Abstract
Leukemia inhibitory factor (LIF) expression in the uterus is essential for embryo implantation in mice. Here we describe the spatial and temporal regulation of LIF signaling in vivo by using tissues isolated from uteri on different days over the implantation period. During this time, LIF receptors are expressed predominantly in the luminal epithelium (LE) of the uterus. Isolated epithelium responds to LIF by phosphorylation and nuclear translocation of signal transducer and activator of transcription (Stat) 3, but not by an increase in mitogen-activated protein kinase levels. The related cytokines Il-6, ciliary neurotrophic factor, as well as epidermal growth factor, do not activate Stat3, although epidermal growth factor stimulates mitogen-activated protein kinase. In vivo Stat3 activation is induced by LIF alone, resulting in the localization of Stat3 specifically to the nuclei of the LE coinciding with the onset of uterine receptivity. The responsiveness of the LE to LIF is regulated temporally, with Stat activation being restricted to day 4 of pregnancy despite the presence of constant levels of LIF receptor throughout the preimplantation period. Uterine receptivity is therefore under dual control and is regulated by both the onset of LIF expression in the endometrial glands and the release from inhibition of receptor function in the LE.
Implantation of the mammalian embryo is an essential step in the establishment of a normal pregnancy. In preparation for implantation, the uterus undergoes continuous synchronized waves of proliferation and differentiation in response to the rise and fall of the ovarian hormones estrogen (E2) and progesterone (P4) (1). In rodents and in humans, the onset of implantation is marked by the physical interaction between the apical surface of the luminal epithelium (LE) and the trophoblast of the hatched blastocyst. Implantation proceeds initially by apposition, followed by adhesion, and finally by penetration of the LE by the trophoblast. The stromal cells underlying the epithelium and adjacent to the site of blastocyst attachment respond by proliferating and differentiating, a process referred to as decidualization. Decidualization serves to sustain the early postimplantation embryo and to restrict trophoblast invasion of the uterus (2).
In the mouse, implantation occurs at a specific time after ovulation, the so-called “implantation window,” which lasts for ≈18–24 h and usually starts early on the fourth day of pregnancy (day 1 ≡ day of plug). Before the start of implantation, the uterus is unresponsive to the blastocyst or to other decidualizing signals. The uterus again becomes refractory to embryo implantation on the fifth day of pregnancy (3). In addition to regulating implantation during a normal reproductive cycle, many mammalian species, either because of lactational or environmental cues, can maintain embryos in a dormant state, delaying implantation until more favorable conditions appear for supporting embryonic growth (4). The control of implantation is therefore primarily maternal with E2 and P4 regulating changes in the expression of adhesion molecules, cytokines, and transcriptional factors, many of which have been implicated in mediating implantation. Because of the complexity and dynamic nature of implantation, the molecular changes are still poorly understood (5).
A variety of cytokines and growth factors have been proposed to regulate implantation (6), and to date, only leukemia inhibitory factor (LIF) has been shown to be essential at initiating the process in mice (7). LIF, a member of the Il-6 family of secreted cytokines, is expressed in the endometrial glands just before the onset of implantation and is secreted into the uterine lumen (8). In LIF-deficient female mice, embryo development is arrested at the appositional phase with the blastocysts attached to the LE. The uterus is, however, unresponsive to the embryo or other decidualizing signals, resulting implantation failure (9). These results may be of general significance to mammals because in many other species, including humans, uterine levels of LIF increase at the onset of implantation, suggesting that LIF may be widely involved in regulating the process (10–14).
LIF acts on cells by binding to the heterodimeric LIF receptor consisting of the two transmembrane proteins, gp130 and the LIF receptor α (LIFRα). On binding, the receptors dimerize and recruit the nonreceptor tyrosine kinases Jak1 or 2 and Tyk2, which phosphorylate the cytoplasmic domains of the receptors. This cascade results in the recruitment and phosphorylation of the latent signal transducer and activator of transcription (Stat) transcription factors by the Jaks. The phosphorylated Stats dimerize and translocate to the nucleus where they act to regulate gene expression. In addition to the Jak–Stat signaling pathway, other pathways, including the mitogen-activated protein kinase (MAPK), protein kinase C (PKC), and PI3-kinase pathways, also are activated by the LIF receptor (15).
