Abstract
Desulfovibrio vulgaris Hildenborough strains with significantly increased tolerance to NaCl were obtained via experimental evolution. A NaCl-evolved strain, ES9-11, isolated from a population cultured for 1200 generations in medium amended with 100 mM NaCl, showed better tolerance to NaCl than a control strain, EC3-10, cultured for 1200 generations in parallel but without NaCl amendment in medium. To understand the NaCl adaptation mechanism in ES9-11, we analyzed the transcriptional, metabolite and phospholipid fatty acid (PLFA) profiles of strain ES9-11 with 0, 100- or 250 mM-added NaCl in medium compared with the ancestral strain and EC3-10 as controls. In all the culture conditions, increased expressions of genes involved in amino-acid synthesis and transport, energy production, cation efflux and decreased expression of flagellar assembly genes were detected in ES9-11. Consistently, increased abundances of organic solutes and decreased cell motility were observed in ES9-11. Glutamate appears to be the most important osmoprotectant in D. vulgaris under NaCl stress, whereas, other organic solutes such as glutamine, glycine and glycine betaine might contribute to NaCl tolerance under low NaCl concentration only. Unsaturation indices of PLFA significantly increased in ES9-11. Branched unsaturated PLFAs i17:1 ω9c, a17:1 ω9c and branched saturated i15:0 might have important roles in maintaining proper membrane fluidity under NaCl stress. Taken together, these data suggest that the accumulation of osmolytes, increased membrane fluidity, decreased cell motility and possibly an increased exclusion of Na+ contribute to increased NaCl tolerance in NaCl-evolved D. vulgaris.
Keywords: D. vulgaris, experimental evolution, salt adaptation, transcriptomics, metabolites assay, PLFA analysis
Introduction
Desulfovibrio vulgaris is a member of the dissimilatory sulfate-reducing bacteria (SRB) widely distributed in anaerobic environments, such as gas pipelines, subsurface tanks, sediments and off-shore petroleum production facilities (Postgate, 1984). Much research has focused on D. vulgaris as a model to explore the SRB that have important roles in the biogeochemical cycling of sulfur, carbon and nitrogen and potentially the bioremediation of toxic heavy metals and radionuclides. In addition, SRB have been identified as culprits in the biocorrosion of ferrous metal installations in the petroleum industry and concrete structures in wastewater collection systems (Muyzer and Stams, 2008; Zhou et al., 2011). Effective reduction of highly toxic Cr (VI) or U (VI) to less toxic Cr (III) or U (IV) by D. vulgaris Hildenborough has been reported (Lovley et al., 1993; Lovley and Phillips, 1994; Elias et al., 2004), and D. vulgaris was found to exist in uranium (VI) contaminated field site (Chang et al., 2001). High NaCl concentration is also present in several DOE (Department of Energy) sites contaminated with toxic metals. Therefore, evolving NaCl-tolerant D. vulgaris strains and identifying the NaCl adaptation mechanisms could have important consequences for bioremediation strategies and the prediction of biocorrosion for the petroleum industry.
Several aspects of the NaCl stress responses in microorganisms have been studied. Intracellular accumulation of cocktails of organic solutes has been shown to be one of the major strategies to cope with osmotic stress caused by high concentrations of NaCl (Roberts, 2005). In the model Gram-negative bacterium, Escherichia coli, glutamate (Glu), proline, glycine betaine (GB), ectoine and trehalose accumulated during osmotic stress (Jebbar et al., 1992; Strøm and Kaasen, 1993; McLaggan et al., 1994). Glu was found to be the primary osmolyte in other microorganisms, such as Salmonella typhimurium (Botsford et al., 1994) and Rhizobium meliloti (Botsford and Lewis, 1990). GB and proline have been reported to be important osmolytes in Listeria monocytogenes (Ko et al., 1994) and Bacillus subtilis (Whatmore et al., 1990), respectively. GB and trehalose were the major compatible solutes in the moderately halophilic SRB Desulfovibrio halophilus (Welsh et al., 1996). In D. vulgaris, GB and ectoine were found to be important for relieving NaCl stress caused by exposure to 250 mM NaCl for 4 h (Mukhopadhyay et al., 2006). By contrast, accumulation of Glu (eightfold) and alanine (Ala, 1.8-fold) was detected in D. vulgaris exposed to 250-mM NaCl stress for about 100 h (He et al., 2010). However, the osmoprotectants that counteract the osmotic stress caused by NaCl during evolutionary adaptation of D. vulgaris are unknown. Other NaCl tolerance mechanisms include changes of membrane lipid composition to compensate for the decrease in membrane fluidity. Levels of unsaturated fatty acids in membrane lipids increased in B. subtilis under osmotic stress (López et al., 2000; Los and Murata, 2004). An increase of branched fatty acids in Gram-positive halophilic and halotolerant bacteria or unsaturated fatty acids in Gram-negative and Gram-positive halophilic bacteria increases the membrane fluidity when grown in media with increasing salt concentrations (Kates, 1986; Russell, 1989). In D. vulgaris, an increase of unsaturated branched fatty acid i17:ω9c was detected in response of D. vulgaris to NaCl shock (Mukhopadhyay et al., 2006). Any changes in phospholipid fatty acid (PLFA) composition during evolutionary NaCl adaptation of D. vulgaris remain to be determined.
Salt-stress tolerance is a polygenic trait especially attractive for evolutionary studies (Dhar et al., 2011). Considering the complexity of interactions between natural environments and microorganisms, it is challenging to understand how microorganisms respond to stresses and then adapt genetically. The response of D. vulgaris to NaCl shock (Mukhopadhyay et al., 2006) and 100-h exposure (He et al., 2010) have been reported. In addition, experimental evolution of microorganisms under controlled laboratory conditions have been established (Elena and Lenski, 2003). Therefore, we aimed to reveal the possible adaptation mechanism of D. vulgaris developed through experimental evolution under NaCl stress by transcriptomic, metabolite and PLFA analyses.
Towards this goal, NaCl-adapted strains of D. vulgaris was obtained after culturing for 1200 generations in medium amended with 100 mM NaCl. Examination of single-colony isolates from the NaCl-evolved cultures indicated that the accumulation of organic osmolytes, increased membrane fluidity, increased exclusion of Na+ and decreased cell motility contribute to the increased NaCl tolerance.
Materials and methods
Bacterial strains and growth conditions
A clonal isolate of D. vulgaris Hildenborough (ATCC 29579) was used as the ancestor (An) to found 12 populations for experimental evolution. Six populations (EC, population Nos. 1–6) were subsequently propagated in control environment (defined medium LS4D with 60 mM lactate as electron donor and 50 mM sulfate as electron acceptor, no supplemented NaCl, total [Na+] is about 210 mM; Mukhopadhyay et al., 2006) and six populations (ES, population Nos. 7–12) propagated in a constant NaCl stress environment (LS4D with additional 100 mM NaCl, total [Na+] is about 310 mM) for 1200 generations. The cell cultures were grown at 37 °C anaerobically and serially transferred every 48 h with a 1–100 dilution. Single-colony isolates (strains designated as EC3-10 and ES9-11) from the evolved populations EC No.3 or ES No.9, respectively, were obtained by plating and single-colony isolation.
Analysis of growth data
Growth phenotypes of the ancestral strain (An) and evolved strains ES9-11 and EC3-10 in control medium LS4D or NaCl stress medium (LS4D+250 mM NaCl) were examined with three replicates for each strain. Growth rate and yield were obtained independently from the growth curve of each replicate. Yield was the maximum OD600 (optical density measured at the wavelength of 600 nm) reached by the culture and the growth rate was 2.303 × the slope of the linear portion of the growth curve obtained by plotting log10(OD600) with time.
Biomass production
Freezer stocks of the ancestral strain An and the evolved strains, ES9-11 and EC3-10, were inoculated into 10 ml LS4D with 1–100 dilution and grown to stationary phase. These cultures (1 ml) were then inoculated into the production vessels containing 100 ml of LS4D supplemented with different concentrations of NaCl (0, 100 or 250 mM). All samples were prepared in triplicates. The mid-exponential phase cultures (OD600∼0.4 with exception of OD600∼0.25 for the An strain grown in LS4D+250 mM NaCl) were harvested by centrifugation at 6000 g for 10 min at 4 °C with 50 ml cell culture in each Falcon tube and the biomass was frozen in liquid nitrogen and kept at −80 °C for transcript and metabolite assays.
Isolation of total RNA, genomic DNA and fluorescence labeling
Isolation, purification and fluorescence labeling of total cellular RNA were performed as described previously (Chhabra et al., 2006). Genomic DNA was isolated from D. vulgaris with CTAB method (Zhou et al., 1996). Total RNA and genomic DNA were labeled with fluorescent dye Cy5 or Cy3, respectively. Cy5-labeled cDNA and Cy3-labeled gDNA were dried and stored at −20 °C before hybridization.
Microarray hybridization and data analysis
The D. vulgaris whole-genome oligonucleotide (70mer) microarray covering 3482 of the 3531 protein-coding sequences of the D. vulgaris genome (He et al., 2006) was used for transcriptomics analysis. Array hybridizations were carried out with TECAN hybridization station (TECAN HS4800, TECAN Group Ltd, Durham, NC, USA). Array hybridization and data analysis were performed as described previously (Mukhopadhyay et al., 2006; Zhou et al., 2010). Microarray data have been deposited in the NCBI GEO database under accession number GSE39675.
The similarities of transcriptional profiling among different samples were analyzed with detrended correspondence analysis and AnoSim. The transcript level of each open reading frame was calculated as the ratio of signal intensity of Cy5 to Cy3, and the genes detected in at least two out of three replicates were kept for statistical analysis.
Extraction and analysis of metabolites by LC-MS (liquid chromatography-mass spectrometry) or CE-MS (capillary electrophoresis-mass spectrometry)
Metabolites were extracted via a methanol/water/chloroform extraction procedure and analyzed with CE-MS and LC-MS (Baidoo et al., 2008; He et al., 2010). All solvents were of HPLC (high-performance liquid chromatography) or greater grade (Honeywell Burdick and Jackson, Muskegon, MI, USA). Formic acid (>98% v/v) was purchased from Riedel-de Haën (Seelze, Germany). Chemical standards for amino acids were purchased from Sigma-Aldrich (St Louis, MO, USA). The stock solutions (200 μM) for amino acids were prepared with methanolwater (50:50, v/v). The lyophilized extracts from freeze-dried biomass (pellet from 50 ml of cell culture with OD600∼0.4 as described above) were reconstituted in 500 μl of methanol–water (50:50, v/v) solution or diluted to ensure that metabolite concentrations were within the ranges of the calibration curves.
