1 phosphorylation (Supplementary Information, Fig.S8d). However, SIK2 can increase YAP-mediated transcription in a GAL4-DBD-TEAD4 luciferase readout, which has been widely used to assess Hpo pathway activity in mammalian cells20 (Supplementary Information, Fig.S8e). These data suggest that human SIK kinases functionally inhibit Hpo signalling, but it is unclear whether the exact mechanism is conserved.
Discussion
In this study we combine the protein-protein interaction detection method split TEV and RNAi screening in Drosophila cell culture to identify Hpo pathway modulators. Split TEV was first developed in mammalian cells55, and subsequently shown to be a valuable tool to monitor phosphorylation-dependent interactions of proteins56. Here, we applied split TEV for the first time in Drosophila cell culture, both for constitutive (Hpo dimerisation) and regulated (Yki/14.3.3) interactions. The success of our screening approach suggests that split TEV will prove invaluable in mapping signalling pathways by providing functional readouts that can be combined with RNAi or pharmacological approaches.
Our results identify Sik kinases as Hpo upstream regulators. In particular, activated Sik2 can induce overgrowth and activation of Yki transcriptional activity (Figs. 3, 4), while depletion of Sik2/3 in the wing leads to undergrowth (Fig.5). Interestingly, Sik3 can also antagonise Hpo signalling, but in an isoform-specific manner. The relative contribution of Sik2 and Sik3 to growth in various tissues needs to be addressed to appreciate the possible redundancy between these two kinases, and potentially other AMPK family kinases. A recent report shows that LKB1, which regulates all AMPK family members66, may inhibit YAP activity independently of the core Hpo cassette, suggesting a complex interplay between LKB1/AMPKs and the Hpo pathway67.
The effect of Sik2/3 on Hpo signalling is mediated, at least in part, by Sav phosphorylation on S413 (Figs. 7,8). Our data suggest that Sav phosphorylation by Sik reduces its ability to efficiently scaffold the Hpo/Wts core kinase complex, thereby reducing Yki inhibitory phosphorylation (Fig.8c-e). In agreement with this notion, Sav-S413A displayed an enhanced ability to reduce growth in vivo (Fig.8f-j).Siks play a major role in inhibiting gluconeogenesis in the liver in response to high glucose levels through inhibitory phosphorylation of the transcriptional co-activator CRTC2 (CREB-regulated transcription coactivator 2)/TORC2, and activatory phosphorylation of the Histone Deacetylase HDAC4, a function which appears to beconserved in Drosophila60,62,65,68.
The Drosophila Siks have mostly been studied in the context of energy balance in the brain69 and fat body (FB – the Drosophila equivalent of the liver and adipose tissue), where they prevent the mobilisation of fat and glycogen stores by antagonising CRTC and Foxo-activated transcription62,65,69. The Siks are under hormonal control by Insulin receptor (InR) signalling via the downstream kinase Akt, which phosphorylates and activates Sik2/3, and the glucagon homologue Adipokinetic Hormone (AKH), which signals through a G-protein coupled receptor (GPCR) and PKA, which phosphorylates and inhibits Siks65,69. Under fasting conditions, low Insulin and high AKH activity combine to shut down Sik3, thereby promoting gluconeogenesis and inducing mobilisation of FB lipid stores to restore circulating glucose levels65. Our work suggests a role for Sik2/3 in growth control during development. Analogously to InR signalling in Drosophila, which promotes both nutrient storage in the FB and developmental growth of peripheral tissues, the Sik kinases might couple Hpo pathway activity to nutrient or energy availability, ensuring that Yki is only able to drive tissue growth under favourable conditions. Recent work has shown that Drosophila InR signalling can promote cell proliferation through Yki70. Furthermore, mammalian liver cells stimulated with glucagon or the PKA activator Forskolin display reduced levels of phosphorylated YAP and increased YAP activity, an effect that may conceivably be mediated by the SIKs71.
