Abstract
Chronic stress is a risk factor for several neuropsychiatric diseases, such as depression and psychosis. In response to stress glucocorticoids (GCs) are secreted that bind to mineralocorticoid and glucocorticoid receptors, ligand-activated transcription factors that regulate the transcription of gene networks in the brain necessary for coping with stress, recovery, and adaptation. Chronic stress particularly affects the dentate gyrus (DG) subregion of the hippocampus, causing several functional and morphological changes with consequences for learning and memory, which are likely adaptive but at the same time make DG neurons more vulnerable to subsequent challenges. The aim of this study was to investigate the transcriptional response of DG neurons to a GC challenge in male rats previously exposed to chronic restraint stress (CRS). An intriguing finding of the current study was that having a history of CRS had profound consequences for the subsequent response to acute GC challenge, differentially affecting the expression of several hundreds of genes in the DG compared with challenged nonstressed control animals. This enduring effect of previous stress exposure suggests that epigenetic processes may be involved. In line with this, CRS indeed affected the expression of several genes involved in chromatin structure and epigenetic processes, including Asf1, Ash1l, Hist1h3f, and Tp63. The data presented here indicate that CRS alters the transcriptional response to a subsequent GC injection. We propose that this altered transcriptional potential forms part of the molecular mechanism underlying the enhanced vulnerability for stress-related disorders like depression caused by chronic stress.
Chronic stress is known to be a risk factor for the development of stress-related disorders, such as depression and psychosis (1, 2). During stress activation of the hypothalamic-pituitary-adrenal axis results in release of glucocorticoids (GCs) by the adrenal cortex, which activate the brain mineralocorticoid and glucocorticoid receptors (3). These ligand-activated nuclear receptors regulate the transcription of gene networks necessary for coping with stress, recovery, and adaptation (4). The dentate gyrus (DG) subregion of the hippocampus is the entry point for information processing in the hippocampus and is particularly sensitive to the effects of chronic stress (5, 6). Upon chronic stress exposure, DG neurons display a variety of functional and morphological changes, with consequences for learning and memory. Long-term potentiation, a lasting synaptic strengthening that likely underlies learning and memory formation, is suppressed in the DG by chronic stress exposure (7–9), as is the case with the formation of new neurons in the adult brain by neurogenesis in the subgranular zone of the DG (10–13). Changing GC levels also have profound consequences for the migration, positioning, and functional connectivity of the newborn neurons (14). Because the continuous addition of new neurons to the hippocampal circuitry via adult DG neurogenesis has been implicated in aspects of memory function, suppression of this process likely negatively influences cognitive processing (15–17). Although antidepressants increase neural progenitor cells, the role of neurogenesis in the pathogenesis of depression is disputed (13, 18, 19). The DG is also the site of substantial rapid changes in histone marks in response to acute stress, as an index of epigenetic effects on gene expression (20–23).
GC action is highly context dependent and depends on both the cellular and environmental context and the activation state of neural cells, culminating in differences in activity of intracellular pathways and thus availability of cofactors and cross talk partners for glucocorticoid and mineralocorticoid receptors in hippocampal cells (24). This context specificity has consequences for the set of genes that is transcriptionally modulated by GCs under different conditions and underlies the flexibility of the stress response, allowing us to deal with and adapt to changing situations. We previously studied the transcriptional response to chronic stress exposure in the DG subregion of the hippocampus (25). Despite the clearly defined changes in synaptic transmission and structural plasticity in the DG, the effects on gene expression were very subtle. Consistent with this observation, some of the changes in hippocampal function after chronic stress are not obvious under baseline conditions and only become apparent when GC receptors (GR) is subsequently activated, such as the enhanced synaptic excitation of DG cells with respect to α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor-mediated synaptic responses in the DG, the larger Ca2+ currents (23), and the responsiveness of the mammalian target of rapamycin (mTOR) pathway (26).
The aim of the current study was to gain more insight in the context dependency of GC action in the hippocampus under baseline conditions and after chronic stress exposure. Because some of the effects of chronic stress require GC action to become apparent, we generated expression profiles of the rat hippocampal DG region in animals with and without a history of chronic restraint stress (CRS) and with or without subsequent GC challenge. Based on the context dependency of GC action, we hypothesized that the transcriptional response to GR action in the DG would differ depending on stress history.
Materials and Methods
Animals and treatment
Adult male Sprague Dawley rats (70 d of age) were obtained from Charles River (Germantown, Maryland) and maintained on a 12-hour light, 12-hour dark cycle (lights on at 8 am) with ad libitum access to food and water. For microarray analysis, rats were either handled for 21 days (nonstressed handled animals are hereafter referred to as controls) or subjected to CRS for 6 hours per day during 21 days (22). On day 22, half of the rats received a GC challenge, which consisted of an injection with corticosterone (injection, sc 5 mg/kg, in propylene glycol, hereafter referred to as GC challenge or GCs), and were killed 3 hours later. The other half of the rats were left undisturbed and did not receive a vehicle injection to avoid eliciting a stress response. The unchallenged rats were killed at the same time point as the injected rats. This resulted in 4 experimental groups (all n = 6) for the microarray analysis: 1) control, 2) control + GCs, 3) CRS, and 4) CRS + GCs. After decapitation, brains were rapidly dissected and snap frozen in isopentane (cooled in ethanol placed on pulverized dry ice) and stored at −80°C for later use.
In a separate experiment, body weight and relative thymus weight were determined in control and CRS animals as a bioassay reflecting stress and GC exposure over the 21-day period. A clear decrease in body weight gain and relative thymus weight was observed upon CRS (26). Animal care was conducted in accordance with the Rockefeller University Animal Care Committee.
Laser microdissection (LMD)
LMD was performed as previously described (27). Briefly, coronal brain sections (8 μm) containing the rostral rat hippocampus were mounted on polyethylene naphthalate (PEN) membrane slides (1440-1000; PALM, Bernried, Germany) and stored at −80°C until further use. On the day of LMD, the slides were briefly stained with hematoxylin (10%) and dehydrated in 70%, 95%, and 100% ethanol. The DG subregion of the hippocampus was laser microdissected on a PALM MicroLaser System (PALM). Per rat the entire DG region from a total of 8 hippocampal sections per brain hemisphere was dissected and pooled to constitute a sample for subsequent linear amplification and microarray hybridization.
RNA isolation and linear amplification
Immediately after collecting the sample, 100 μL of TRIzol reagent (15596-026; Invitrogen Life Technologies, Carlsbad, California) was added to the collected tissue fragments, and RNA was isolated according to the manufacturer's instructions. Linear acrylamide (5 μL of 5 mg/mL) (AM9520; Ambion, Austin, Texas) was added as a carrier in the isopropanol precipitation. The RNA pellet was dissolved in 10 μL DEPC-H20, and RNA quality and quantity were checked by analyzing 1 μL of RNA on the Agilent 2100 Bioanalyzer using the RNA 6000 Pico LabChip kit (5065-4473; Agilent Technologies, Palo Alto, California).
Ten nanograms of total RNA were used as input in the MessageAmp II aRNA kit (AM1751; Ambion) and subjected to 1 round of linear amplification. A total of 100 ng of amplified RNA from the first round was subjected to a second round of linear amplification using the MessageAmp II Biotin-enhanced kit (AM1791; Ambion).