Cell motility assay
Mid-exponential phase cells (5 μl, OD600∼0.4) of the ancestral strain An and the evolved strains, ES9-11 and EC3-10 (precultured from glycerol stocks and grown for one growth cycle with LS4D as described above), were applied on the surface of soft agar (0.4% (w/v)) plates (LS4D or LS4D+250 mM NaCl). The plates were cultivated at 37 °C anaerobically for 4 days and the colony diameters were measured.
PLFA assays
Pellets from 50 ml of late-exponential phase cultures were collected and frozen in liquid nitrogen. The PLFA assay was conducted by Microbial ID Inc. (Newark, DE, USA). Briefly, fatty acids were extracted and methylated and then analyzed on a gas chromatograph equipped with a flame ionization detector. Peaks were named by comparing the calibration mixture with Sherlock software (Microbial ID Inc.). Unsaturation index (UI) of PLFAs was calculated as UI=((C:1 × 1)+(C:2 × 2)+(C:3 × 3)+(C:4 × 4))/100, where C:1, C:2, C:3 and C:4 represent fatty acid's proportion (%) with 1, 2, 3 and 4 double bonds, respectively) (Ruess et al., 2007).
Results
Increased NaCl tolerance in evolved D. vulgaris
An additional 100 mM NaCl in growth medium LS4D (containing ca. 210 mM Na+) slightly decreased the final biomass yield and extended the lag phase of D. vulgaris. By contrast, a significant decrease of final biomass yield (about 50%) and growth rate were observed when grown in LS4D amended with 250 mM NaCl (He et al., 2010). We interpreted these observations to define the 100-mM NaCl amendment to be a low stress and 250-mM NaCl, a high stress condition. Therefore, LS4D+100 mM NaCl was used as the stress condition for experimental evolution and LS4D+250 mM NaCl was used for evaluation of NaCl tolerance in evolved D. vulgaris.
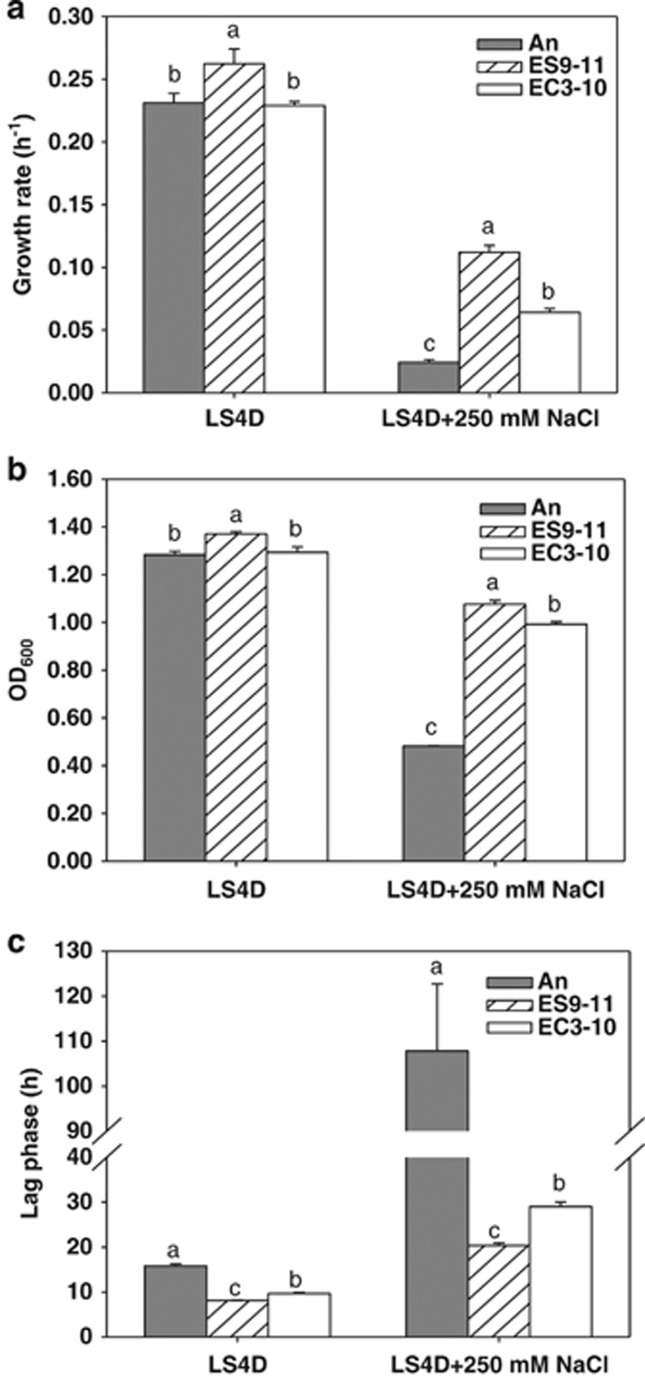
Significant improvement of tolerance to NaCl was observed in evolved D. vulgaris populations that had been cultured in unamended medium before challenging with 250 mM NaCl (Supplementary Figure S1). In order to identify the adaptation mechanisms underlying the improved tolerance phenotype, single-colony isolates ES9-11 from population ES No.9 evolved in LS4D+100 mM NaCl and EC3-10 from population EC No.3 evolved in LS4D were further investigated. The strains were each chosen as isolates with the best growth performance in LS4D+250 mM NaCl from 15 randomly picked colonies from their respective populations. As shown in Figure 1 and Supplementary Figure S2, in LS4D+250 mM NaCl, significant increases (P<0.0001) of growth rate and final biomass yield, but shortened lag phase, were observed for evolved strains compared with the ancestral strain, An, which had been maintained as a freezer stock. The growth rate of NaCl-evolved ES9-11 increased about fourfold (Figure 1a), and the final biomass yield was about twofold than that of ancestral An (Figure 1b), which were also significantly higher than that of control-evolved EC3-10 (growth rate, P<0.0001; final biomass, P=0.0022; Figures 1a and b). The lag phase (20.3±0.6 h) in ES9-11 was significantly shorter than in EC3-10 (29.0±1.0 h, P=0.0002) and in An (107.8±14.9 h, P=0.0005) (Figure 1c). Similarly, in LS4D, better growth of ES9-11 was observed with significantly increased growth rate, final biomass and shortened lag phase compared with An (P-values 0.0213, 0.0022 and <0.0001, respectively) or EC3-10 (P-values 0.0065, 0.0034 and 0.0009 respectively). By contrast, the growth of EC3-10 was similar to An except for a significantly (P<0.0001) shortened lag phase (Figure 1). These data demonstrated that strain ES9-11 evolved under NaCl stress appeared to be more tolerant to a challenge of higher NaCl than strain EC3-10 that was evolved in medium without NaCl amendment.
Figure 1.
Improvement of NaCl tolerance in evolved D. vulgaris strains. The growth rates (a), biomass yields (b) and lag phases (c) of NaCl-evolved ES9-11, control-evolved EC3-10 and ancestral strain An in standard defined medium (LS4D) and high NaCl stress (LS4D+250 mM NaCl) are shown. Averages of three biological replicates are shown. Error bars indicate s.d. The significance of differences among three strains is shown at the P<0.05 level (t-test).
Transcriptional profiling changes in evolved D. vulgaris
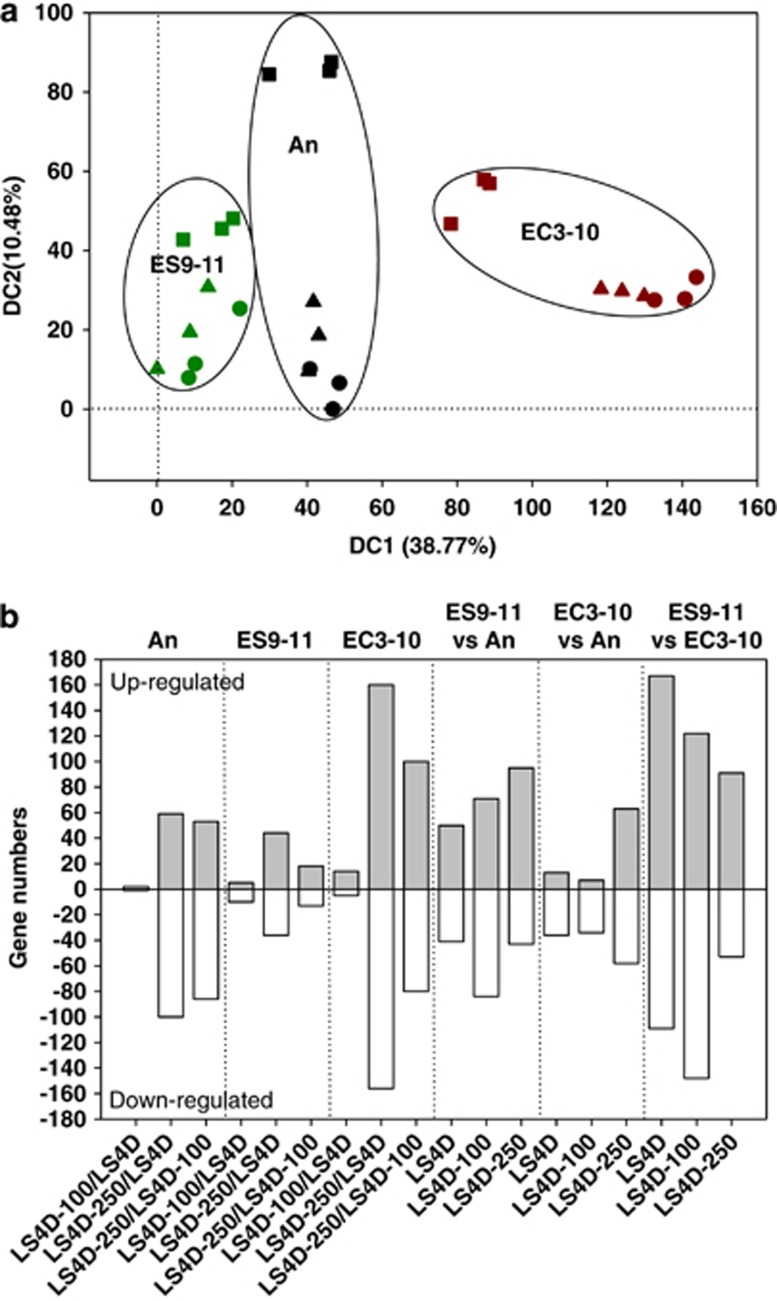
The significant improvement of NaCl tolerance in NaCl-evolved ES9-11 encouraged us to explore the gene transcription changes. We hypothesized that genetic changes accumulated during evolution will be reflected in gene transcription profiling. The overall similarity of the transcription profiling among ancestral strain, NaCl-evolved ES9-11 and control-evolved EC3-10 is presented using a detrended correspondence analysis plot (Figure 2a). The three strains are separated by Axis 1, which explained 38.77% of the total variance. Low NaCl stress did not stimulate substantial changes in any of the strains, but the additional 250 mM NaCl in the medium induced the largest changes in the transcriptional profile of the ancestral strain, intermediate changes in the control-evolved EC3-10 and minimal changes in the NaCl-evolved ES9-11. Statistical tests with AnoSim demonstrated the significant differences among the three strains: An vs ES9-11 (R=0.876, P=0.001), An vs EC3-10 (R=0.592, P=0.001), and two evolved strains ES9-11 and EC3-10 (R=0.807, P=0.001).