Recent work has established the SIK kinases as candidate oncogenes in ovarian and lung cancer72–74. We showed that SIK2 promotes YAP-dependent transcription in human cells (Supplementary Information, Fig.S8e). However, since Sav-S413 is not conserved, the molecular mechanism by which SIK2 regulates the mammalian Hpo pathway may differ and should be investigated further. Interestingly, YAP, the Yki orthologue, has also been reported to function as an ovarian cancer oncogene75,76. SIK2 inhibitors may therefore prove attractive candidates to boost Hpo pathway activity in ovarian tumour cells, though this strategy may be less effective in tumours harbouring YAP amplifications77.
Methods
Cloning of expression constructs
For split TEV fusions, the pAWF and pAWH vectors from the Drosophila Gateway Vector Collection were modified by introducing an NTEV or a CTEV fragment to yield pAWF-NTEV or pAWH-CTEV vectors. The NTEV fragment (amino acids 1-118 of the TEV protease), followed by a flexible linker (GGGGSGGGGS), was amplified by PCR and introduced into the pAWF vector by using the SacI restriction enzyme. The CTEV fragment (amino acids 119-221, with a S219P point mutation for a more stable TEV protease variant 78), also followed by the flexible linker, was PCR-amplified and ligated into the pAWH vector using the enzymes SacI and SacII. The ORF for 14.3.3 epsilon (CG31196-PA) was amplified from Drosophila embryonic cDNA. The ORFs for Sik2 (CG4290, clone RH42017) and Sik3-PA (CG42856-PA, clone LD07105) were from the DGRC clone collection. Sik3-PB was cloned from cDNA generated from larval fly preparations. For a detailed procedure, see Supplementary Information, Fig.S5. All ORFs were amplified by PCR using primers bearing flanking attB1 and attB2 sequences and subjected to BP reactions using the pDONR/Zeo vector (Invitrogen) to generate Entry vectors. Expression vectors were obtained via LR reactions using the pAFW, pAWF, pAHW and pAWH vectors from the DGRC Gateway collection. Vectors for Hpo, Wts, Mats, Sav, Rassf, Kibra, Ex and Mer were previously described28,57. Point mutations and, if required, ORF corrections (i.e. for RH42017, Sik2) were generated by using site-directed mutagenesis (SDM, Agilent Technologies). The following primers were used for SDM: Sik2-S1032A (constitutively active), 5′-CGCGAAGGAAGACGGGCGGCTGATGGCCTGGTGGCC-3′; Sik2-D263A (kinase-dead), 5′-GGCATAGTGCATCGGGCTCTCAAGGCGGAGAACC-3′; Sik3-S563A (constitutively active), 5′-CATTTGGGCGTCGTGCCGCAGACGGCGGGGCAAATC-3′; Sik3-D163A (kinase-dead), 5′-GGTGGTGCATCGCGCTCTCAAGGCCGAGAATGTCC-3′; Sav-S162A, 5′-GGATGCGCCGTCAGCAGGCGTCGCCACTGCTGC-3′; Sav-S413A, 5′-GCTGCAGAAGGTACCAGCACAGCAATCGCTCACG-3′
For transgenic flies, the pPW vector from the DRGC Gateway collection and a Gateway-compatible pUAST-attB vector 79 allowing site-specific integration into the fly genome were used.
Human MST1 was PCR-amplified from image clone 7939613 (Imagenes) and cloned into pENTR207 (Invitrogen). For split TEV fusions in mammalian cells, the previously described N/CTEV-pDEST vectors were used55.
All PCRs were run using proofreading polymerases (Pwo Master, Roche; Pfu Turbo, Stratagene), and all PCR-amplified ORFs were sequence-verified.
Split TEV and GAL4-DBD-Yki luciferase assays
For individual split TEV assays, plasmids encoding N- and CTEV protein fusions along with the cytosolic transcriptional split TEV reporter GV-2ER and pUAS-Fluc were used. For GAL4-DBD-Yki assays, vectors encoding GAL4-DBD-Yki, Ex and the reporter UAS-Fluc were used. For Drosophila cell culture, luciferase assays were run in 96-well formats for 72h using S2R+ cells transfected using FugeneHD with a Renilla luciferase as internal control (pActin5C-hRL). Equal amounts of DNA were transfected into each well (20 ng for each overexpressing plasmid, 30 ng for pUAS-Fluc, and 3 ng for pActin5C-hRL). pAFW was used to adjust DNA amounts.