GeneChip hybridization and data analysis
The biotinylated aRNA was hybridized to GeneChip Rat Genome 230 2.0 Arrays (Affymetrix, Santa Clara, California) containing 31 099 probe sets representing over 28 000 well-substantiated rat genes. Hybridizations were conducted at the Leiden Genome Technology Center (Leiden University). A total of 24 microarrays was hybridized. The data were normalized using Mas 5.0 and subjected to statistical analysis using Linear Models for Microarray Data (28), a package for the R computing environment that allows multiple comparison of experimental groups. To identify genes differentially expressed in response to GCs in the DG of the hippocampus, a 2-way ANOVA was performed with group and treatment as factors, followed by pairwise post hoc comparisons. Genes with P < .01 were considered significant.
WebGestalt (WEB-based GEne SeT AnaLysis Toolkit version 2) was used to identify enriched gene sets among the lists of significant genes representing specific biological processes or molecular functions (http://bioinfo.vanderbilt.edu/webgestalt/) (29, 30). WebGestalt is a toolkit that incorporates information from different centrally and publicly curated databases, including Gene Ontology (GO), KEGG, and WikiPathways. Gene lists containing the probe set identifiers of significant genes were uploaded in WebGestalt, using rnorvegicus_affy_rat230_2 as a reference set. Three different types of enrichment analysis were performed: GO analysis, KEGG, and WikiPathways analysis. The hypergeometric test was used for enrichment evaluation analysis, with a significance level chosen to identify the 10 categories with the most significant P values (default Top 10 setting) and a cutoff for a required minimum of 4 genes per category for the enrichment analysis. Only gene sets in the Top 10 with a raw P value of at least 0.05 were taken into account.
Real-time quantitative PCR (RT-qPCR)
RT-qPCR was performed to confirm differential expression of genes indicated by the microarray analysis. Per group a selection of up-regulated and down-regulated genes was analyzed by qPCR covering different fold changes (FCs) and P values. The selection was not based on gene function. Primers were designed using Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) (for primer sequences, please see Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org). RT-qPCR was performed using a Light Cycler 2.0 Real-Time PCR System (Roche Applied Science, Basel, Switzerland). cDNA synthesis was performed on 400 ng of the second round cRNA using the iScript cDNA Synthesis kit (170-8897; Bio-Rad, Hercules, California). PCR was performed using the LightCycler FastStart DNAPLUS SYBR Green I kit (Roche Applied Science). Dissociation curves were examined for each primer pair and controlled for specificity of the reaction and genomic contamination by checking the no reverse transcriptase and no template control samples.
The standard curve method was used to quantify the expression differences (31). Expression levels of the validated genes were normalized against the expression levels of tubulin, beta 2A class IIa, which was shown to be highly stable and not to be affected by CRS or GCs (Supplemental Figure 1). Normalized expression levels were analyzed in GraphPad Prism 6 (GraphPad Software, Inc, San Diego, California) by 2-way ANOVA with group and treatment as factors in combination with post hoc testing to assess significant differential expression of GC-responsive genes. Pair wise comparisons were performed using a 1-tailed unpaired t test. Significance was accepted at P < .05.
Results
Acute GC challenge robustly affects the DG transcriptome in both control and stressed animals
Two-way ANOVA identified a total of 945 genes with significantly different expression levels in the DG region of the hippocampus when comparing all 4 groups (P < .01) and 2249 genes if a P value threshold of less than 0.05 was applied. The full list of 945 genes is accessible in Supplemental Table 2. Subsequent post hoc testing yielded a total of 525 genes (P < .01) that were differentially affected by GC challenge in control animals (Supplemental Table 3). These 525 genes consisted of almost equal numbers of up-regulated (291 genes; 55% of total) and down-regulated (234 genes; 45% of total) genes.
In animals with a stress history, 576 genes (P < .01) responded to GC challenge (Supplemental Table 4), of which 331 (57%) were up-regulated and 245 (43%) down-regulated.
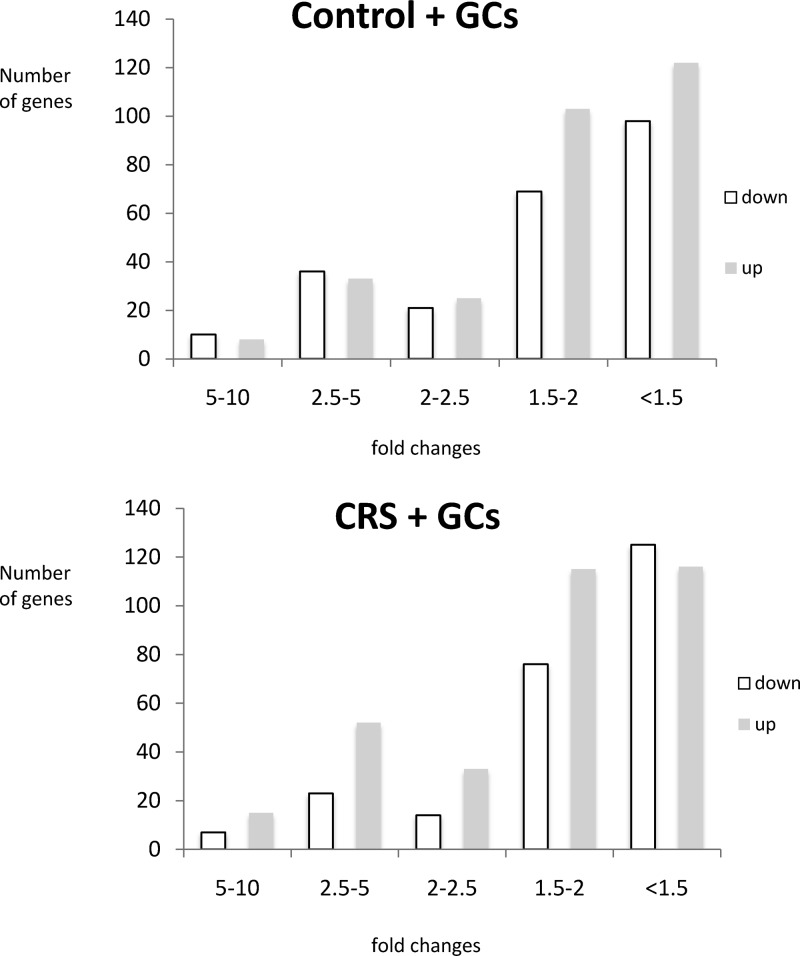
If the threshold was relaxed to P < .05, 733 and 765 genes were significantly affected by GC challenge in control and stress animals, respectively. The expression changes induced by the GC challenge were highly similar with regard to magnitude of change in control animals and animals with a stress history, with 75% of the genes showing a FC smaller than 2-fold. A minority of genes had a FC above 2.5, and none of the genes had a FC above 10 (Figure 1).
Figure 1.
Bar charts showing the distribution of FCs among the genes up-regulated (gray bars) and down-regulated (white bars) by GC challenge in control animals (control + GCs, top chart) and animals that were previously stressed (CRS + GCs, lower chart).
A total of 234 genes (P < .01) were affected by CRS compared with control in the DG region of the hippocampus (Supplemental Table 5), of which 113 (48%) up-regulated and 121 (52%) down-regulated.