Figure 2.
Distinct gene transcriptional profiling of evolved D. vulgaris strains. (a) The gene transcriptional profiling of NaCl-evolved ES9-11, control-evolved EC3-10 and ancestral strain An in medium with or without NaCl amendment are separated on detrended correspondence analysis plot. Each symbol represents a replicate sample. Circle, LS4D; triangle, LS4D+100 mM NaCl; square, LS4D+250 mM NaCl. (b) Numbers of differentially expressed genes (with cutoff values for log2 R(treatment/control) and Z score as 1.5) in medium with or without NaCl amendment. LS4D-100, LS4D+100 mM NaCl; LS4D-250, LS4D+250 mM NaCl.
The number of genes with significant changes (cutoff values for log2 R(treatment/control) and Z score were 1.5) also demonstrated significant differences among the three strains. As shown in Figure 2b, for strain An, 100-mM NaCl stress had almost no affect on transcription, whereas 250-mM NaCl stress had a major impact. A similar result, but dampened, was seen for the NaCl-evolved ES9-11, while a similar result but exaggerated was seen for the control-evolved EC3-10. Consistent with the minimal changes induced by 250-mM NaCl stress (Figure 2a), the smallest number of differentially expressed genes was detected in ES9-11 under 250-mM NaCl stress. Compared with An, in all the culture conditions, there were more differentially expressed genes in ES9-11 than in EC3-10. In addition, greater numbers of differentially expressed genes were found in ES9-11 compared with EC3-10 in all the culture conditions (Figure 2b). The results indicated that there were more gene expression changes in NaCl-evolved ES9-11 than in control-evolved EC3-10 under no-NaCl stress or NaCl stress conditions. Most of the differentially expressed genes in ES9-11 belonged to the functional categories of amino-acid metabolism and transport, inorganic ion transport, cell motility, energy production and conversion, signal transduction mechanisms and transcription as described below (Tables 1, 2, 3).
Table 1. Expression changes of genes involved in amino-acid synthesis and transport or inorganic ion transport and metabolism in strain ES.
| DVU Nos. | Annotation |
LS4D |
LS4D-100 |
LS4D-250 |
|||
|---|---|---|---|---|---|---|---|
| ES vs An | ES vs EC | ES vs An | ES vs EC | ES vs An | ES vs EC | ||
| Amino-acid synthesis and transport | |||||||
| DVU0593 | L-lysine exporter, putative | 2.75 (4.13) | 2.39 (3.96) | 2.36 (4.36) | 1.48 (2.87) | 4.29 (2.60) | 2.51 (3.54) |
| DVU2615 | Bacterial extracellular solute-binding protein, family 3 | 2.45 (4.20) | 3.28 (4.70) | 2.34 (4.34) | 3.78 (5.72) | 3.51 (2.43) | 4.16 (4.59) |
| DVU1378 | Ketol-acid reductoisomerase (ilvC) | 0.05 (0.08) | 0.18 (0.28) | 0.45 (0.73) | 0.41 (0.66) | 1.10 (2.07) | 1.26 (2.36) |
| DVU3291 | Glutamate synthase, iron–sulfur cluster-binding subunit, putative | 0.94 (0.99) | −0.49 (−0.59) | 0.99 (0.81) | −0.30 (−0.27) | 1.43 (1.42) | 0.77 (0.71) |
| DVU2341 | Amino-acid ABC transproter, permease protein, His/Glu/Gln/Arg/opine family | 0.99 (1.89) | −0.08 (−0.15) | 1.12 (2.13) | 0.01 (0.02) | 3.12 (1.89) | 1.53 (2.77) |
| DVU2342 | Amino-acid ABC transporter, periplasmic amino-acid-binding protein | 0.57 (0.97) | 0.18 (0.3) | 0.96 (1.86) | 0.09 (0.18) | 2.19 (3.40) | 1.50 (2.64) |
| DVU2343 | Amino-acid ABC transporter, ATP-binding protein, glnQ | −0.24 (−0.35) | NA(NA) | 1.23 (1.98) | 0.77 (1.36) | 1.07 (1.98) | NA |
| DVU2740 (livF) | High-affinity branched-chain amino-acid ABC transporter, ATP-binding protein | NA | NA | 0.15 (0.16) | 0.17 (0.16) | 0.69 (1.06) | 0.20 (0.18) |
| DVU2741 (livG) | High-affinity branched-chain amino-acid ABC transporter, ATP-binding protein | −0.14 (−0.21) | 0.04 (0.05) | 1.04 (1.06) | 0.36 (0.38) | 0.51 (0.72) | 0.20 (0.29) |
| DVU2742 (livM) | High-affinity branched-chain amino-acid ABC transporter, permease protein | 0.25 (0.31) | 1.05 (1.56) | 0.46 (0.71) | 0.79 (1.22) | 0.93 (0.96) | 0.33 (0.42) |
| DVU2743 (livH) | High-affinity branched-chain amino-acid ABC ransporter, permease protein | −0.35 (−0.3) | −1.24 (−1.33) | −0.80 (−1.36) | −1.83 (−2.55) | 1.47 (0.93) | 1.55 (1.06) |
| DVU2744 | High-affinity branched-chain amino-acid ABC transporter, perisplasmic amino-acid-binding protein | 0.48 (0.83) | 0.10 (0.16) | 0.14 (0.24) | −0.12 (−0.20) | 2.26 (2.16) | 0.89 (1.41) |
| DVU1827 | Acetylornithine deacetylase/succinyl-diaminopimelate desuccinylase family protein | −0.48 (−0.79) | −0.76 (−1.28) | −1.05 (−1.48) | −1.35 (−1.87) | −1.14 (−1.82) | −0.45 (−0.65) |
| DVU0115 | Shikimate 5-dehydrogenase (aroE) | −0.28 (−0.25) | −0.04 (−0.04) | 0.20 (0.17) | 0.25 (0.20) | −1.45 (−1.55) | −1.16 (−0.85) |
| DVU2492 | N-(5-phosphoribosyl)anthranilate isomerase (trpF-2) | −0.08 (−0.10) | 0.20 (0.25) | −0.31 (−0.29) | −0.52 (−0.52) | −1.52 (−1.52) | −1.27 (−1.46) |
| DVU3223 | Aspartate aminotransferase (aspB) | −0.51 (−0.82) | −1.08 (−1.82) | −0.38 (−0.47) | −0.71 (−0.88) | −1.40 (−1.59) | −0.46 (−0.42) |
| Inorganic ion transport and metabolism | |||||||
| DVU2571 | Ferrous iron transport protein B (feoB) | 2.26 (4.11) | 2.28 (4.14) | 2.69 (4.82) | 2.73 (4.77) | −0.25 (−0.31) | 2.98 (4.11) |
| DVU2572 | Ferrous iron transport protein A, putative | 1.45 (2.56) | 2.26 (3.99) | 1.87 (3.08) | 2.30 (3.69) | −0.58 (−0.71) | 2.61 (2.99) |
| DVU2573 | Hypothetical protein | 1.39 (2.59) | 2.42 (4.59) | 1.52 (2.56) | 2.18 (3.75) | 0.05 (0.09) | 1.85 (2.95) |
| DVU2574 | Ferrous ion transport protein, putative | 1.16 (2.16) | 2.65 (4.58) | 1.17 (2.03) | 2.00 (3.54) | 0.37 (0.54) | 1.42 (2.63) |
| DVU1778 | Cation efflux family protein | 0.80 (1.50) | 0.96 (1.75) | 0.91 (1.71) | 1.04 (1.97) | 2.18 (2.45) | 0.91 (1.67) |
| DVU2477 | Phosphate ABC transporter, periplasmic phosphate-binding protein, putative | −1.81 (−2.37) | −2.20 (−2.83) | −0.56 (−0.39) | 0.20 (0.14) | −0.89 (−0.61) | −0.32 (−0.36) |
| DVU2478 | Phosphate ABC transporter, permease protein, putative | −1.05 (−0.95) | −0.73 (−0.57) | −2.21 (−3.15) | −0.85 (−0.96) | −0.08 (−0.06) | −0.16 (−0.17) |
| DVU2479 | Phosphate ABC transporter, permease protein, putative | −0.38 (−0.50) | −0.74 (−1.06) | 0.25 (0.23) | −0.22 (−0.19) | −0.76 (−1.04) | −0.45 (−0.65) |
Abbreviations: An, ancestor DvH; ATP, adenosine triphosphate; DVU, Desulfovibrio vulgaris; EC, strain isolated from population No.3 evolved under control condition; ES, strain isolated from population No.9 evolved under salt-stress condition; LS4D-100, LS4D+100 mM NaCl; LS4D-250, LS4D+250 mM NaCl; NA, not applicable.
Log2R ratios are shown with Z score in parentheses. Boldface indicates more than twofolds of expression changes.