To understand further how LIF acts in regulating embryo implantation, we determined the distribution of LIF receptors in the uterus and the signaling pathways activated by LIF treatment. Here we show that functional LIF receptors are expressed predominantly in the LE. The epithelium responds to LIF by phosphorylation and nuclear translocation of Stat3, but not by increasing MAPK levels. In contrast, Il-6, ciliary neurotrophic factor, as well as epidermal growth factor (EGF), all of which can activate Stat3, have no effect on Stat activation in the epithelium, although EGF stimulated MAPK. In vivo Stat3 activation by LIF results in the relocalization of Stat3 from the cytoplasm to the nuclei of the LE coincident with the onset of receptivity. Surprisingly, the responsiveness of the LE to LIF is temporally regulated, with Stat activation being restricted to day 4 despite constant receptor levels throughout the preimplantation period. Therefore, LIF receptor activity, with respect to Stat3 phosphorylation, is regulated temporally in the LE, and that uterine receptivity is controlled by the onset of LIF expression and a release from inhibition of receptor function.
Materials and Methods
Mice.
LIF-deficient mice were maintained in our existing colony. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. All mice were mated naturally with the assumption that mating occurred around midnight with day 1 of pregnancy being equivalent to day of plug, and implantation taking place on the evening of day 4. To stage the time of pregnancy, the uteri or oviducts were flushed to determine the developmental stage of the embryos. Early day 3 (9 a.m.), the embryos were at the 8-cell stage and located near the utero–tubal junction. Early day 4 (9 a.m.) cavitating blastocysts were surrounded still by a zona and were present in the uterine lumen. By day 4 in the afternoon, all blastocysts had hatched. The morning of day 5 was defined by the inability to flush blastocysts from the uterus with obvious implantation sites.
Cytokines.
Recombinant human and mouse LIF, Il-6, and ciliary neurotrophic factor were purchased from R & D systems, EGF was purchased from Sigma, and HIL-6, a fusion protein between IL-6 and IL-6 receptor, was a kind gift from S. Rose-John (Carl–Albrechts University, Kiel, Germany).
Isolation of the LE.
LE free of stromal and blood cells with very limited glandular epithelial contamination was isolated from the uterus by using mild enzymatic digestion and mechanical suction (16). Sheets of LE were collected, washed successively in PBS, and RNA or protein isolated. Gravity sedimentation was used to separate large pieces of LE from contaminating blood cells. For cytokine treatment, LEs were incubated in serum-free Opti-MEM (BRL) for 3 h followed by the addition of 100 ng/ml cytokine for 20–30 min, lysed, centrifuged, and protein extracts aliquoted.
Gel Retardation Assay.
LE nuclear extracts were prepared with minor modifications (17). Nuclear extract (5 μg) was assayed by using 32P-labeled DNA oligomers of CAAT enhancer-binding protein (C/EBP)-β and Stats1–5 consensus-binding sequences (Santa Cruz Biotechnology) according to the manufacturer's instructions (Promega). The gels were exposed by using a PhosphorImager (Molecular Dynamics) or to film. Supershifts were achieved by using DNA oligomers with two tandem repeat consensus Stat3-binding sites and 1 μg of Stat3 antibody (Santa Cruz Biotechnology) with the same amount of pre-immune rabbit antibody as control.
Western Blot.
Blotting was performed by using standard procedures. The primary antibodies were rabbit (Rb) anti-mouse-P-Ty-Stat3, Stat3, P-MAPK, and MAPK (New England Biolabs), or a monoclonal antibody to Stat3 (Transduction Laboratories, Lexington, KY). Peroxidase-conjugated anti-rabbit or anti-mouse IgG antibody was used to detect binding. Specific bands were visualized by enhanced chemiluminescence (ECL Plus; Amersham Pharmacia), using a DDC camera (Stratagene), and then exposed with film (Kodak). Signal quantification was performed by the National Institutes of Health IMAGE v1.62 software with the ratios between phosphorylated target protein/target protein being calculated and statistically analyzed.