In general, the effects of CRS on the hippocampal DG transcriptome were much more subtle than the effects of the acute GC challenge, both in terms of number of affected genes as well as P value distribution. None of the significant genes affected by CRS survived correction for multiple testing, with the smallest false discovery rate (FDR)-adjusted P value starting at 0.41 and reaching 0.99 at the 14th most significant gene in rank. Among the significant genes responding to acute GC challenge in the DG, 55 were highly significant with an FDR-adjusted P < .01 and 123 P < .05 in the control animals and 50 (FDR-adjusted P < .01) and 137 (FDR-adjusted P < .05) in the animals with a stress history. Plotting the distribution of both unadjusted and FDR-adjusted P values of the highest scoring 1000 genes per comparison showed that indeed the P values were much less robust in the gene list affected by CRS than in the acute GC-challenge groups, with a slight shift towards higher statistical significance in the stressed animals that received a GC challenge (Supplemental Figure 2).
Different genes respond to GC challenge depending on previous history of animals
Although at a first glance the transcriptional response to acute GC challenge seemed quite comparable in control animals and animals with a previous stress history, at least in terms of number of genes and FCs, we subsequently looked at whether there was overlap in identity of differentially expressed genes. Approximately 30% (258 genes) of the total number of genes differentially expressed by acute GC challenge were in common to control and stressed animals if a P value threshold of 0.01 was applied. If the threshold was relaxed to P < .05, the overlap increased to approximately 58%, implying that depending on the previous history of the animals (ie, stress or no stress), a subsequent challenge with GCs results in a considerably different transcriptional response in the DG of the hippocampus (Supplemental Figure 3).
Validation of microarray results
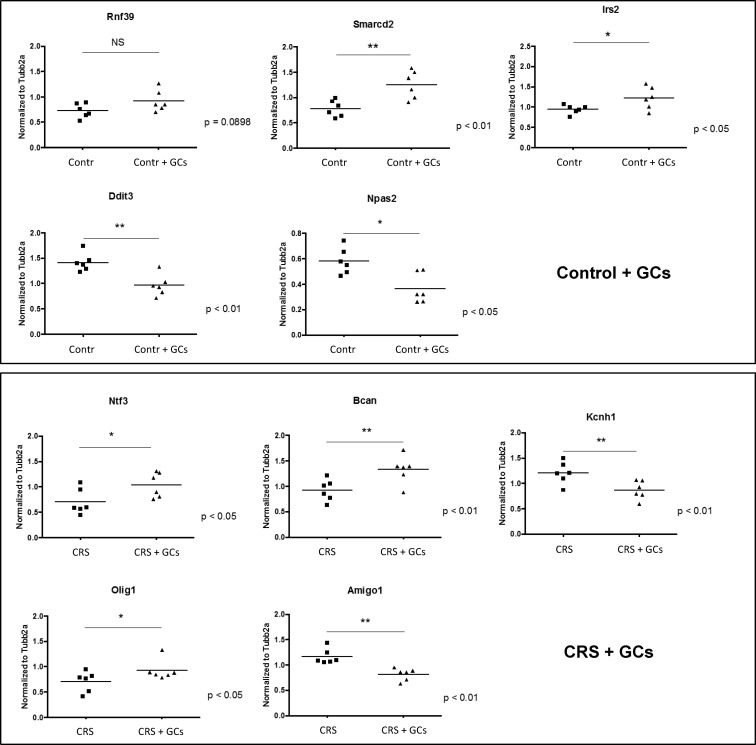
Using RT-qPCR, we validated a selection of the transcriptional changes induced by acute GC challenge, as indicated by the microarray analysis. A total of 21 genes was selected for validation, including both genes uniquely affected by GC challenge (P < .01) in either the control or stress animals as well as genes in common to both groups. The percentage of successfully validated genes was high, with only 3 of the in total 31 RT-qPCRs not yielding a significant outcome (Table 1), of which 2 were just above the significance threshold of 0.05. The genes responsive to GC challenge in either control or stressed animals are depicted in Figure 2.
Table 1.
RT-qPCR Validation of Genes Regulated by CORT Challenge in the DG Region of the Hippocampus, According to Microarray Analysis
| Probe Set Id | Gene Name | Gene Symbol | Control+CORT FC (microarray) | P | Stress+CORT FC (microarray) | P | Control+CORT FC (RT-qPCR) | P | Stress+CORT FC (RT-qPCR) | P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1370159_at | SWI-SNF-related, matrix-associated, actin-dependent regulator of chromatin | Smarcd2 | 1.5 | ↑ | 3.7E-03 | 1.6 | ↑ | .0023 | ||||||
| 1368662_at | Ring finger protein 39 | Rnf39 | 1.3 | ↑ | 6.6E-04 | 1.3 | ↑ | .0444 | ||||||
| 1373210_at | Laminin, β 1 | Lamb1 | 1.4 | ↑ | 2.0E-03 | 1.4 | ↑ | .0978 | ||||||
| 1371091_at | Insulin receptor substrate 2 | Irs2 | 1.3 | ↑ | 1.2E-03 | 1.3 | ↑ | .0211 | ||||||
| 1369590_a_at | DNA-damage inducible transcript 3 | Ddit3 | 1.5 | ↓ | 2.2E-03 | 1.5 | ↓ | .0013 | ||||||
| 1383439_at | Neuronal PAS domain protein 2 | Npas2 | 1.5 | ↓ | 3.3E-04 | 1.6 | ↓ | .0034 | ||||||
| 1387267_at | Neurotrophin 3 | Ntf3 | 1.4 | ↑ | 7.6E-03 | 1.5 | ↑ | .0215 | ||||||
| 1369831_at | Brevican | Bcan | 1.6 | ↑ | 1.5E-04 | 1.4 | ↑ | .0083 | ||||||
| 1387200_at | Oligodendrocyte transcription factor 1 | Olig1 | 1.6 | ↑ | 2.7E-04 | 1.3 | ↑ | .0439 | ||||||
| 1368061_at | Potassium voltage-gated channel, subfamilyH (eag-related), member 1 | Kcnh1 | 1.3 | ↓ | 1.6E-03 | 1.4 | ↓ | .0078 | ||||||
| 1383499_at | Adhesion molecule with Ig-like domain 1 | Amigo1 | 1.3 | ↓ | 1.0E-03 | 1.4 | ↓ | .0006 | ||||||
| 1371363_at | Glycerol-3-phosphate dehydrogenase 1 | Gpd1 | 2.3 | ↑ | 4.1E-06 | 2.1 | ↑ | 2.0E-05 | 3.0 | ↑ | .0007 | 2.3 | ↑ | .0046 |
| 1368478_at | Dopamine receptor D1a | Drd1a | 2.6 | ↓ | 3.6E-10 | 2.2 | ↓ | 7.9E-09 | 3.6 | ↓ | .0011 | 3.4 | ↓ | <.0001 |
| 1370491_a_at | Histidine decarboxylase | Hdc | 2.3 | ↑ | 1.3E-05 | 4.8 | ↑ | 4.5E-10 | 2.8 | ↑ | .0009 | 8.0 | ↑ | <.0001 |
| 1370408_at | Putative small membrane protein NID67 | Nid67 | 1.4 | ↑ | 2.6E-03 | 1.4 | ↑ | 1.5E-03 | 1.1 | ↑ | .1577 | 1.4 | ↑ | .0018 |
| 1371615_at | Diacylglycerol O-acyltransferase 2 | Dgat2 | 1.5 | ↑ | 1.4E-06 | 1.3 | ↑ | 1.0E-04 | 1.4 | ↑ | .0005 | 1.3 | ↑ | .0731 |
| 1387169_at | Transducin-like enhancer of split 3, homolog of Drosophila E(spl) | Tle3 | 1.3 | ↑ | 5.7E-03 | 2.0 | ↑ | 4.9E-07 | 1.3 | ↑ | .0048 | 1.8 | ↑ | .0027 |
| 1392189_at | Regulatory factor X, 4 (influences HLA class II expression) | Rfx4 | 1.4 | ↓ | 2.1E-03 | 1.5 | ↓ | 3.9E-04 | 1.5 | ↓ | .0021 | 1.6 | ↓ | .0006 |
| 1369067_at | Nuclear receptor subfamily 4, group A, member 3 | Nr4a3 | 3.0 | ↓ | 1.7E-06 | 2.1 | ↓ | 1.9E-04 | 2.6 | ↓ | .0009 | 2.6 | ↓ | <.0001 |
| 1368013_at | DNA-damage-inducible transcript 4-like | Ddit4l | 2.9 | ↓ | 1.8E-07 | 2.3 | ↓ | 8.4E-06 | 3.8 | ↓ | .0003 | 3.5 | ↓ | .0005 |
| 1368678_at | Brain-derived neurotrophic factor | Bdnf | 2.0 | ↓ | 1.2E-04 | 1.8 | ↓ | 6.1E-04 | 2.8 | ↓ | <.0001 | 1.8 | ↓ | .0077 |
From left to right the probe set id, gene name, and gene symbol are indicated followed by the FC and P value according to the microarray analysis in the control animals and stressed animals, respectively. Arrows indicate whether the gene is up-regulated (↑) or down-regulated (↓) by the CORT challenge. The last four columns on the right indicate the P values obtained in the RT-qPCR validation in the control animals and stressed animals, respectively.