Table 2. Expression changes of selected genes involved in cell motility in strain ES.
| DVU Nos. | Annotation |
LS4D |
LS4D-100 |
LS4D-250 |
|||
|---|---|---|---|---|---|---|---|
| ES vs An | ES vs EC | ES vs An | ES vs EC | ES vs An | ES vs EC | ||
| DVU0170 | Methyl-accepting chemotaxis protein | 2.20 (4.27) | 3.38 (4.58) | 1.51 (2.83) | 2.85 (4.00) | 2.54 (2.53) | 2.74 (3.34) |
| DVU3155 (dcrH) | Methyl-accepting chemotaxis protein DcrH | 1.87 (3.62) | 2.09 (2.96) | 1.79 (3.31) | 2.66 (4.21) | 2.64 (2.03) | 3.80 (5.67) |
| DVU3182 (dcrA) | Methyl-accepting chemotaxis protein DcrA | 1.00 (1.61) | 0.73 (1.23) | 0.95 (1.59) | 0.78 (1.31) | 2.10 (1.61) | 1.77 (1.56) |
| DVU1904 (cheW-2) | Chemotaxis protein CheW | 0.86 (1.61) | 1.63 (2.85) | 0.59 (1.10) | 1.43 (2.48) | 1.66 (3.03) | 0.99 (1.83) |
| DVU1263 | Type IV prepilin-like proteins leader peptidase (pppA) | 1.28 (2.30) | 1.46 (2.80) | 1.13 (2.14) | 1.35 (2.48) | 2.54 (1.55) | 2.45 (2.10) |
| DVU0307 | Flagella basal body rod domain protein | −2.20 (−2.22) | −2.57 (−2.60) | −2.03 (−2.36) | −2.58 (−3.06) | −2.48 (−3.54) | −3.44 (−4.94) |
| DVU2444 | Flagellin | −4.54 (−7.12) | −4.91 (−7.55) | −4.50 (−5.34) | −4.99 (−5.84) | −3.85 (−6.07) | −5.00 (−8.11) |
| DVU2445 | Hypothetical protein | −4.20 (−6.79) | −4.59 (−7.23) | −3.69 (−4.94) | −4.47 (−5.66) | −3.54 (−3.64) | −4.79 (−4.94) |
| DVU2948 | Bacterial flagellin N-terminal domain protein | −1.92 (−2.04) | −2.82 (−3.24) | −2.02 (−2.68) | −2.60 (−3.44) | −2.01 (−1.38) | −3.11 (−4.60) |
| DVU1441 | Flagellin | −2.13 (−2.19) | −2.30 (−2.37) | −1.86 (−2.00) | −2.19 (−2.36) | −3.16 (−4.80) | −3.26 (−4.83) |
| DVU1442 | Flagellin FlaG, putative | −0.59 (−1.14) | −0.93 (−1.64) | −1.04 (−1.04) | −1.47 (−1.53) | −1.56 (−1.02) | −1.76 (−1.18) |
| DVU1443 (flgE) | Flagellar hook protein FlgE | −1.68 (−1.55) | −2.29 (−2.14) | −2.70 (−1.90) | −3.14 (−2.23) | −3.63 (−2.73) | −4.18 (−3.16) |
| DVU0310 (fliI) | Flagellum-specific ATP synthase FliI | −0.27 (−0.45) | −0.61 (−1.07) | −0.78 (−1.09) | −0.92 (−1.43) | −0.31 (−0.26) | −1.23 (−1.55) |
| DVU0311 | Flagellar assembly protein FliH, putative | −0.53 (−0.78) | −0.58 (−0.99) | −0.54 (−0.57) | −0.86 (−0.92) | −0.94 (−1.01) | −0.66 (−0.51) |
| DVU0312 (fliG) | Flagellar motor switch protein FliG | −0.92 (−1.40) | −0.80 (−1.17) | −0.24 (−0.32) | −0.17 (−0.22) | −0.69 (−0.96) | −0.7 (−0.99) |
| DVU0313 (fliF) | Flagellar M-ring protein FliF | −1.35 (−1.38) | −1.83 (−1.86) | −0.75 (−0.68) | −1.05 (−0.96) | −0.69 (−1.19) | −0.91 (−1.64) |
| DVU0314 (fliE) | Flagellar basal body component FliE | −1.00 (−1.12) | −1.55 (−1.69) | −1.17 (−1.4) | −1.31 (−1.59) | −1.63 (−2.56) | −1.15 (−1.53) |
| DVU0315 (flgC) | Flagellar basal-body rod protein FlgC | −1.55 (−2.02) | −1.98 (−2.43) | −1.59 (−1.51) | −1.65 (−1.55) | −2.13 (−2.21) | −2.07 (−2.10) |
| DVU0316 (flgB) | Flagellar basal-body rod protein FlgB | −0.20 (−0.32) | 0.02 (0.03) | 0.25 (0.37) | 0.26 (0.37) | −0.29 (−0.37) | −0.79 (−0.93) |
| DVU0512 | Flagellar basal-body rod protein, putative | −0.24 (−0.30) | −0.61 (−0.76) | 0.35 (0.48) | 0.27 (0.37) | −2.08 (−2.01) | −1.85 (−1.78) |
| DVU0513 (flgG) | Flagellar basal-body rod protein FlgG | −0.25 (−0.33) | −1.32 (−2.05) | −1.61 (−1.20) | −1.71 (−1.26) | −1.66 (−0.93) | −1.94 (−1.16) |
| DVU0514 | FlgA family protein | 0.37 (0.48) | 0.15 (0.19) | 0.49 (0.51) | 0.23 (0.23) | −1.26 (−1.01) | −0.88 (−0.63) |
| DVU0515 (flgH) | Flagellar L-ring protein FlgH | −0.56 (−1.00) | −0.73 (−1.3) | −0.78 (−1.19) | −0.57 (−0.86) | −1.46 (−2.32) | −1.58 (−2.49) |
| DVU0516 (flgI) | Flagellar P-ring protein FlgI | −0.46 (−0.68) | −0.22 (−0.36) | −0.68 (−1.08) | −0.94 (−1.37) | −1.76 (−2.05) | −2.15 (−1.85) |
| DVU0517 | Peptidase, M23/M37 family | −1.87 (−2.13) | −2.43 (−2.68) | −1.35 (−1.75) | −2.10 (−2.73) | −1.27 (−0.83) | −2.32 (−2.19) |
| DVU0518 | Hypothetical protein | −3.06 (−3.97) | −3.67 (−4.77) | −2.91 (−3.9) | −3.46 (−4.96) | −2.86 (−1.88) | −4.41 (−5.72) |
| DVU0519 | Flagellar hook-associated protein FlgK, putative | −1.79 (−3.14) | −1.76 (−3.16) | −1.28 (−1.47) | −1.26 (−1.40) | −1.85 (−1.56) | −1.99 (−1.69) |
| DVU0520 | Flagellar hook-associated protein FlgL, putative | −0.74 (−0.83) | −1.31 (−1.48) | −0.25 (−0.27) | −0.6 (−0.67) | −0.15 (−0.11) | −1.30 (−1.90) |
| DVU0521 (csrA) | Carbon storage regulator | −0.14 (−0.26) | −0.16 (−0.29) | −0.35 (−0.64) | −0.26 (−0.47) | 0.45 (0.71) | −0.48 (−0.87) |
| DVU0522 | Conserved hypothetical protein | −0.51 (−0.92) | −0.52 (−0.94) | −0.06 (−0.09) | −0.23 (−0.4) | 0.21 (0.37) | −0.25 (−0.36) |
| DVU0523 (flgM) | Negative regulator of flagellin synthesis FlgM | −1.60 (−2.61) | −1.14 (−1.96) | −2.08 (−1.93) | −1.99 (−1.85) | −0.62(−0.48) | −1.69 (−1.43) |
| DVU0524 | Hypothetical protein | −1.85 (−3.23) | −1.41 (−2.28) | −1.18 (−2.08) | −1.78 (−3.40) | −1.17 (−1.95) | −2.19 (−3.46) |
Abbreviations: An, ancestor DvH; ATP, adenosine triphosphate; DVU, Desulfovibrio vulgaris; EC, strain isolated from population No.3 evolved under control condition; ES, strain isolated from population No.9 evolved under salt stress condition; LS4D-100, LS4D+100 mM NaCl; LS4D-250, LS4D+250 mM NaCl.
Log2R ratios are shown with Z score in parentheses. Boldface indicates more than twofolds of expression changes.
Table 3. Expression changes of genes involved in energy production and conversion or signal transduction and transcription in strain ES.
| DVU Nos. | Annotation |
LS4D |
LS4D-100 |
LS4D-250 |
|||
|---|---|---|---|---|---|---|---|
| ES vs An | ES vs EC | ES vs An | ES vs EC | ES vs An | ES vs EC | ||
| DVU3107 | Cytochrome c family protein | 1.17 (2.06) | 2.08 (2.77) | 1.20 (1.82) | 2.21 (3.13) | 1.47 (1.72) | 2.35 (3.75) |
| DVU0624 | NapC/NirT cytochrome c family protein | 0.34 (0.64) | 0.54 (1.05) | 0.56 (1.09) | 0.61 (1.16) | 1.00 (1.97) | 0.24 (0.47) |
| DVU0625 | Cytochrome c nitrite reductase, catalytic subunit NfrA, putative | 0.16 (0.27) | 0.58 (0.93) | 0.25 (0.38) | 0.34 (0.56) | 1.35 (1.97) | 1.06 (1.63) |
| DVU0702 | Cytochrome c family protein | 0.16 (0.3) | 0.43 (0.78) | −0.06 (−0.08) | 0.21 (0.32) | 1.24 (1.85) | 0.02 (0.03) |
| DVU1817 | Cytochrome c-553 (cyf) | 0.52 (0.93) | 0.69 (1.23) | 0.68 (1.18) | 0.88 (1.61) | 1.31 (2.51) | 0.48 (0.90) |
| DVU0253 | Oxidoreductase, FAD/iron–sulfur cluster-binding domain protein | 0.72 (1.28) | 2.24 (3.75) | 0.56 (0.92) | 2.14 (3.8) | 1.30 (2.22) | 1.31 (1.96) |
| DVU2645 | Na+/H+ antiporter family protein | 1.53 (1.96) | 1.30 (1.29) | 1.28 (1.16) | 0.79 (0.63) | 0.22 (0.16) | −0.29 (−0.20) |
| DVU2103 | Iron–sulfur cluster-binding/ATPase domain protein | 1.76 (1.40) | 2.16 (2.93) | 0.98 (1.68) | 1.89 (3.12) | 0.26 (0.19) | 0.93 (0.91) |
| DVU2104 | Iron–sulfur cluster-binding/ATPase domain protein | 1.50 (0.92) | 3.15 (2.44) | 0.84 (0.58) | 1.17 (0.60) | 1.59 (1.61) | −0.46 (−0.33) |
| DVU0531 | hmc operon protein 6 | 0.59 (0.76) | 1.05 (1.21) | 1.25 (2.00) | 1.08 (1.25) | 1.49 (1.40) | −0.17 (−0.22) |
| DVU0532 | hmc operon protein 5 | 2.34 (1.94) | 1.40 (1.56) | 1.43 (1.13) | 0.83 (0.73) | 2.46 (2.33) | 0.07 (0.09) |
| DVU0533 | hmc operon protein 4 | 1.90 (1.25) | 0.44 (0.50) | 1.64 (2.01) | 0.70 (0.85) | 3.03 (2.16) | −0.44 (−0.50) |
| DVU0534 | hmc operon protein 3 | 0.43 (0.52) | −0.32 (−0.45) | 0.96 (1.38) | 0.33 (0.47) | 2.71 (1.57) | −0.1 (−0.12) |