For mammalian cell culture, luciferase assays were performed in 96-well formats using HEK293 cells transfected with Lipofectamine LTX and a mix of Renilla luciferase plasmids (CMV-RL, TK-RL, SV40-RL at 1:2:10 molar proportions) as internal control. Equal amounts of DNA were transfected into each well (20 ng for all plasmids, including pUAS-Fluc, and 1 ng for Renilla mix. pcDNA3 was used to adjust DNA amounts). After 24h of incubation, cells were lysed using 1×PassiveLysis buffer (Promega), followed by the Dual Luciferase® Reporter Assay (Promega). The readings for both channels (firefly, Renilla) were recorded using an EnVision® Multilabel Reader (PerkinElmer) and a PHERAstar Plus reader (BMG Labtech).
Cell culture
S2 cells were maintained in Schneider’s medium (Sigma) supplemented with 10% FCS, S2R+ cells were maintained in Shields and Sang M3 Insect Medium (Sigma) supplemented with 10% FCS, and GlutaMAX (Invitrogen) at 25°C at atmospheric levels of CO2. For passaging, cells were washed in PBS, trypsinised for 3 mins, resuspended in full medium, spun and sparsely plated. HEK293 cells were grown in high glucose DMEM medium (Invitrogen) supplemented with 10% FCS, and GlutaMAX at 37°C at 5% CO2. All media were supplemented with 50 μg penicillin and 50 μg streptomycin.
Genome-wide RNAi screening
The Ambion Silencer® Drosophila RNAi Library, which contains 13059 unique dsRNAs targeting 92% of known protein-coding genes, was used and, as previously described57, replated from its original 96-well plate format onto white 384-well plates (Corning) using a Beckman Biomek FX robot(220 ng of dsRNA/well in 5 μl PBS).
Controls were manually added into empty wells of this 384-well format: DsRed was used as a baseline control, yki as a functional control, wts and mats as negative controls, and mts and rassf as positive controls. The genome-wide RNAi screen was run in triplicates, using plasmids encoding Yki-NTEV, 14.3.3eps-CTEV, GV-2ER (each 10ng/well), UAS-Fluc (20ng/well) and a Renilla luciferase under the baculoviral OpIE2 promoter (pIZ-Rluc, 3ng/well) to normalise for transfection efficiency and cell number. For the screen, S2R+ cells were batch-transfected with all plasmids (Yki-NTEV, 14.3.3eps-CTEV, GV-2ER, UAS-Fluc, pIZ-RL) using FugeneHD (Roche), and 6000 cells were plated per 384-well using a Matrix WellMate (Thermo Scientific). After 72h of incubation, cells were lysed and processed for the Dual Luciferase® Reporter Assay (Promega).
Analysis of the screening data
Readings for firefly and Renilla luciferases were processed and analysed using R and the CellHTS2 bioconductor package (http://www.bioconductor.org/packages/devel/bioc/html/cellHTS2.html). Both firefly and Renilla data were firstly normalised by dividing each value by the plate median of sample wells to account for plate-to-plate variation. Interpretation of firefly/Renilla ratios suggested a bias towards wells with extreme effects on cell number (either increased or decreased Renilla values). To correct for this, loess regression was used to define an expected relationship between the values for Renilla only and the firefly/Renilla ratio, and Renilla values were then corrected by subtracting this expected value57. The corrected firefly/Renilla ratios were then divided by the experimental median absolute deviation of ratios to calculate a robust Z-score, indicating the number of standard deviations away from the screen mean. Triplicate plates were summarised by taking the median of the replicate Z-scores, and hits were defined as those genes with median Z-scores greater than +3 or less than −3.
For the secondary screening process, all hits were subjected to a second round of Yki-NTEV/14.3.3eps-CTEV split TEV and GAL4-DBD-Yki/Ex assays, using new dsRNA amplicons for each candidate. To normalise for promoter-dependent off-targets, we used an Actin promoter-driven Renilla luciferase (pAFW-hRL) in the secondary assays. In addition, we removed all hits which displayed a Renilla value that was below 25% of the mean Renilla value across a given plate. In the following cross-correlation process (i.e. for pathway inhibitors: high values in split TEV Yki/14.3.3 assay and low values in GAL4-DBD-Yki/Ex assay), further potential off-target hits were removed, yielding 46 final candidates (see Supplementary Information, Fig.3 and Supplementary Information, Table 1 for details).