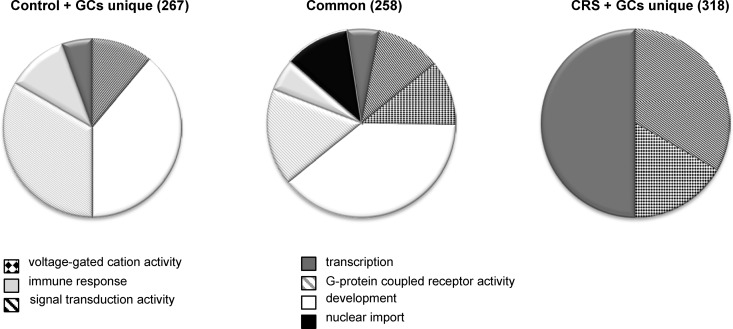
Figure 2.
Pie charts of overrepresented GO terms among the 576 genes that were differentially expressed upon GC challenge. The differentially expressed genes were divided in a group that responded to GCs in both controls and CRS animals (center) or only in controls (left) or in CRS animals (right). The pie charts represent the GO terms that were overrepresented in the 3 groups of GC-responsive genes and show that after CRS, GC challenge gives rise to a different gene signature than in control animals.
Pathway analysis
The genes differentially expressed in response to the GC challenge were subjected to pathway analysis to identify enriched gene sets. The responsive genes were divided into 3 groups: the 258 genes in common to control and stressed animals as well as the 267 and 318 genes that uniquely responded to GC challenge in the control and the stressed animals, respectively (Supplemental Figure 3). These common and unique genes are listed in separate worksheets of Supplemental Tables 3 and 4. The 258 GC-responsive genes in common to control and stressed animals contained 30 enriched GO terms using a P value cutoff of .05, of which 16 also had an FDR-adjusted P < .05. The most prominent enriched biological processes were several GO terms representing development, nuclear import, G protein-coupled receptor (GPCR) activity, signal transduction, voltage-gated cation channel activity, cytokine binding, and chromatin binding (Figure 3 and Supplemental Table 5). The 267 genes uniquely responsive to GC challenge in control animals contained 26 enriched GO terms using a P value cutoff of 0.05, of which 6 also had an FDR-adjusted P < .05. Despite the fact that the individual genes were different, the enriched GO terms showed considerable similarity to the enriched GO terms in the overlap group, with GPCR activity and signal transduction being enriched in both groups and several enriched GO terms related to development (Supplemental Table 6). Class A rhodopsin-like GPCRs was significantly overrepresented according to WikiPathways (30) in both the 258 shared genes and the 267 genes unique to the control animals.
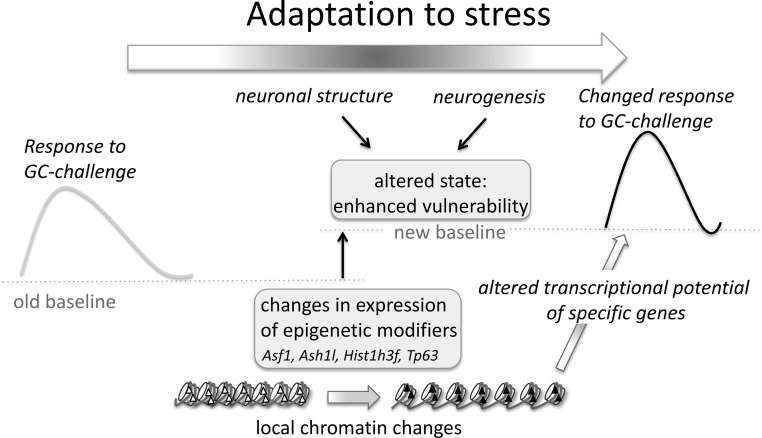
Figure 3.
Model of how chronic stress exposure induces changes in expression of epigenetic modifiers, which underlie a new steady state that is more vulnerable to subsequent challenges. In this model, adaptive changes occur in response to chronic stress, which include changes in the expression of epigenetic modifiers. These epigenetic modifiers give rise to local chromatin changes that underlie the altered transcriptional response of specific genes to a subsequent GC challenge. Together with other stress-induced changes, such as changes in neuronal structure and neurogenesis, these changes in chromatin underlie an altered state of enhanced vulnerability that arises due to the chronic stress exposure.
A completely different picture emerged from the pathway analysis of the 318 differentially expressed genes upon GC challenge unique to the stressed animals (Figure 3 and Supplemental Table 6). Despite the fact that this group contained the most genes (ie, 318, vs 258 and 267 in the overlap and control groups, respectively), the number of enriched GO terms was a lot lower than observed in the other groups, with only 7 significant GO terms using a P cutoff of 0.05 and none left if applying an FDR-adjusted P < .05. Three of the 7 enriched GO terms concerned transcriptional regulation. In addition, signal transduction and voltage-gated cation channel activity were among the enriched GO terms. When extending the pathway analysis to include WikiPathways, 2 pathways emerged that were not identified in the other groups: apoptosis mechanisms and the TNF-α/nuclear factor (NF)-κB signaling pathway.
Discussion
Chronic stress is a risk factor for several neuropsychiatric diseases, such as depression and psychosis, in which the hippocampus has been proposed to play an important role (2). Hippocampal neurons are particularly sensitive to the effects of chronic stress (5), but what exactly the molecular underpinnings of this enhanced vulnerability are, in particular when exposed to challenges, such as new stressors, remains unknown. The aim of this study was to investigate the transcriptional response of DG granule neurons to a GC challenge in animals with and without a history of chronic stress, so in the context of either healthy or vulnerable neurons.