| DVU0535 | hmc operon protein 2 | NA | NA(NA) | 2.03 (3.77) | 2.26 (3.11) | 0.04 (0.05) | 0.41 (0.31) |
| DVU0536 | High-molecular-weight cytochrome C (hmcA) | 0.98 (1.56) | 1.75 (2.27) | 1.28 (1.61) | 1.51 (1.38) | 1.15 (1.50) | −0.30 (−0.40) |
| DVU0305 (fd II) | Ferredoxin II | 0.41 (0.64) | 2.13 (3.46) | 1.61 (2.42) | 1.21 (1.96) | 2.06 (3.64) | 0.32 (0.58) |
| DVU0384 | Flavoredoxin (flr) | 0.33 (0.61) | 0.24 (0.43) | 0.67 (1.28) | 0.59 (1.14) | 1.69 (2.33) | 0.78 (1.47) |
| DVU2451 | L-lactate permease family protein | 0.85 (1.67) | 0.94 (1.77) | 1.31 (2.45) | 1.26 (2.19) | 1.72 (3.18) | 0.22 (0.41) |
| DVU2683 | L-lactate permease family protein | 0.54 (0.94) | 0.20 (0.34) | 1.17 (1.86) | 0.13 (0.18) | 1.39 (1.32) | −0.11 (−0.2) |
| DVU1780 | Conserved hypothetical protein | −1.74 (−2.88) | −1.86 (−2.09) | −1.75 (−3.31) | −1.39 (−2.66) | −0.85 (−0.72) | −1.55 (−1.47) |
| DVU1781 | Conserved hypothetical protein | −3.91 (−6.41) | −3.4 (−5.38) | −4.00 (−7.15) | −2.89 (−5.22) | −3.93 (−3.98) | −4.17 (−3.98) |
| DVU1782 | Iron–sulfur cluster-binding protein | −3.16 (−4.39) | −3.27 (−4.03) | −3.01 (−4.88) | −3.23 (−5.58) | −3.04 (−1.98) | −3.14 (−2.64) |
| DVU1783 | Cysteine-rich domain protein | −2.76 (−4.53) | −2.84 (−4.42) | −1.72 (−2.20) | −1.53 (−2.00) | −2.89 (−4.25) | −1.97 (−2.64) |
| DVU2286 | Hydrogenase, CooM subunit, putative | −0.75 (−1.38) | −1.04 (−1.86) | −1.14 (−1.98) | −1.50 (−2.58) | −0.83 (−1.25) | −0.73 (−1.06) |
| DVU2287 | Hydrogenase, CooK subunit, selenocysteine-containing, putative | NA | NA | NA | NA | NA | NA |
| DVU2288 | Hydrogenase, CooL subunit, putative | −0.90 (−1.5) | −1.65 (−2.72) | −1.27 (−2.06) | −1.85 (−2.91) | −0.97 (−1.58) | −0.58 (−0.90) |
| DVU2289 | Hydrogenase, CooX subunit, putative | −0.48 (−0.85) | −1.22 (−2.18) | −0.98 (−1.68) | −1.29 (−2.13) | −0.70 (−1.17) | −0.56 (−0.88) |
| DVU2290 | Hydrogenase, CooU subunit, putative | −1.08 (−2.07) | −1.41 (−2.67) | −1.32 (−2.31) | −1.36 (−2.31) | −0.98 (−1.55) | −0.69 (−1.06) |
| DVU2291 | Carbon monoxide-induced hydrogenase CooH, putative | −0.60 (−1.13) | −0.97 (−1.82) | −0.79 (−1.41) | −1.12 (−2.04) | −0.71 (−1.24) | −0.63 (−1.04) |
| DVU2292 | Hydrogenase nickel insertion protein HypA (hypA) | −1.02 (−1.74) | −1.43 (−2.27) | −1.05 (−1.93) | −1.16 (−2.11) | −0.83 (−1.44) | −0.25 (−0.41) |
| DVU2293 | Iron–sulfur protein CooF (cooF) | −0.75 (−1.42) | −0.68 (−1.28) | −0.67 (−1.27) | −0.74 (−1.40) | −0.53 (−0.95) | −0.24 (−0.43) |
| Signal transduction and transcription | |||||||
| DVU0598 | Carbon-starvation protein A, putative | 3.56 (6.62) | 1.04 (2.03) | 4.49 (7.27) | 1.14 (1.53) | 7.20 (4.18) | 0.49 (0.82) |
| DVU0599 | Carbon-starvation protein A, putative | 4.02 (7.36) | 0.03 (0.05) | 4.56 (8.15) | 0.08 (0.16) | 5.14 (6.35) | 0.40 (0.77) |
| DVU0529 | Transcriptional regulator, rrf2 protein, putative | 1.35 (1.68) | 2.10 (2.02) | 1.73 (2.54) | 1.89 (2.09) | 2.28 (2.46) | 0.50 (0.53) |
| DVU0530 | Response regulator, rrf1 protein | 0.81 (1.12) | 0.95 (1.16) | 1.73 (2.34) | 0.81 (1.25) | 1.99 (1.90) | 0.09 (0.14) |
| DVU2114 (atoC) | Sigma-54-dependent transcriptional regulator/response regulator | 0.88 (1.53) | 1.31 (2.45) | 0.93 (1.38) | 1.22 (2.07) | 1.89 (2.37) | 1.21 (1.77) |
| DVU0942 (fur) | Ferric uptake regulator | 0.49 (1.26) | 1.40 (2.05) | 0.78 (0.86) | 1.05 (1.67) | 1.88 (1.22) | 0.43 (0.75) |
| DVU0629 | Transcriptional regulator, TetR family | 0.60 (1.12) | 1.46 (2.45) | 0.74 (1.21) | 1.30 (2.44) | 1.55 (2.10) | 0.55 (0.98) |
| DVU0630 | Hypothetical protein | 0.88 (1.62) | 1.13 (2.02) | 1.19 (2.22) | 1.21 (2.24) | 1.86 (1.17) | 1.00 (1.32) |
| DVU0138 | Response regulator | 0.58 (1.03) | 1.29 (2.28) | 0.41 (0.73) | 1.12 (2.04) | 1.54 (3.07) | 0.86 (1.56) |
| DVU1516 | Hypothetical protein | 1.60 (1.16) | 1.31 (1.07) | 1.90 (1.74) | 1.64 (1.5) | −0.15 (−0.13) | 0.30 (0.25) |
| DVU1517 | Transcriptional regulator cII, putative | 1.75 (1.56) | 1.79 (1.79) | 1.63 (1.20) | 1.76 (1.54) | 0.71 (0.56) | −0.54 (−0.4) |
| DVU0006 | Universal stress protein family | 0.75 (1.47) | 1.39 (2.66) | 0.58 (0.97) | 1.22 (2.09) | 1.40 (2.55) | 0.74 (1.39) |
| DVU3187 (hup-4) | DNA-binding protein HU | 0.12 (0.24) | 1.07 (1.95) | 0.13 (0.20) | 0.79 (1.30) | 1.84 (3.15) | 0.93 (1.79) |
| DVU2686 | Peptidase, S24 family | 0.66 (1.20) | 1.60 (2.51) | 0.61 (0.94) | 0.91 (1.52) | 1.60 (2.39) | 0.44 (0.73) |
| DVU1995 (rsbV) | Anti-anti-sigma factor | 0.74 (1.31) | 1.14 (1.57) | 0.02 (0.03) | 0.28 (0.51) | 1.62 (1.11) | 2.29 (2.10) |
| DVU1967 | Transcriptional regulator, rrf2 protein, putative | 1.66 (2.15) | 0.84 (1.01) | 0.43 (0.35) | 1.04 (1.15) | 0.49 (0.49) | −0.11 (−0.10) |
| DVU1968 | Oxidoreductase, putative | 0.11 (0.13) | 1.44 (2.05) | 1.18 (1.04) | 1.41 (1.23) | −0.40 (−0.34) | −0.96 (−0.90) |
| DVU1969 | Hypothetical protein | 1.81 (2.03) | 0.73 (0.64) | 1.44 (1.49) | 1.93 (2.16) | −0.85 (−1.03) | −1.27 (−1.07) |
| DVU1970 | Response regulator | 1.16 (0.89) | 1.08 (1.25) | 1.74 (1.17) | 1.42 (1.22) | 0.09 (0.08) | −0.14 (−0.16) |
| DVU0408 | Response regulator/sensory box/GGDEF domain/EAL domain protein | 0.34 (0.66) | 0.40 (0.76) | 0.19 (0.35) | 0.08 (0.14) | 2.39 (1.53) | 1.54 (1.44) |
| DVU0720 | HAMP domain protein | 0.05 (0.09) | 0.51 (0.96) | 0.12 (0.18) | 0.24 (0.43) | 2.39 (2.14) | 0.94 (1.20) |
| DVU0606 | Transcriptional regulator, ArsR family/methyltransferase, UbiE/COQ5 family | 0.47 (0.91) | −0.19 (−0.37) | 0.76 (1.18) | 0.39 (0.69) | 1.64 (2.90) | −0.18 (−0.32) |
| DVU1285 | Response regulator | 0.58 (1.08) | 0.97 (1.55) | 0.60 (0.95) | 0.87 (1.65) | 1.68 (1.23) | 0.27 (0.41) |
| DVU1690 | Transcriptional regulator, TetR family | −0.04 (−0.05) | 0.81 (1.34) | −0.10 (−0.19) | 0.01 (0.01) | 1.74 (3.06) | 0.58 (1.08) |
Abbreviations: An, ancestor DvH; ATP, adenosine triphosphate; DVU, Desulfovibrio vulgaris; EC, strain isolated from population No.3 evolved under control condition; ES, strain isolated from population No.9 evolved under salt-stress condition; FAD, flavin adenine dinucleotide; HAMP, Histidine kinases, Adenyl cyclases, Methyl-accepting proteins and Phosphatase; HU, heat unstable; LS4D-100, LS4D+100 mM NaCl; LS4D-250, LS4D+250 mM NaCl; NA, not applicable.
Log2R ratios are shown with Z score in parentheses. Boldface indicates more than twofolds of expression changes.
Amino-acid synthesis and transport
Under no-NaCl stress, 100-mM NaCl or 250-mM NaCl stress conditions, ES9-11 had a significant increase in expression of DVU0593 encoding L-lysine exporter and DVU2615 encoding a family 3 bacterial extracellular solute-binding protein (Table 1). Under 250-mM NaCl stress, ES 9-11 had increased expression of genes DVU2341-DVU2343 in a three-gene operon encoding His/Glu/Gln/Arg/opine family amino-acid ABC transporter, of genes DVU2743 and DVU2744 in a five-gene operon encoding high-affinity branched-chain amino-acid ABC transporter, of DVU1378 encoding ketol-acid reductoisomerase (ilvC) involved in the biosynthesis of isoleucine or valine and of DVU3291 encoding the iron–sulfur cluster-binding subunit of Glu synthase. In addition, under NaCl stress conditions, ES-9-11 had a significant decrease in the expression of DVU1827 encoding acetylornithine deacetylase/succinyl-diaminopimelate desuccinylase family protein involved in the biosynthesis of lysine (Table 1).