RNA Analysis.
The plasmids used were mLIF (mouse full-length LIF) in pGEM4; mLIFRα in pBSSK (with 121–900 nucleotides of extracellular domain); mgp130 in pBSSK (including 2,340–2,480 nucleotides corresponding to the cytoplasmic domain); and mEP2 (prostaglandin E2 receptor) in pGEMTeasy (757–1,158 nucleotides).
Northern Analysis.
Whole uterine or LE RNA (100 μg) were electrophoresed in a formaldehyde agarose gel. RNA was transferred onto Hybond-N membranes (Amersham Pharmacia) and hybridized with 32P-labeled antisense RNA probes.
Ribonuclease Protection Assay.
Total RNAs were prepared from days 3–5 LE by using RNeasy according to the manufacturer's instructions. Total RNA (5 μg) was hybridized with 32P-labeled antisense probes of LIF, gp130, LIFRα, and ribosomal protein L19, and 2 μg for prostaglandin E2 receptor, respectively, at 45°C overnight. After treatment with ribonuclease mixture (RNaseA/T1; Ambion, Austin, TX), the RNA was precipitated and electrophoresed by PAGE and autoradiographed.
In Situ Hybridization.
Sense and antisense probes were transcribed and labeled with 33P. Uteri from mice on the 4th day of pregnancy were fixed with 4% (wt/vol) paraformaldehyde and embedded in paraffin. Cross-sections (10 μm) were deparaffinized and rehydrated with RNase-free water and processed as described (8).
Immunostaining.
LIFRα and gp130 were detected in frozen sections with rabbit anti-mouse gp130 antibody (0.4 μg/ml C-20; Santa Cruz Biotechnology), rabbit anti-LIFRα, and rabbit IgG (Vector Laboratories) at the same concentration as the negative control. Antibody binding was visualized with biotin-conjugated goat anti-rabbit antibody and peroxidase-conjugated Avidin D (ABC kit; Vector Laboratories) with 3-amino-9-ethylcarbazole (AEC) or diaminobenzidine (DAB) as the substrate. For Stat3 localization, rabbit anti-Stat3 (New England Biolabs) was used on paraffin sections. Treatment of all uterine sections from LIF-null and wild-type animals was performed by using identical conditions. Sections were counterstained with fast green and mounted with Permount (Fisher).
LIF Binding Assay.
Purified recombinant mouse LIF was iodinated by chloramine T to a specific activity of 13 μCi/μg (1 Ci = 37 GBq). Binding experiments were performed as follows. Pooled uterine epithelial tissue was freshly obtained from 7 day-3, -4, or -5 mice to standardize the number of cells in each preparation, and Fg embryonic carcinoma cells were used as a positive control. The epithelial membrane suspension was mixed continually and distributed equally between low-binding siliconized tubes (Marsh, Rochester, NY) containing increasing nanogram amounts of iodinated LIF (1.6 × 104 cpm) with or without a 1,000-fold excess of unlabeled LIF in a total volume of 300 μl in phosphate buffer containing 0.1% BSA. Tubes were incubated with shaking at 37°C for 45 min, and the pellets were washed 3 times and centrifuged after each washing at 325 × g for 5 min. Radioactivity bound to membrane pellets was measured as described (18).
Results
LIF Target Tissues in the Uterus.