Identification of genes and pathways in DG
In the current study, we identified genes and pathways that are responsive to a GC challenge and/or chronic stress in the DG region of the hippocampus. The reliability of the obtained data was underscored by the high percentage of successfully validated expression changes using RT-qPCR, as well as by the presence of several genes previously described in literature to be under control of GCs and/or stress, such as brain-derived neurotrophic factor, histidine decarboxylase, Kruppel-like factor (Klf)9, Klf11, metallothionein 1A and 2A, fibroblast growth factor 2 and 10, adrenoceptor alpha 1D, and GR itself (32–39). Besides known GC-responsive genes, we identified many novel genes responsive to GCs and/or stress. Interestingly, we identified many novel GC-responsive genes that were exclusively responsive to acute GC challenge in the context of a history of chronic stress.
We previously described how a single GC injection had a clear effect on the hippocampal expression of genes involved in cellular metabolism and energy production, including glycolytic enzymes, ATPases and ATP synthases, and mitochondrial components (4). However, in the current study, effects on energy metabolism did not stand out as overrepresented GO terms, both after chronic stress exposure and GC challenge. This is likely due to the fact that we generated expression profiles from the DG region of the hippocampus that was isolated by LMD, allowing very precise isolation of specific neuronal subpopulations, with a relatively low percentage of coisolated glial cells compared with isolating whole hippocampus (27). It has been shown that glucose is primarily taken up by astrocytes and that transport and metabolism of glucose preferentially take place in the glial cell compartment compared with neurons (40, 41).
Impact of chronic stress on DG neurons
Chronic stress has profound effects on DG neurons, including suppression of adult neurogenesis and functional changes in Ca2+ currents, leading to larger Ca2+ current amplitudes if animals with a previous history of stress are reexposed to a new stressor (42). After chronic stress, we identified several transcriptional changes of genes involved in synaptic transmission and learning and memory, including cocaine- and amphetamine-regulated transcript prepropeptide, unc-13 homolog C, and ADAM metallopeptidase domain 10 (43–45) (Supplemental Table 5).
Chronic stress has also been shown to decrease new cell proliferation in the adult rat DG and to differentially affect apoptotic cell death in the subgranular zone and granule cell layer of the DG (46). Consistent with this, we observed differential expression of several genes involved in cell cycle and/or apoptosis in response to CRS, including stromal antigen 3, cyclin-dependent kinase inhibitor 3, anaphase-promoting complex subunit 4, apoptosis-inducing factor, mitochondrion-associated 2, myc-like oncogene, s-myc protein, and tumor protein p63 (Tp63) (Supplemental Table 5).
Consistent with our previous study using the chronic unpredictable stress paradigm, CRS exposure itself had fairly modest effects on the DG transcriptome (25). An explanation for this could be that earlier transient waves of gene expression have occurred, which have resulted in adaptive changes in neuronal structure, morphology, and function induced by chronic stress exposure. This may represent a new steady state or baseline that is part of the adaptive response to chronic stress and which, although only modestly different from the situation under basal nonstressed conditions, gives rise to an altered response pattern under challenged conditions (Figure 4). Such a new steady state could be the equivalent of Selye's phase of resistance (47) during chronic stress, which is characterized by increased allostatic load and vulnerability to subsequent stressors (1).
Figure 4.
RT-qPCR validation of genes responsive to GC challenge in either control (top panel) or CRS (lower panel) animals. *P < .05; **P < .01. The full gene names of the genes depicted here are listed in Table 1.
Chronic stress alters the responsivity of the DG
Several observations support this idea of an altered baseline after chronic stress exposure. For example, in hippocampal slices from rats with basal corticosteroid levels, no effect of previous stress exposure was observed on AMPA receptor-mediated synaptic responses of dentate granule cells. However, AMPA receptor-mediated synaptic responses from chronically stressed rats were significantly enhanced after corticosterone administration, pointing to an enhanced synaptic flow and enhanced excitation of projection areas of the DG when corticosterone levels rise (48). Furthermore, chronic stress exposure resulted in functional changes in Ca2+ currents, leading to larger Ca2+ current amplitudes, only if hippocampal slices from animals with a previous history of stress were treated with corticosterone 1-4 hours before recording (42). These findings suggest that after chronic stress exposure, there is an increased calcium load when DG granule neurons are reexposed to an acute stressor, potentially resulting in increased vulnerability of the cells. Consistent with this increased calcium load, several transcripts encoding proteins involved in calcium ion binding and intracellular signaling resulting in elevation of cytosolic calcium ion concentration were differentially expressed after chronic stress in this study, including EF-hand calcium binding domain 2, cadherin 1, protocadherin 17, and S100 calcium-binding protein A4 and prostaglandin E receptor 3 (Supplemental Table 5).
Indeed, an intriguing finding of the current study is that having a history of stress exposure had profound consequences for the subsequent response to acute GC challenge, yielding a distinct gene expression profile in the DG region of the hippocampus compared with that of challenged nonstressed animals. Because the GC challenge occurred only 24 hours after the CRS, it cannot be ruled out that this still represents a delayed effect of the procedure rather than an enduring effect of CRS. Future experiments should focus on whether this changed transcriptional response is reversible and, if so, how long it persists. In spite of this, a plausible explanation for the altered transcriptional response to GC challenge would be that epigenetic mechanisms, affecting chromatin structure and hence transcriptional potential of target genes, have been affected by the chronic stress exposure (Figure 4).
Changes in the epigenome by chronic stress
It is becoming increasingly clear that environmental factors can lay down or erase epigenetic marks, such as histone modifications and DNA methylation, resulting in local changes in gene expression, with consequences for risk for developing depressive disorders (49). The stress system is prone to programming by environmental factors, such as maternal care and prenatal and early life stress, resulting in alterations in stress responsiveness that persist throughout the lifespan (50–52).
Stress has also been shown to trigger epigenetic modifications in the adult brain. Forced swim stress and novelty stress produced a significant increase in phospho-acetylation of histone H3, at serine 10 and lysine 14 (H3S10p-K14ac), in the DG of the hippocampus, a combination of histone marks that is associated with a transcriptionally active chromatin state (20, 21, 53). Hunter et al (22) reported a complex, rapid, and subregion-specific effect within the hippocampus on several histone marks, including the repressive H3K9me3 and H3K27me3 marks as well as the active H3K4me3 mark in response to restraint stress. A subsequent study showed that the repressive histone H3K9me3 mark was targeted to transposable elements in the genome and may serve as a means of containing potential genomic instability induced by stress (23).
In line with these observations, in the current study, 3 weeks of restraint stress affected the expression of genes belonging to GO terms representing transcriptional regulation, DNA binding, and transcription factor activity. Several genes were differentially expressed that are involved in chromatin structure and epigenetic processes, including antisilencing factor 1 (Asf1), absent, small, homeotic disk 1 like (Ash1l), histone cluster 1, H3f (Hist1h3f), and Tp63. Asf1 is a ubiquitous eukaryotic histone-binding protein involved in both the assembly and disassembly of nucleosomes in cellular systems (54). Ash1l is a histone methyltransferase that has been shown to occupy most, if not all, active genes (55, 56). Hist1h3f encodes a member of the histone H3 family of core nucleosomal proteins involved in the make-up of chromatin structure. Tp63 is an upstream regulator of several chromatin-remodeling proteins (57, 58). It is therefore feasible that as a consequence of the differential expression of Asf1, Ash1l, Hist1h3f, and Tp63 induced by the chronic stress exposure, local chromatin remodeling has taken place, affecting the transcriptional potential of individual genes that collectively contribute to the altered state of enhanced vulnerability of hippocampal neurons (Figure 4). This may provide an explanation for the different transcriptional response observed in the DG of animals with and without a previous history of stress, when faced with a subsequent GC challenge.