Inorganic ion transport and metabolism
Under no-NaCl stress or 100-mM NaCl stress, ES9-11 had significant increases in the expression of genes DVU2571-DVU2574 involved in iron transport, but lack of further increase of expression under 250-mM NaCl stress (Table 1). Under 250-mM NaCl stress, strain ES9-11 had a significant increase in expression of DVU1778 encoding cation efflux family protein.
Cell motility
In all the conditions tested, ES9-11 had significant increases in expression of chemotaxis genes such as methyl-accepting chemotaxis protein (MCP) genes DVU3155 (dcrH) and DVU0170 and a general decrease in expression of 22 flagellar system related genes (Table 2). Under 250-mM NaCl stress, ES9-11 had significant increases in expression of DVU3182 (dcrA) and DVU1904 (cheW-2) compared with An (Table 2).
Energy production and conversion
Again under all the tested conditions, ES9-11 had significant increases in expression of genes involved in electron transfer system such as DVU0531-DVU0536 (hmc operon) and a cytochrome c gene DVU3107 compared with An (Table 3). Significant increases in the expression of a few other cytochrome genes under 250-mM NaCl stress were also seen in ES9-11. Under no-NaCl stress, ES9-11 had a significant increase in expression of DVU2645 encoding Na+/H+ antiporter family protein. Under no-NaCl stress and 100-mM NaCl stress, strain ES9-11 had a generally decreased expression of hydrogenase genes in the coo operon (DVU2286-2293). In addition, ES9-11 tended to increase the expression of two L-lactate permease family protein genes DVU2451 and DVU2683 under NaCl stress compared with An (Table 3).
Signal transduction and transcription
When compared with An, ES9-11 had significant increases in expression of genes DVU0598 and DVU0599 encoding putative carbon-starvation proteins in all the conditions (Table 3). Under no-NaCl stress or 10- mM NaCl stress conditions, ES9-11 had a significant increase in expression of DVU1967 encoding a transcriptional regulator. Under 250-mM NaCl stress, ES9-11 had significant increases in expression of genes DVU2114 (atoC, a sigma-54-dependent transcriptional regulator/response regulator), DVU0942 (fur, ferric uptake regulator), DVU0629 (transcriptional regulator, TetR family), DVU0138 (response regulator), DVU3187 (hup-4, DNA-binding protein HU (heat unstable)), DVU1995 (rsbV, anti-anti-sigma factor) and DVU0408 encoding a response regulator/sensory box/GGDEF (Gly-Gly-Asp-Glu-Phe) domain/EAL (Glu-Ala-Leu) domain protein.
Transcriptional changes of genes involved in other gene categories such as ‘lipid transport and metabolism' or ‘replication, recombination and repair' are listed in Supplementary Table S1. These data were interpreted to indicate that ES9-11 adaptation to NaCl had occurred at the level of gene expression, potentially leading to increased biosynthesis or transport of amino acids, cation effluxes and energy production but decreased motility.
Accumulation of organic solutes in evolved D. vulgaris
Metabolite measurements were carried out to determine whether the transcriptional changes were reflected at the metabolite level. The total abundances of 23 detected metabolites (Supplementary Table S2) were much higher in NaCl-evolved ES9-11 and control-evolved EC3-10 relative to the ancestral strain when grown in control medium (Figure 3a). However, under NaCl stress conditions, ES9-11 showed the capacity to greatly increase intracellular metabolites relative to An, in contrast to EC3-10 (Figure 3a). Glu and Ala were the two most abundant amino acids in ES9-11 and An under all the tested culture conditions (Supplementary Table S2). Significant abundance increases of Glu induced by NaCl stress was observed in ES9-11, but no significant abundance changes of Ala were induced by NaCl stress in ES9-11 and decreased Ala abundance under NaCl stress conditions were seen in EC3-10 (Figure 3b). Under no-NaCl stress, significant (P<0.05) abundance increases of 12 metabolites, including Glu, Ala, Asp, Val, Met, Thr, Ile, Leu, Gly, Pyruvic acid, Phe and agmatine, and significant abundance decrease of Lys were detected in ES9-11 (Supplementary Table S2). Glu is likely the most important contributor to osmoprotection under NaCl stress. Other less abundant metabolites such as Asp and Val could contribute little osmoprotection (Figure 3c). Interestingly, abundances of Gln, Gly and GB in ES9-11 increased under 100-mM NaCl stress only. The abundance changes of metabolites such as Glu and Lys corresponded well with the transcriptional changes (Table 1). The metabolite data suggested important roles of Glu, possibly Ala, Asp and Val in relieving NaCl stress, while other organic solutes such as Gln, Gly and GB might contribute under low NaCl stress only.
Figure 3.
Accumulation of organic solutes in evolved D. vulgaris strains. The sums of detected organic solutes are much higher in NaCl-evolved ES9-11 in all culture conditions (a). Abundance changes of two abundant amino acids Glu and Ala are shown in (b). Changes of less abundant organic solutes such as Asp, Leu, Val and Lys (c), and induction of Gln, Gly and GB under low NaCl stress (d) are shown. Error bars indicate s.d. The significance of abundance changes is shown at the P<0.05 level (t-test). LS4D-100, LS4D+100 mM NaCl; LS4D-250, LS4D+250 mM NaCl.
Decreased cell motility in evolved D. vulgaris
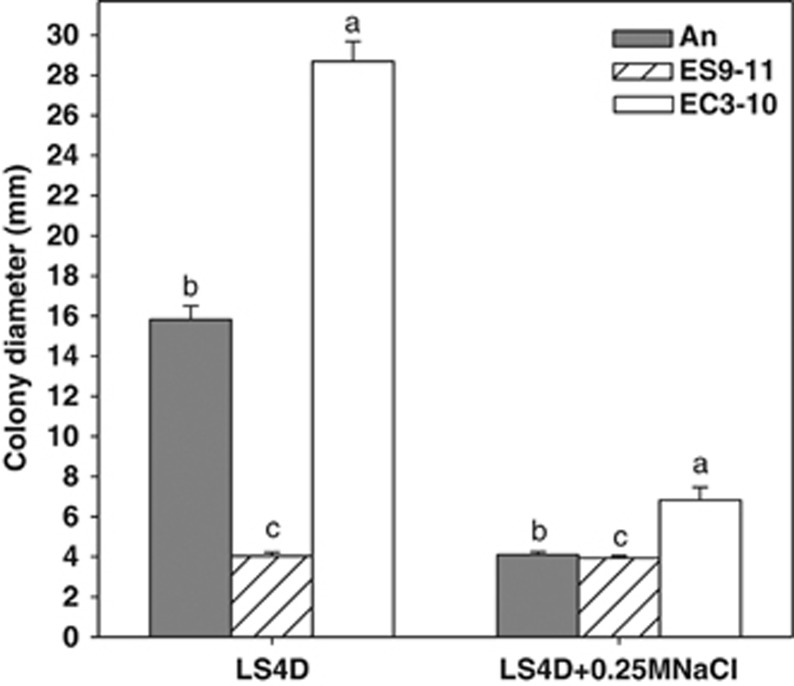
To test whether decreased expression of genes involved in flagellar assembly impacted cell motility, cell motility was evaluated by measuring the colony diameter after 4 days of incubation on soft agar. Decreases of cell motility induced by 250-mM NaCl stress were observed for NaCl-evolved ES9-11 (P=0.0216), control-evolved EC3-10 (P<0.0001) and An (P<0.0001), and motility of ES9-11 was the lowest under no-stress and NaCl-stress conditions (P<0.0001) (Figure 4), consistent with decreased expressions of flagellar system genes in ES9-11 in all the culture conditions. The fastest growth but lowest motility in ES9-11 under no-NaCl stress and 250-mM NaCl stress suggests the possibility of more energy flow towards osmoprotection with less energy available for other functions, such as motility.
Figure 4.
Changes of cell motility in evolved D. vulgaris strains under no-NaCl stress or 250-mM NaCl stress conditions. A t-test was performed to assess the significance of difference among strains (P<0.0001).