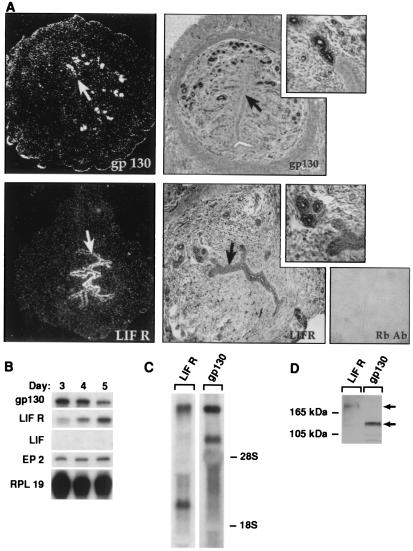
To identify the target tissues for LIF in the uterus, the expression patterns of the LIF receptor components, gp130 and LIFRα, were determined during the preimplantation period. In situ hybridization of sections of mouse uterus from the 4th day of pregnancy revealed LIFRα mRNA is expressed highly in the LE with only background levels detectable in the stroma (Fig. 1A). Gp130 was expressed highly in glandular epithelium (GE) and at lower levels in the LE. Immunostaining, using antibodies against the C termini of both receptors, showed similar patterns of expression for both the LIF receptor and gp130 with some evidence for expression of the receptors in the blood vessels and capillaries, as well as the myometrium (Fig. 1A).
Figure 1.
LE is a LIF target tissue in the uterus. (A) In situ hybridization and immunostaining localizes LIFRα mRNA and protein to the LE and gp130 mRNA and protein predominantly in GE (arrows). (B) Analysis of purified LE by ribonuclease protection assay identifies LIFRα and gp130 mRNA transcripts and (C) multiple transcripts detected by Northern blot. (D) LIF receptor (190-kDa) and gp130 (125-kDa) protein are detected by Western blot analysis of LE protein extracts. The purity of LE preparation was confirmed by the absence of the LIF message that is expressed in the GE and the presence the prostaglandin E2 receptor message, which is specific for the LE (B). RPL19, ribosomal protein L19.
To confirm these results, intact LE was purified as monolayer sheets by using a combination of dissection and enzymatic procedures, resulting in a homogenous source of material. Ribonuclease protection assays demonstrated that both gp130 and LIFRα messages are present in LE throughout the implantation period (days 3–5 postcoitus; Fig. 1B). As markers for assessing the purity of the epithelial preparations, prostaglandin E2 receptor served as a positive control (19), and LIF, which is induced in GE on day 4 of pregnancy, acted as a negative control (8). Northern blot analysis with 100 μg of total RNA isolated from purified LE confirmed the ribonuclease protection assay results (Fig. 1C). As expected, multiple RNA transcripts, generated by alternate splicing or polyadenylation, were detected by Northern analysis on whole uteri and purified LE for both receptor components, consistent with previous reports (20, 21). Western blots confirmed that the transcripts for both receptors are translated (Fig. 1D), showing that both components of the LIF receptor are present in the LE.
The LE Responds to LIF.
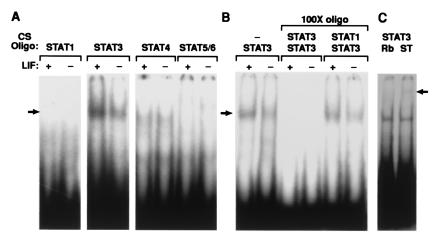
The LIFRα activates several signaling pathways in a variety of cell types, including the Jak–Stat, MAPK, and PI3-kinase pathways (15), whereas gp130 participates in the activation of Stats 1, 3, and 5b (22). Intact LE was isolated from day-4 uteri, allowed to recover for 2–3 h, and then treated with or without 100 ng of recombinant LIF. Nuclear extracts were prepared, and gel-shift assays determined that among the Stats, Stat3 was the sole member to be activated in the LE (Fig. 2A). Heterodimers with other Stat family members were not induced (Fig. 2B), which was confirmed by supershift assay (Fig. 2C). Another transcription factor, C/EBP-β (NF-IL-6), that can be up-regulated by activation of MAPK by gp130 (23), was not induced (data not shown).
Figure 2.
LIF activates Stat3 but not other Stats in LE. (A) Nuclear extracts were prepared from day-4 LE with (+) or without (−) LIF treatment. 32P-labeled DNA oligomers (CS Oligo) for the different consensus Stat-binding sites were used to assay the DNA-binding activities, revealing the specific activation of Stat3. (B) Stat3-specific binding activity was confirmed by it being out-competed with 100-fold unlabeled Stat3, but not by Stat1 oligomers and (C) antibody to Stat3 generated a supershifted band (arrow). Rb, rabbit Ab; ST, Stat Ab.