Chronic stress alters transcriptional response to GC challenge
Several transcription factors displayed an altered transcriptional potential to GC challenge in stressed animals, including TOB1 transducer of ERBB2 (Tob1), nuclear receptor subfamily 1, group D, member 1 (NR1D1), and Klf11, which have been linked to depression and may contribute to the enhanced vulnerability of the brain after chronic stress exposure. In the brain, Tob1 plays a role in hippocampus-dependent learning and memory and was recently found to be down-regulated in postmortem brain tissue from patients with major depressive and bipolar disorder (59–61). Nr1d1 is a nuclear receptor that suppresses gene transcription and is involved in circadian clock regulation. Several studies have suggested that Nr1d1 may play a role in bipolar disorder, in which deregulation of circadian rhythm is frequently observed (62, 63). Nr1d1 was recently shown to mediate circadian regulation of inflammatory cytokines, thus forming a link between the clock and inflammatory pathways (64). Klf11 is a transcription factor activated in the brain by stress and elevated GCs (33). Klf11 regulates transcription of the dopamine D2 receptor gene and of monoamine oxidase A (MAO A), a catalytic enzyme that metabolizes the 3 major monoamines (serotonin, norepinephrine, and dopamine) in the brain (65). Elevated levels of MAO A may underlie the imbalance in monoamines observed in the depressed brain (66). It has been hypothesized that the GC-Klf11-MAO A pathway may play a crucial role in the pathophysiology of stress-related disorders (33).
Alterations in key cellular processes by chronic stress
Besides transcription factors, genes affecting a wide variety of processes, such as neuronal plasticity, cell survival, and inflammation, were differentially affected by the GC challenge in control and stressed animals. Overrepresentation of the WikiPathways “apoptosis mechanisms” and “TNF-α/NF-κB signaling pathway” among the GC-responsive genes uniquely identified in the stressed animals support this. Indeed, chronic stress has been shown to increase the susceptibility of certain populations of neurons to apoptotic cell death, whereas conversely, several antidepressants promote neuroprotection (67). Chronic stress reduces neurogenesis and cell-cycle progression in the DG and causes remodeling of hippocampal circuitry (46, 68–71). Previously, we found reduced NF-κB signaling in the mouse hippocampus after exposure to a chronic psychosocial stressor (72). Proinflammatory cytokines, such as TNF-α and signaling pathways, which modulate NF-κB activity, play an active role in the regulation of hippocampal synaptic plasticity (73). Cytokine-induced impairments in synaptic and structural plasticity have been suggested to play a role in the pathophysiology of depression (74–78).
We recently showed that some of the genes differentially affected by acute GC challenge in the DG region of the hippocampus belong to the mTOR pathway, which plays a central role in translational control and long-lasting synaptic plasticity (26). In neurons, the mTOR pathway modulates local translation of proteins at the synapse and, therefore, is critical for different forms of synaptic plasticity, including long-term potentiation and long-term depression (79, 80). Interestingly, an acute GC challenge significantly reduced mTOR protein levels in the hippocampus of animals with a chronic stress history but not in unstressed controls (26). Therefore, differential regulation of this pathway upon chronic stress exposure may be one of the routes via which chronic stress is known to affect hippocampal synaptic plasticity and enhance vulnerability to stress-related disorders. The mTOR pathway has been linked to depression and its activation to novel mechanisms underlying rapid antidepressant effects (81, 82). In postmortem brains of major depressive disorder patients, a significant reduction in mTOR and some of its downstream targets involved in translation initiation were identified (83). Consequently, mTOR and its upstream (eg, FK506 binding protein 5 and GR) and downstream signaling partners could be important targets for the development of novel antidepressants.
Conclusion
The data presented here indicate that chronic stress exposure alters the transcriptional response to a subsequent GC challenge, affecting the expression of several hundreds of genes in the DG region of the hippocampus. We propose that this altered transcriptional potential forms part of the molecular mechanism underlying the enhanced vulnerability for stress-related disorders like depression caused by chronic stress.
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research Grant 836.06.010 (MEERVOUD) (to N.A.D.), TI Pharma Grant T5-209, and Human Frontier Science Program Grant RGP39. E.R.d.K. was supported by the Royal Netherlands Academy of Science. B.S.M. was supported by the National Institutes of Health Grant MH41256.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AMPA
- 2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propanoic acid)
- Asf1
- antisilencing factor 1
- Ash1l
- absent, small, homeotic disk 1 like
- CRS
- chronic restraint stress
- DG
- dentate gyrus
- FC
- fold change
- FDR
- false discovery rate
- GC
- glucocorticoid
- GO
- Gene Ontology
- GPCR
- G protein-coupled receptor
- GR
- GC receptor
- Hist1h3f
- histone cluster 1, H3f
- Klf
- Kruppel-like factor
- LMD
- laser microdissection
- MAO A
- monoamine oxidase A
- mTOR
- mammalian target of rapamycin
- NF
- nuclear factor
- NR1D1
- nuclear receptor subfamily 1, group D, member 1
- RT-qPCR
- real-time quantitative PCR
- Tp63
- tumor protein p63.