These hits were then subjected to a pS168-Yki western blot analysis using individual dsRNAs targeting each candidate. Inhibitors displayed enhanced levels of pS168-Yki staining, and conversely, activators showed decreased levels. Controls for each setup were mer (for activators) and cka (for inhibitors).
Co-immunoprecipitations and western blots
S2 and S2R+ cells were transfected with indicated plasmids using the Effectene transfection kit (Promega). In the case of RNAi treatments, cells were incubated with 15 μg dsRNA/well 24h prior to transfection. After 48h of expression, cells were lysed in 1% Triton-X lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton-X 100, 1 mM EGTA) supplemented with 0.1 M NaF, phosphatase inhibitor cocktails 2 and 3 (Sigma) and protease inhibitor cocktail (Roche). Cell extracts were spun for 10 mins at 4°C at 14000 rpm. FLAG-tagged proteins were purified using anti-FLAG M2 affinity gel (Sigma). After 2h of incubation, the FLAG immunoprecipitates were washed four times in lysis buffer. Lambda (λ) phosphatase (NEB) treatment was performed in lysates for 30 min using 2000 units. Protein gels were run and blotted using the Mini-PROTEAN Tetra Electrophoresis System (Biorad). Detection of purified and associated proteins was performed by western blot analysis using chemiluminescence (Western Lightning® Plus-ECL, PerkinElmer). Densitometric analysis of western blots was done using ImageJ, using a protocol from lukemiller.org (http://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/). Significance was calculated using 1-way ANOVA in GraphPad Prism, followed by Bonferroniposthoc tests. Western blots were probed with antibodies against pS168-Yki28, Yki80, pS413-Sav (generated by Eurogentec SA, Seraing, Belgium, using the phospho-peptide LQKVPS(PO3H2)QQSL), FLAG (mouse M2, Sigma, 3165), Myc (mouse 9E10, Santa Cruz Biotechnology, sc-40), HA (rat 3F10, Roche Applied Science, 11867423), ERα (rabbit HC-20, Santa Cruz Biotechnology, sc-543), α-tubulin (mouse, Sigma, T9026), using dilutions of 1:1000 for all antibodies.
Immunostainings
Wing imaginal discs from third instar larvae were dissected, fixed for 25 min in 4% PFA, washed three times in PBS supplemented with 0.1% Triton-X (PBS-T), permeabilised for 15 min in PBS supplemented with 0.3% Triton-X, washed once with PBS-T, and pre-blocked in PBS-T containing 10% normal goat serum (NGS) for 30 min. Discs were incubated in the primary antibody overnight diluted in PBS-T containing 10% NGS, followed by three washes with PBS-T, and an incubation with a secondary antibody in PBS-T/10% NGS for 3h at RT. After four further washes, discs were mounted in Vectashield mounting medium with DAPI (Vectorlabs). Dilutions for the antibodies against β-galactosidase (Promega, Z278B), Ex (a kind gift from Allen Laughon), Dlg (DSHB collection, monoclonal 4F3) and Rhodamine Red-X (Jackson ImmunoResearch, 111-295-003) were 1:500.
Analysis of the Drosophila wing
Adult wings were mounted using Euparal mounting medium (Agar Scientific) and imaged using a Zeiss Axio-plan 2 imaging system (using Optovar optics, 1.6×) and a 2.5× objective (Plan-NEOFLUAR, Zeiss) connected to a Leica DFC420C camera. Wing sizes were quantified by measuring the posterior area divided by total wing area using ImageJ. Significance was determined using the 1-way ANOVA statistic tool in GraphPad Prism, followed by Bonferroni posthoc tests.
Supplementary Material
Acknowledgements
We would like to gratefully thank A.Kerai for help with re-screening candidates; T.Gilbank, S. Maloney and F. Earl for maintaining flies; L. Muthusamy for sequencing and helping with plating RNAi libraries; the Vienna Drosophila RNAi Centre for fly stocks; O. Shaham and B. Thompson for comments on the manuscript. We are very grateful to P. Gaspar for help with fly genetics, F. Josue and P. Langton for advice, and to A. Teleman and A. Celik for sharing reagents and exchanging unpublished data prior to publication. We thank K.-A. Nave (Göttingen) for on-going support. M.C.W. and M.J.R. acknowledge the support by the Bundesministerium für Wirtschaft und Forschung (Grants 03EFT6N1 and 16V0008). M.C.W. was supported by an EMBO Long-Term Fellowship. The Tapon laboratory is supported by Cancer Research UK.