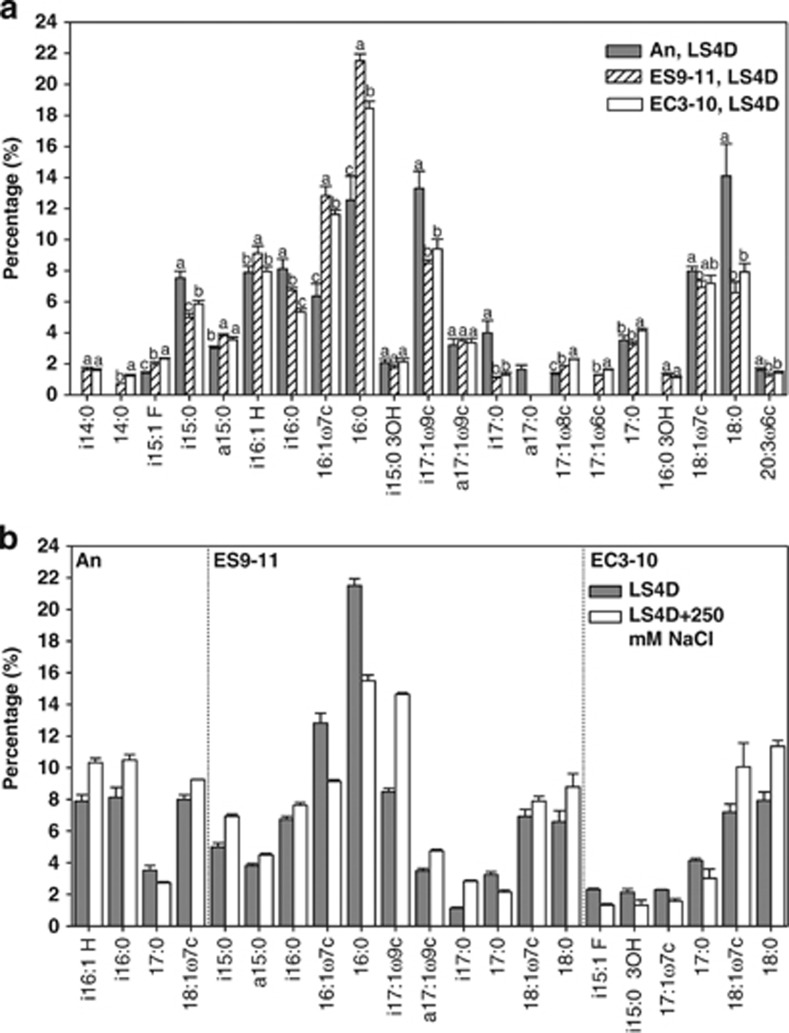
Changes of PLFA composition in evolved D. vulgaris
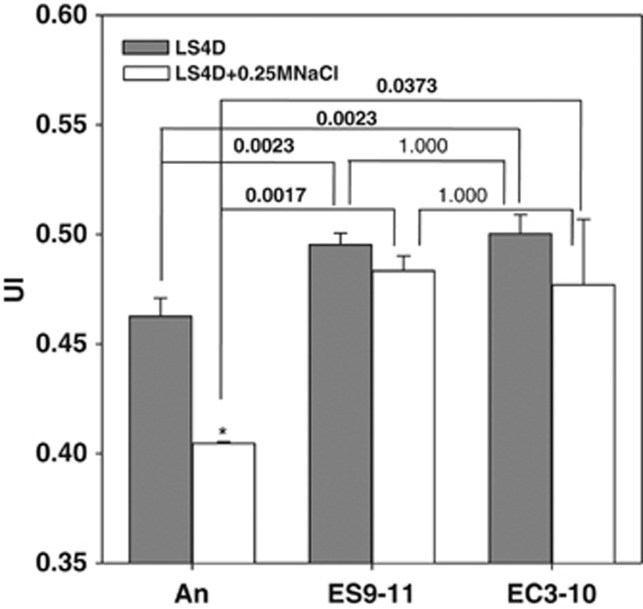
PLFA profiling was investigated to test whether the expression changes of genes involved in lipid metabolism was reflected in the cell membrane PLFA composition. Under no-NaCl stress, there was a significant increase of the UI of PLFAs in NaCl-evolved ES9-11 and control-evolved EC3-10 compared with An. Under 250-mM NaCl stress, unlike the significant (P=0.004) decrease of UI in An, no significant changes of UI were found in ES9-11 or EC3-10 (Figure 5). PLFA composition changes under no-NaCl stress or 250-mM NaCl stress conditions were shown in Figure 6. Under no-NaCl stress, increases of unsaturated 16:1 ω7c and saturated 16:0 were found in ES9-11 and EC3-10, but significant higher percentages of fatty acids 16:1 ω7c (P=0.0414) and 16:0 (P=0.0011) were found in ES9-11 than EC3-10; increase of unsaturated i16:1 H was only detected in ES9-11; decreases of fatty acids i15:0, i16:0, i17:1 ω9c, i17:0 and 18:0 were detected in ES9-11 and EC3-10, but percentages of i15:0 (P=0.0123) and i16:0 (P=0.001) were significantly different between ES9-11 and EC3-10 (Figure 6a). Under 250-mM NaCl stress, different PLFA composition changes were found in ES9-11, EC3-10 or An (Figure 6b). In addition to the increase of 18:1ω7c and decrease of 17:0 seen in all the strains, unique changes in ES9-11 included increased i17:1 ω9c, a17:1 ω9c, i15:0 and decreased 16:1 ω7c and 16:0. In conclusion, unsaturated i16:1 H and 16:1 ω7c might be the major fatty acids contributing to the increase of UI under no-NaCl stress. Branched PLFAs, including i17:1 ω9c, a17:1 ω9c and i15:0, might have important roles in keeping proper membrane fluidity under NaCl stress.
Figure 5.
Changes of UI of PLFA in evolved D. vulgaris strains under no-NaCl stress or 250-mM NaCl stress conditions. Bold font indicates P<0.05. Error bars indicate s.d. *P<0.05.
Figure 6.
PLFA composition changes in evolved D. vulgaris strains. Under no-NaCl stress, there are many PLFA composition changes in NaCl-evolved ES9-11 and control-evolved EC3-10 compared with ancestral strain An (a). The significance of abundance changes is shown at the P<0.05 level (t-test). Under 250-mM NaCl stress, composition changes (P<0.05) of PLFAs are different in strains ES9-11, EC3-10 or An (b).
Discussion
Experimental evolution with microbes has been employed to address evolution-related theories and study genetically simple traits (Elena and Lenski, 2003; Kawecki et al., 2012). NaCl tolerance is a complex trait involving several pathways (Warringer et al., 2003; Dhar et al., 2011). This study provides an example of selection of NaCl tolerance trait through experimental evolution. Three main physiological components of NaCl stress response, including adaptation time to NaCl stress, growth rate and efficiency of growth in NaCl stress (Warringer et al., 2003), have been improved in evolved D. vulgaris. Mechanisms underlying the phenotypic adaptation include significant changes in favor of NaCl tolerance in gene expression, metabolites abundance and PLFA profile.
Data from this study suggest that accumulation of organic solutes is important for NaCl adaptation during evolution. Upon exposure to NaCl stress, physiological changes are triggered by the rapid efflux of cellular water. Immediate responses such as accumulation of intracellular K+ and Glu were found in E. coli (McLaggan et al., 1994); long-term osmoadaptative responses are accumulation of non-ionic compatible solutes, which interfer little with most cytoplasmic enzymes (Sleator and Hill, 2002). Glu appears to be the most important osmopreotectant in evolved D. vulgaris. Glu was the most abundant organic solute in ES9-11 and its abundance significantly increased under NaCl stress. By contrast, no significant abundance changes of Ala were induced by NaCl stress. Glu is a commonly accumulated microbial osmolyte (Botsford and Lewis, 1990; Botsford et al., 1994; McLaggan et al., 1994); however, an osmoregulatory role for Ala during NaCl stress has rarely been reported (Thomas and Shanmugasundaram, 1991). Accumulation of Ala was observed in D. vulgaris exposed to 250-mM NaCl stress for about 100 h (He et al., 2010), but further study is required to confirm the role of Ala in osmoprotection during evolution. Addition of Lys in medium did not improve growth of D. vulgaris under NaCl stress (He et al., 2010), significantly decreased abundance of Lys was observed in NaCl-evolved ES9-11, confirming that Lys is not an osmoprotectant in NaCl stress response of D. vulgaris. GB production was lower in NaCl-evolved ES9-11 than the ancestral strain under 250-mM NaCl stress, which was opposite to its increased abundance in D. vulgaris exposed to 250 mM NaCl for 4 h (Mukhopadhyay et al., 2006). However, abundance of Gln, Gly and GB increased under 100-mM NaCl stress. Accumulation of different sets of organic solutes under low or high NaCl stresses has been found in the facultative anaerobe Erwinia chrysanthemi (Goude et al., 2004). It is likely that different sets of organic solutes were used as osmoprotectants to cope with low or high NaCl stresses in D. vulgaris. Further studies are required to identify other non-ionic compatible solutes important for osmoadaptation of D. vulgaris to NaCl stress.
PLFA composition changes is another important aspect of NaCl adaptation in D. vulgaris. It is well known that NaCl stress increases membrane rigidity and causes membrane PLFA composition changes in bacteria (Kates, 1986; Los and Murata, 2004). Possible role of unsaturated branched PLFA i17:1 ω9c in D. vulgaris exposed to 250-mM NaCl stress was reported (Mukhopadhyay et al., 2006). In NaCl-evolved ES9-11, under no-NaCl stress, unsaturated PLFAs such as 16:1 ω9c and i16:1 H appeared to be the major contributors for increased UI; under 250-mM NaCl stress, in addition to i17:1 ω9c, percentages of branched PLFAs such as a17:1 ω9c and i15:0 significantly increased, confirming the roles of branched PLFAs in maintaining proper membrane fluidity adaptation of D. vulgaris to NaCl stress.
Other physiological aspects important for adaptation to NaCl stress were suggested by gene expression changes in NaCl-evolved ES9-11, such as: increased energy conversion and detoxification of Na+ possibly resulted from increased expressions of two lactate permease family protein genes and Na+/H+ antiporter gene, respectively; and role of fur in gene regulation. These gene expression changes were not seen in NaCl stress responses in literature. Possible role of fur in osmotic stress of D. vulgaris was suggested in previous deletion mutant study (Bender et al., 2007) and Fur was found essential for growth of Helicobacter pylori in NaCl-challenged cells (Gancz and Merrell, 2011). Further study is needed to identify the role of fur in NaCl adaptation of D. vulgaris. Some gene expression changes were consistent with previous results, namely NaCl shock response and short-term adaptation to NaCl stress in D. vulgaris or other bacteria. For example, the repression of flagella assembly genes (Steil et al., 2003; Liu et al., 2005; Mukhopadhyay et al., 2006), increased expression of iron-uptake genes (Hoffmann et al., 2002; Steil et al., 2003; He et al., 2010), cation efflux genes (Mukhopadhyay et al., 2006) and hmc operon genes (Dolla et al., 2000). Lacking of increased expression levels of genes related to general stress responses, such as heat-shock genes (He et al., 2010; Cameron et al., 2012) important for stress-induced protein folding and other damages, suggests the adaptation to NaCl stress in NaCl-evolved ES9-11.
Certain level of NaCl tolerance improvement was observed in control-evolved EC3-10. The [Na+] in LS4D is about 210 mM, which is higher compared with the medium used by ATCC (about 90 mM). Higher [Na+] in LS4D may be responsible for enhancing NaCl tolerance in EC3-10. Therefore, ionic strength of the culture medium is an important factor to be considered in experiment design, especially in experimental evolution.