References
- 1. McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62:3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Kloet ER, Joëls M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475 [DOI] [PubMed] [Google Scholar]
- 3. Reul JM, de Kloet ER. Two receptor systems for corticosterone in rat brain: microdistribution and differential occupation. Endocrinology. 1985;117:2505–2511 [DOI] [PubMed] [Google Scholar]
- 4. Datson NA, van der Perk J, de Kloet ER, Vreugdenhil E. Identification of corticosteroid-responsive genes in rat hippocampus using serial analysis of gene expression. Eur J Neurosci. 2001;14:675–689 [DOI] [PubMed] [Google Scholar]
- 5. Maras PM, Baram TZ. Sculpting the hippocampus from within: stress, spines, and CRH. Trends Neurosci. 2012;35:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lucassen PJ, Heine VM, Muller MB, et al. Stress, depression and hippocampal apoptosis. CNS Neurol Disord Drug Targets. 2006;5:531–546 [DOI] [PubMed] [Google Scholar]
- 7. Alfarez DN, Joëls M, Krugers HJ. Chronic unpredictable stress impairs long-term potentiation in rat hippocampal CA1 area and dentate gyrus in vitro. Eur J Neurosci. 2003;17:1928–1934 [DOI] [PubMed] [Google Scholar]
- 8. Bodnoff SR, Humphreys AG, Lehman JC, Diamond DM, Rose GM, Meaney MJ. Enduring effects of chronic corticosterone treatment on spatial learning, synaptic plasticity, and hippocampal neuropathology in young and mid-aged rats. J Neurosci. 1995;15:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krugers HJ, Goltstein PM, van der Linden S, Joëls M. Blockade of glucocorticoid receptors rapidly restores hippocampal CA1 synaptic plasticity after exposure to chronic stress. Eur J Neurosci. 2006;23:3051–3055 [DOI] [PubMed] [Google Scholar]
- 10. Cameron HA, Tanapat P, Gould E. Adrenal steroids and N-methyl-D-aspartate receptor activation regulate neurogenesis in the dentate gyrus of adult rats through a common pathway. Neuroscience. 1998;82:349–354 [DOI] [PubMed] [Google Scholar]
- 11. Czéh B, Michaelis T, Watanabe T, et al. Stress-induced changes in cerebral metabolites, hippocampal volume, and cell proliferation are prevented by antidepressant treatment with tianeptine. Proc Natl Acad Sci USA. 2001;98:12796–12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gould E, Daniels DC, Cameron HA, McEwen BS. Expression of adrenal steroid receptors by newly born cells and pyknotic cells in the dentate gyrus of the postnatal rat. Mol Cell Neurosci. 1992;3:44–48 [DOI] [PubMed] [Google Scholar]
- 13. Lucassen PJ, Meerlo P, Naylor AS, et al. Regulation of adult neurogenesis by stress, sleep disruption, exercise and inflammation: implications for depression and antidepressant action. Eur Neuropsychopharmacol. 2010;20:1–17 [DOI] [PubMed] [Google Scholar]
- 14. Fitzsimons CP, van Hooijdonk LW, Schouten M, et al. Knockdown of the glucocorticoid receptor alters functional integration of newborn neurons in the adult hippocampus and impairs fear-motivated behavior [published online August 28, 2012]. Mol Psychiatry. doi:10.1038/mp.2012.123 [DOI] [PubMed] [Google Scholar]
- 15. Aimone JB, Wiles J, Gage FH. Potential role for adult neurogenesis in the encoding of time in new memories. Nat Neurosci. 2006;9:723–727 [DOI] [PubMed] [Google Scholar]
- 16. Dupret D, Revest JM, Koehl M, et al. Spatial relational memory requires hippocampal adult neurogenesis. PLoS One. 2008;3:e1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. Molecular and cellular approaches to memory allocation in neural circuits. Science. 2009;326:391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boldrini M, Underwood MD, Hen R, et al. Antidepressants increase neural progenitor cells in the human hippocampus. Neuropsychopharmacology. 2009;34:2376–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czéh B, Lucassen PJ. What causes the hippocampal volume decrease in depression? Are neurogenesis, glial changes and apoptosis implicated? Eur Arch Psychiatry Clin Neurosci. 2007;257:250–260 [DOI] [PubMed] [Google Scholar]
- 20. Bilang-Bleuel A, Ulbricht S, Chandramohan Y, De Carli S, Droste SK, Reul JM. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: involvement in a glucocorticoid receptor-dependent behavioural response. Eur J Neurosci. 2005;22:1691–1700 [DOI] [PubMed] [Google Scholar]
- 21. Chandramohan Y, Droste SK, Reul JM. Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the N-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. J Neurochem. 2007;101:815–828 [DOI] [PubMed] [Google Scholar]
- 22. Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106:20912–20917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hunter RG, Murakami G, Dewell S, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA. 2012;109:17657–17662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Datson NA, Morsink MC, Meijer OC, de Kloet ER. Central corticosteroid actions: search for gene targets. Eur J Pharmacol. 2008;583:272–289 [DOI] [PubMed] [Google Scholar]
- 25. Datson NA, Speksnijder N, Mayer JL, et al. The transcriptional response to chronic stress and glucocorticoid receptor blockade in the hippocampal dentate gyrus. Hippocampus. 2012;22:359–371 [DOI] [PubMed] [Google Scholar]
- 26. Polman JA, Hunter RG, Speksnijder N, et al. Glucocorticoids modulate the mTOR pathway in the hippocampus: differential effects depending on stress history. Endocrinology. 2012;153:4317–4327 [DOI] [PubMed] [Google Scholar]
- 27. Datson NA, Meijer L, Steenbergen PJ, et al. Expression profiling in laser-microdissected hippocampal subregions in rat brain reveals large subregion-specific differences in expression. Eur J Neurosci. 2004;20:2541–2554 [DOI] [PubMed] [Google Scholar]
- 28. Smyth G. Limma: linear models for microarray data. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, New York: Springer; 2005;397–420 [Google Scholar]
- 29. Duncan D, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 2010;11:1–120043860 [Google Scholar]
- 30. Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 32. Bagamasbad P, Ziera T, Borden SA, et al. Molecular basis for glucocorticoid induction of the kruppel-like factor 9 gene in hippocampal neurons. Endocrinology. 2012;153:5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grunewald M, Johnson S, Lu D, et al. Mechanistic role for a novel glucocorticoid-KLF11 (TIEG2) protein pathway in stress-induced monoamine oxidase A expression. J Biol Chem. 2012;287:24195–24206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith MA, Makino S, Kvetnansky R, Post RM. Stress and glucocorticoids affect the expression of brain-derived neurotrophic factor and neurotrophin-3 mRNAs in the hippocampus. J Neurosci. 1995;15:1768–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zahnow CA, Panula P, Yamatodani A, Millhorn DE. Glucocorticoid hormones downregulate histidine decarboxylase mRNA and enzyme activity in rat lung. Am J Physiol. 1998;275:L407–L413 [DOI] [PubMed] [Google Scholar]
- 36. Frank MG, Der-Avakian A, Bland ST, Watkins LR, Maier SF. Stress-induced glucocorticoids suppress the antisense molecular regulation of FGF-2 expression. Psychoneuroendocrinology. 2007;32:376–384 [DOI] [PubMed] [Google Scholar]
- 37. Kelly EJ, Sandgren EP, Brinster RL, Palmiter RD. A pair of adjacent glucocorticoid response elements regulate expression of two mouse metallothionein genes. Proc Natl Acad Sci USA. 1997;94:10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He W, Meng T, Wu M, Shi B, Lu SJ, Li CH. Perturbation of Fgf10 signal pathway in mouse embryonic palate by dexamethasone and vitamin B12 in vivo. J Pediatr Surg. 2010;45:2030–2035 [DOI] [PubMed] [Google Scholar]
- 39. Shimojo M, Hiroi N, Yakushiji F, Ueshiba H, Yamaguchi N, Miyachi Y. Differences in down-regulation of glucocorticoid receptor mRNA by cortisol, prednisolone and dexamethasone in HeLa cells. Endocr J. 1995;42:629–636 [DOI] [PubMed] [Google Scholar]