Footnotes
Accession number The RNAi screening data were deposited into GenomeRNAi under the accession numer: GR00218-S.
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem. J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 2.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development (Cambridge, England) 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantalacci S, Tapon N, Léopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 5.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 7.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 8.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 9.Kango-Singh M, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 10.Tapon N, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Lai Z-C, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF Family Protein Scalloped Mediates Transcriptional Output of the Hippo Growth-Regulatory Pathway. Developmental cell. 2008 doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, et al. The TEAD/TEF Family of Transcription Factor Scalloped Mediates Hippo Signaling in Organ Size Control. Developmental cell. 2008 doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goulev Y, et al. SCALLOPED Interacts with YORKIE, the Nuclear Effector of the Hippo Tumor-Suppressor Pathway in Drosophila. Current biology: CB. 2008 doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 18.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Current biology: CB. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 20.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 22.McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development (Cambridge, England) 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- 23.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nature cell biology. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 24.Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Current biology: CB. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 25.Poon CLC, Lin JI, Zhang X, Harvey KF. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Boggiano JC, Vanderzalm PJ, Fehon RG. Tao-1 phosphorylates Hippo/MST Kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badouel C, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Oh H, Reddy BVVG, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paramasivam M, Sarkeshik A, Yates JR, 3rd, Fernandes MJG, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan SW, et al. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 37.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nature genetics. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 38.Mao Y, et al. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development (Cambridge, England) 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- 39.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Willecke M, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Chen C-L, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varelas X, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Li L, Tumaneng K, Wang C-Y, Guan K-L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silvis MR, et al. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi C, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev. Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Densham RM, et al. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol. Cell. Biol. 2009;29:6380–6390. doi: 10.1128/MCB.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 52.Fernández BG, et al. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138:2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 53.Sansores-Garcia L, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada K-I, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 55.Wehr MC, et al. Monitoring regulated protein-protein interactions using split TEV. Nature methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- 56.Wehr MC, Reinecke L, Botvinnik A, Rossner MJ. Analysis of transient phosphorylation-dependent protein-protein interactions in living mammalian cells using split-TEV. BMC biotechnology. 2008;8:55. doi: 10.1186/1472-6750-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 59.Horike N, et al. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 2003;278:18440–18447. doi: 10.1074/jbc.M211770200. [DOI] [PubMed] [Google Scholar]
- 60.Dentin R, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 61.Okamoto M, Takemori H, Katoh Y. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol. Metab. 2004;15:21–26. doi: 10.1016/j.tem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 62.Choi S, Kim W, Chung J. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC) J. Biol. Chem. 2011;286:2658–2664. doi: 10.1074/jbc.C110.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 64.Lizcano JM, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang B, et al. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2012 doi: 10.1038/onc.2012.431. doi:10.1038/onc.2012.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mihaylova MM, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang B, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008;7:434–444. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Straßburger K, Tiebe M, Pinna F, Breuhahn K, Teleman AA. Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev. Biol. 2012;367:187–196. doi: 10.1016/j.ydbio.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Yu F-X, et al. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmed AA, et al. SIK2 Is a Centrosome Kinase Required for Bipolar Mitotic Spindle Formation that Provides a Potential Target for Therapy in Ovarian Cancer. Cancer Cell. 2010;18:109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Charoenfuprasert S, et al. Identification of salt-inducible kinase 3 as a novel tumor antigen associated with tumorigenesis of ovarian cancer. Oncogene. 2011;30:3570–3584. doi: 10.1038/onc.2011.77. [DOI] [PubMed] [Google Scholar]
- 74.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hall CA, et al. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70:8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, et al. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 77.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 79.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colombani J, Polesello C, Josué F, Tapon N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Current biology: CB. 2006;16:1453–1458. doi: 10.1016/j.cub.2006.05.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.