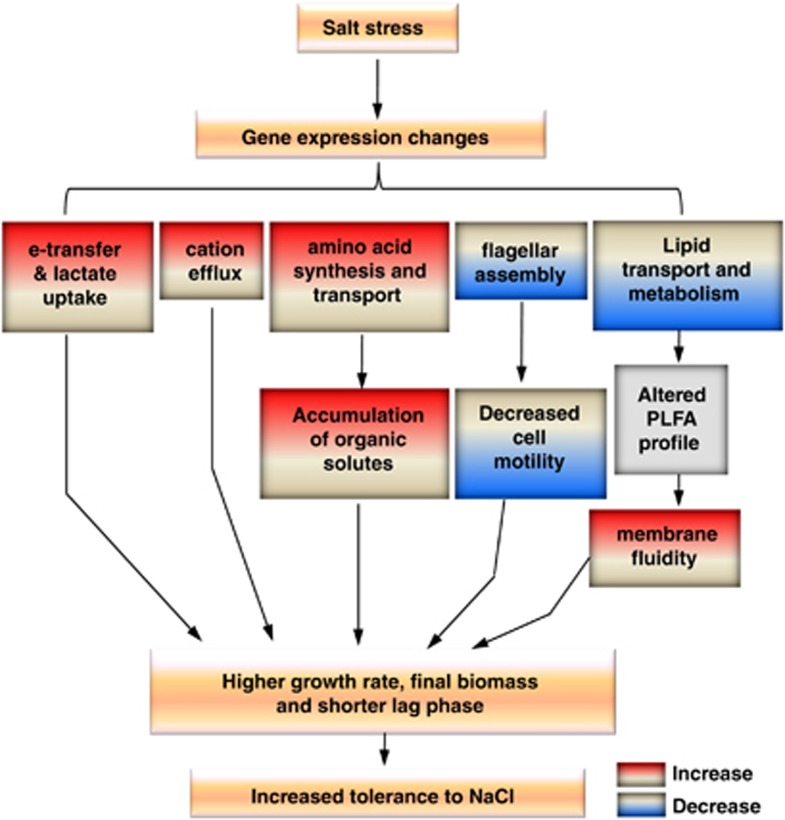
A conceptual model of NaCl adaptation was proposed based on the data with single-colony isolates from D. vulgaris evolved under low NaCl stress (Figure 7). Constant exposure to low NaCl stress induced dramatic transcriptional changes of functional genes and regulatory genes. Altered gene expressions (basal changes) when cultured in medium without NaCl amendment suggests the possibility of genetic adaptation. The basal increase of Na+/H+ antiporter expression and the increased expression of a cation efflux protein encoding gene in response to NaCl stress might enable efficient ionic detoxification. Basal increases and increased expression of amino-acid synthesis and transport genes lead to intracellular accumulation of compatible solutes. Also, increased expression of lactate-uptake genes and genes involved in energy conversion ensure that sufficient energy is produced for cellular activities. In addition, decreased expression of flagellar assembly genes resulting in decreased cell motility allows more energy available for osmoprotection. Changes of genes involved in lipid metabolism may contribute to the significant changes of PLFA composition. The basal increase of unsaturated or unsaturated branched PLFA under no-NaCl stress and the unique increase of unsaturated or unsaturated branched PLFA under NaCl stress increases the membrane fluidity, which is critical for proper cellular function. Expression changes of regulatory genes under no-stress and NaCl-stress conditions ensure the rapid adjustment of gene expression to meet the challenges of NaCl stress. As a result, the inhibitory effect of NaCl stress on growth of D. vulgaris is conquered to a great extent: an increased growth rate and final biomass but shortened lag phase were observed for the NaCl-evolved strain ES9-11 under NaCl stress.
Figure 7.
A conceptual model of adaptation to NaCl stress in evolved D. vulgaris. Gene expression changes induced by NaCl stress, accumulation of organic solutes, increased membrane fluidity, decreased cell motility, potentially increased exclusion of Na+ and increased energy metabolism contribute to increased tolerance to NaCl in evolved D. vulgaris.
Acknowledgments
This work conducted by ENIGMA—Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov)—a Scientific Focus Area Program at Lawrence Berkeley National Laboratory, was supported by the Office of Science, Office of Biological and Environmental Research, US Department of Energy under Contract No. DE-AC02-05CH11231.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Baidoo EEK, Benke PI, Neususs C, Pelzing M, Kruppa G, Leary JA, et al. Capillary electrophoresis-fourier transform ion cyclotron resonance mass spectrometry for the identification of cationic metabolites via a pH-mediated stacking-transient isotachophoretic method. Anal Chem. 2008;80:3112–3122. doi: 10.1021/ac800007q. [DOI] [PubMed] [Google Scholar]
- Bender KS, Yen H-CB, Hemme CL, Yang Z, He Z, He Q, et al. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 2007;73:5389–5400. doi: 10.1128/AEM.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford JL, Alvarez M, Hernandez R, Nichols R. Accumulation of glutamate by Salmonella typhimurium in response to osmotic stress. Appl Environ Microbiol. 1994;60:2568–2574. doi: 10.1128/aem.60.7.2568-2574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford JL, Lewis TA. Osmoregulation in Rhizobium meliloti: production of glutamic acid in response to osmotic stress. Appl Environ Microbiol. 1990;56:488–494. doi: 10.1128/aem.56.2.488-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A, Frirdich E, Huynh S, Parker CT, Gaynor EC. Hyperosmotic stress response of Campylobacter jejuni. J Bacteriol. 2012;194:6116–6130. doi: 10.1128/JB.01409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Peacock AD, Long PE, Stephen JR, McKinley JP, Macnaughton SJ, et al. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl Environ Microbiol. 2001;67:3149–3160. doi: 10.1128/AEM.67.7.3149-3160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra SR, He Q, Huang KH, Gaucher SP, Alm EJ, He Z, et al. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J Bacteriol. 2006;188:1817–1828. doi: 10.1128/JB.188.5.1817-1828.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R, SÄGesser R, Weikert C, Yuan J, Wagner A. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol. 2011;24:1135–1153. doi: 10.1111/j.1420-9101.2011.02249.x. [DOI] [PubMed] [Google Scholar]
- Dolla A, Pohorelic BKJ, Voordouw JK, Voordouw G. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch Microbiol. 2000;174:143–151. doi: 10.1007/s002030000183. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Elias DA, Suflita JM, McInerney MJ, Krumholz LR. Periplasmic cytochrome c3 of Desulfovibrio vulgaris is directly involved in H2-mediated metal but not sulfate reduction. Appl Environ Microbiol. 2004;70:413–420. doi: 10.1128/AEM.70.1.413-420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancz H, Merrell DS. The Helicobacter pylori ferric uptake regulator (Fur) is essential for growth under sodium chloride stress. J Microbiol. 2011;49:294–298. doi: 10.1007/s12275-011-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goude R, Renaud S, Bonnassie S, Bernard T, Blanco C. Glutamine, glutamate, and {alpha}-glucosylglycerate are the major osmotic solutes accumulated by Erwinia chrysanthemi strain 3937. Appl Environ Microbiol. 2004;70:6535–6541. doi: 10.1128/AEM.70.11.6535-6541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Huang KH, He Z, Alm EJ, Fields MW, Hazen TC, et al. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl Environ Microbiol. 2006;72:4370–4381. doi: 10.1128/AEM.02609-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhou A, Baidoo E, He Q, Joachimiak MP, Benke P, et al. Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris Hildenborough to salt adaptation. Appl Environ Microbiol. 2010;76:1574–1586. doi: 10.1128/AEM.02141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Schutz A, Brosius M, Volker A, Volker U, Bremer E. High-salinity-induced iron limitation in Bacillus subtilis. J Bacteriol. 2002;184:718–727. doi: 10.1128/JB.184.3.718-727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebbar M, Talibart R, Gloux K, Bernard T, Blanco C. Osmoprotection of Escherichia coli by ectoine: uptake and accumulation characteristics. J Bacteriol. 1992;174:5027–5035. doi: 10.1128/jb.174.15.5027-5035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates M. Influence of salt concentration on the membrane lipids of halophilic bacteria. FEMS Microbiol Rev. 1986;39:95–101. [Google Scholar]
- Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC. Experimental evolution. Trends Ecol Evol. 2012;27:547–560. doi: 10.1016/j.tree.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Ko R, Smith LT, Smith GM. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J Bacteriol. 1994;176:426–431. doi: 10.1128/jb.176.2.426-431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gao W, Wang Y, Wu L, Liu X, Yan T, et al. Transcriptome analysis of Shewanella oneidensis MR-1 in response to elevated salt conditions. J Bacteriol. 2005;187:2501–2507. doi: 10.1128/JB.187.7.2501-2507.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Murata N. Membrane fluidity and its roles in the perception of environmental signals. Biochim Biophys Acta. 2004;1666:142–157. doi: 10.1016/j.bbamem.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJP. Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl Environ Microbiol. 1994;60:726–728. doi: 10.1128/aem.60.2.726-728.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Widman PK, Woodward JC, Phillips EJ. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl Environ Microbiol. 1993;59:3572–3576. doi: 10.1128/aem.59.11.3572-3576.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CS, Heras H, Garda H, Ruzal S, Sánchez-Rivas C, Rivas E. Biochemical and biophysical studies of Bacillus subtilis envelopes under hyperosmotic stress. Int J Food Microbiol. 2000;55:137–142. doi: 10.1016/s0168-1605(00)00171-9. [DOI] [PubMed] [Google Scholar]
- McLaggan D, Naprstek J, Buurman ET, Epstein W. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J Biol Chem. 1994;269:1911–1917. [PubMed] [Google Scholar]
- Mukhopadhyay A, He Z, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, et al. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J Bacteriol. 2006;188:4068–4078. doi: 10.1128/JB.01921-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Stams AJM. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Micro. 2008;6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- Postgate JR. The Sulfate-Reducing Bacteria. Cambridge University Press: Cambridge, UK; 1984. [Google Scholar]
- Roberts M. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 2005;1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruess L, Schütz K, Migge-Kleian S, Häggblom MM, Kandeler E, Scheu S. Lipid composition of collembola and their food resources in deciduous forest stands—implications for feeding strategies. Soil Biol Biochem. 2007;39:1990–2000. [Google Scholar]
- Russell N. Adaptive modifications in membranes of halotolerant and halophilic microorganisms. J Bioenerg Biomembr. 1989;21:93–113. doi: 10.1007/BF00762214. [DOI] [PubMed] [Google Scholar]
- Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- Steil L, Hoffmann T, Budde I, Volker U, Bremer E. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J Bacteriol. 2003;185:6358–6370. doi: 10.1128/JB.185.21.6358-6370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strøm AR, Kaasen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- Thomas SP, Shanmugasundaram S. Osmoregulatory role of alanine during salt stress in the nitrogen fixing cyanobacterium Anabaena sp. 287. Biochem Int. 1991;23:93–102. [PubMed] [Google Scholar]
- Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Natl Acad Sci USA. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DT, Lindsay YE, Caumette P, Herbert RA, Hannan J. Identification of trehalose and glycine betaine as compatible solutes in the moderately halophilic sulfate reducing bacterium, Desulfovibrio halophilus. FEMS Microbiol Lett. 1996;140:203–207. [Google Scholar]
- Whatmore AM, Chudek JA, Reed RH. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J Gen Microbiol. 1990;136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]
- Zhou A, He Z, Redding-Johanson AM, Mukhopadhyay A, Hemme CL, Joachimiak MP, et al. Hydrogen peroxide-induced oxidative stress responses in Desulfovibrio vulgaris Hildenborough. Environ Microbiol. 2010;12:2645–2657. doi: 10.1111/j.1462-2920.2010.02234.x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou A, et al. How sulfate reducing microorganisms cope with stress: lessons from systems biology. Nat Rev Microbiol. 2011;9:452–466. doi: 10.1038/nrmicro2575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.