- 40. Chuquet J, Quilichini P, Nimchinsky EA, Buzsáki G. Predominant enhancement of glucose uptake in astrocytes versus neurons during activation of the somatosensory cortex. J Neurosci. 2010;30:15298–15303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jakoby P, Schmidt E, Ruminot I, Gutiérrez R, Barros LF, Deitmer JW. Higher transport and metabolism of glucose in astrocytes compared with neurons: a multiphoton study of hippocampal and cerebellar tissue slices [published online January 25, 2013]. Cereb Cortex. doi:10.1093/cercor/bhs309 [DOI] [PubMed] [Google Scholar]
- 42. van Gemert NG, Joëls M. Effect of chronic stress and mifepristone treatment on voltage-dependent Ca2+ currents in rat hippocampal dentate gyrus. J Neuroendocrinol. 2006;18:732–741 [DOI] [PubMed] [Google Scholar]
- 43. Lipstein N, Schaks S, Dimova K, et al. Nonconserved Ca(2+)/calmodulin binding sites in Munc13s differentially control synaptic short-term plasticity. Mol Cell Biol. 2012;32:4628–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Upadhya MA, Nakhate KT, Kokare DM, Singru PS, Subhedar NK. Cocaine- and amphetamine-regulated transcript peptide increases spatial learning and memory in rats. Life Sci. 2011;88:322–334 [DOI] [PubMed] [Google Scholar]
- 45. Schmitt U, Hiemke C, Fahrenholz F, Schroeder A. Over-expression of two different forms of the α-secretase ADAM10 affects learning and memory in mice. Behav Brain Res. 2006;175:278–284 [DOI] [PubMed] [Google Scholar]
- 46. Heine VM, Maslam S, Zareno J, Joëls M, Lucassen PJ. Suppressed proliferation and apoptotic changes in the rat dentate gyrus after acute and chronic stress are reversible. Eur J Neurosci. 2004;19:131–144 [DOI] [PubMed] [Google Scholar]
- 47. Selye H. Stress in Health and Disease. Boston, Massachusetts: Butterworths; 1976 [Google Scholar]
- 48. Karst H, Joëls M. Effect of chronic stress on synaptic currents in rat hippocampal dentate gyrus neurons. J Neurophysiol. 2003;89:625–633 [DOI] [PubMed] [Google Scholar]
- 49. Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol. 2012;233:102–111 [DOI] [PubMed] [Google Scholar]
- 50. Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Murgatroyd C, Patchev AV, Wu Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566 [DOI] [PubMed] [Google Scholar]
- 52. Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854 [DOI] [PubMed] [Google Scholar]
- 53. Gutièrrez-Mecinas M, Trollope AF, Collins A, et al. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2-MSK1-Elk-1 signaling. Proc Natl Acad Sci USA. 2011;108:13806–13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Daganzo SM, Erzberger JP, Lam WM, et al. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol. 2003;13:2148–2158 [DOI] [PubMed] [Google Scholar]
- 55. Gregory GD, Vakoc CR, Rozovskaia T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397:161–168 [DOI] [PubMed] [Google Scholar]
- 57. Fessing MY, Mardaryev AN, Gdula MR, et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol. 2011;194:825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Keyes WM, Pecoraro M, Aranda V, et al. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jin M, Wang XM, Tu Y, et al. The negative cell cycle regulator, Tob (transducer of ErbB-2), is a multifunctional protein involved in hippocampus-dependent learning and memory. Neuroscience. 2005;131:647–659 [DOI] [PubMed] [Google Scholar]
- 60. Kerman IA, Bernard R, Bunney WE, et al. Evidence for transcriptional factor dysregulation in the dorsal raphe nucleus of patients with major depressive disorder. Front Neurosci. 2012;6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bezchlibnyk YB, Wang JF, McQueen GM, Young LT. Gene expression differences in bipolar disorder revealed by cDNA array analysis of post-mortem frontal cortex. J Neurochem. 2001;79:826–834 [DOI] [PubMed] [Google Scholar]
- 62. Etain B, Milhiet V, Bellivier F, Leboyer M. Genetics of circadian rhythms and mood spectrum disorders. Eur Neuropsychopharmacol. 2011;21:S676–S682 [DOI] [PubMed] [Google Scholar]
- 63. Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythms 2009;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gibbs JE, Blaikley J, Beesley S, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Seo S, Lomberk G, Mathison A, et al. Krüppel-like factor 11 differentially couples to histone acetyltransferase and histone methyltransferase chromatin remodeling pathways to transcriptionally regulate dopamine D2 receptor in neuronal cells. J Biol Chem. 2012;287:12723–12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meyer JH, Ginovart N, Boovariwala A, et al. Elevated monoamine oxidase a levels in the brain: an explanation for the monoamine imbalance of major depression. Arch Gen Psychiatry. 2006;63:1209. [DOI] [PubMed] [Google Scholar]
- 67. McKernan DP, Dinan TG, Cryan JF. “Killing the Blues”: a role for cellular suicide (apoptosis) in depression and the antidepressant response? Prog Neurobiol. 2009;88:246–263 [DOI] [PubMed] [Google Scholar]
- 68. Gould E, Tanapat P. Stress and hippocampal neurogenesis. Biol Psychiatry. 1999;46:1472–1479 [DOI] [PubMed] [Google Scholar]
- 69. Mirescu C, Gould E. Stress and adult neurogenesis. Hippocampus. 2006;16:233–238 [DOI] [PubMed] [Google Scholar]
- 70. Oomen CA, Mayer JL, de Kloet ER, Joëls M, Lucassen PJ. Brief treatment with the glucocorticoid receptor antagonist mifepristone normalizes the reduction in neurogenesis after chronic stress. Eur J Neurosci. 2007;26:3395–3401 [DOI] [PubMed] [Google Scholar]
- 71. Gray JD, Milner TA, McEwen BS. Dynamic plasticity: the role of glucocorticoids, brain-derived neurotrophic factor and other trophic factors. Neuroscience. 2013;239:214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feldker DE, Morsink MC, Veenema AH, et al. The effect of chronic exposure to highly aggressive mice on hippocampal gene expression of non-aggressive subordinates. Brain Res. 2006;1089:10–20 [DOI] [PubMed] [Google Scholar]
- 73. Albensi BC, Mattson MP. Evidence for the involvement of TNF and NF-κB in hippocampal synaptic plasticity. Synapse. 2000;35:151–159 [DOI] [PubMed] [Google Scholar]
- 74. You Z, Luo C, Zhang W, et al. Pro- and anti-inflammatory cytokines expression in rat's brain and spleen exposed to chronic mild stress: involvement in depression. Behav Brain Res. 2011;225:135–141 [DOI] [PubMed] [Google Scholar]
- 75. Koo JW, Duman RS. IL-1β is an essential mediator of the antineurogenic and anhedonic effects of stress. Proc Natl Acad Sci USA. 2008;105:751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Monje ML, Toda H, Palmer TD. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765 [DOI] [PubMed] [Google Scholar]
- 77. Goshen I, Kreisel T, Ben-Menachem-Zidon O, et al. Brain interleukin-1 mediates chronic stress-induced depression in mice via adrenocortical activation and hippocampal neurogenesis suppression. Mol Psychiatry. 2008;13:717–728 [DOI] [PubMed] [Google Scholar]
- 78. Khairova RA, Machado-Vieira R, Du J, Manji HK. A potential role for pro-inflammatory cytokines in regulating synaptic plasticity in major depressive disorder. Int J Neuropsychopharmacol. 2009;12:561–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bekinschtein P, Katche C, Slipczuk LN, et al. mTOR signaling in the hippocampus is necessary for memory formation. Neurobiol Learn Mem. 2007;87:303–307 [DOI] [PubMed] [Google Scholar]
- 80. Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li N, Lee B, Liu RJ, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chandran A, Iyo AH, Jernigan CS, Legutko B, Austin MC, Karolewicz B. Reduced phosphorylation of the mTOR signaling pathway components in the amygdala of rats exposed to chronic stress. Prog Neuropsychopharmacol Biol Psychiatry. 2012;40C:240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jernigan CS, Goswami DB, Austin MC, et al. The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1774–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]