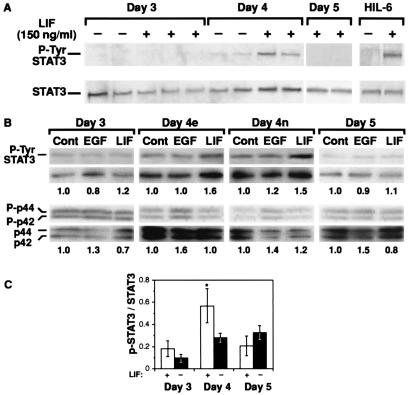
To confirm the gel-sift assays, we used phospho-specific antibodies to determine the ratio of activated tyrosine phosphorylated Stat3 (p-Ty-Stat3) to total Stat3 particularly as Stat3-binding activity increased only slightly and not all of the binding activity was supershifted by Stat3 antibodies (Fig. 2C). LE from day 4 of pregnancy responded to LIF, resulting in an increase in P-Stat3 levels (Fig. 3A), which is in agreement with the up-regulation of Stat3-binding activity. Il-6 and ciliary neurotrophic factor also were tested and both were unable to induce Stat3 phosphorylation (data not shown). However, the soluble IL-6/IL-6 receptor fusion protein, which activates the gp130 homodimer (24), did stimulate Stat3 phosphorylation to a comparable or even enhanced degree to that induced by LIF, confirming the presence of functional gp130 in the LE (Fig. 3A).
Figure 3.
LIF activation of Stat3 in LE is regulated temporally. Purified LE was treated with LIF and the total p-Tyr-Stat3 and Stat3 protein levels determined by Stat3 phospho-specific and Stat3 antibodies on the same blot. (A) Both LIF and HIL-6 fusion protein (150 ng/ml) increase p-Tyr-Stat3 levels. Increased Stat3 phosphorylation after LIF treatment occurs in LE from day-4 LE, but not from days 3 or 5. (B) Such temporal restriction in activation is observed for LIF but not EGF. EGF (100 ng/ml) activates MAPK by a 30–60% increase as shown by the ratios of phosphorylated MAPK to MAPK (the numbers under each lane are the ratios of P-MAPK to MAPK). (C) Analysis of the temporal differences of LIF activation are shown with the ratios of p-Tyr-Stat3 to total Stat3 being higher in day-4 LIF-treated LE (n = 4); this is statistically significant (*, P < 0.035, Student's t test) from that of day-4 untreated LE (n = 4). LIF did not induce Stat3 activation before or after day 4 (n = 4 for day 3 and n = 3 for day 5), revealing that responsiveness of the LE to LIF is regulated temporally.
EGF and other family members, such as amphiregulin, transforming growth factor (TGF)-α, and heparin-binding (HB)-EGF, that activate erbB receptors resulting in Stat activation (25, 26), also have been implicated in regulating proliferation of the LE, as well as possibly mediating embryo implantation (27). Treatment of epithelia from days 3–5 with EGF, compared with LIF, however, did not result in any significant increase in the levels of Stat3 phosphorylation (Fig. 3B).
The MAPK pathway also is activated by LIF in a variety of cell types and has an important effect in modulating Stat3 activity (28–30). Purified LE from days 3–5 was treated with either EGF or LIF to assay MAPK phosphorylation. On day 3, levels for both phosphorylated MAPK proteins were elevated even in controls. By days 4 and 5, however, the levels of phosphorylation had decreased. On all days EGF stimulated an increase (between 30–60%) of phosphorylated MAPK (Fig. 3B). LIF treatment had little or no effect on MAPK levels over the same period.
Temporal Regulation of Stat3 Phosphorylation by LIF.
When luminal epithelia from different days of pregnancy were treated with LIF, it was apparent that Stat activation is regulated temporally. Only LE from early day 4 responded significantly and consistently to LIF, as measured by the ratio of p-Tyr-Stat3 to Stat3 (an increase of 40–60%), whereas epithelia from either early days 3 or 5 did not respond or did so inefficiently (Fig. 3 B and C) with these differences being statistically significant (Fig. 3C).
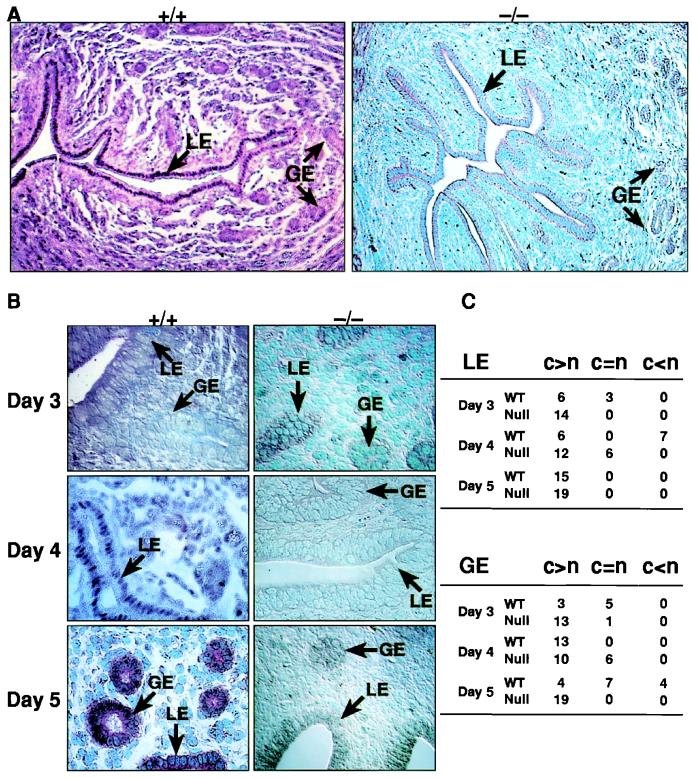
To confirm these observations, sections of uterus from mice at different days of pregnancy were immunostained with an anti-Stat3 antibody (Fig. 4 A and B) to determine the cellular and subcellular localization in the uterus. On day 3 in wild-type mice, Stat3 was localized primarily to the cytoplasm in both the luminal and glandular epithelia with little or no signal detectable in the stroma. By day 4, Stat3 was found consistently in the nuclei throughout the entire LE in all uteri examined and not the GE (Fig. 4A). By day 5, Stat3 had relocalized to an exclusively cytoplasmic distribution in the LE present between implantation sites. In more than half the sections from early day 4, Stat3 clearly was present in the nucleus, whereas on other days (days 3 and 5), Stat3 was predominantly cytoplasmic. Only a few sections from day-5 uteri showed Stat3 nuclear localization, and this was primarily in the GE (Fig. 4 B and C) and in cells that were undergoing decidualization (data not shown). Nuclear localization of Stat3 in day-4 LE was not detected in any LIF-deficient mice (Fig. 4 A–C), supporting our findings that LIF is the principal mediator of Stat3 activation in vivo. However, some day-4 sections of LIF-null mice did show a slight increase in staining of Stat3 in both nucleus and cytoplasm, which agrees with the presence of detectable levels of Stat3-binding activity in LIF-null animals.
Figure 4.
Nuclear translocation of Stat3 is attenuated in LIF −/− mice. (A) Stat3 immunoreactivity in uterine sections reveals a uniform nuclear localization of Stat3 to the entire LE on day 4 in +/+ but not −/− mice. (B) Higher-powered views are shown for days 3–5 of Stat3 localization to the LE and GE nuclei. (C) A summary table of the stained uterine sections is shown where the number of uteri showing predominant cytoplasmic localization (c > n) of Stat3 or nuclear localization (c < n) is presented in both the LE and GE. A few day-3 +/+ sections showed occasional nuclear translocation of Stat3 in both LE and GE. In day-3 LIF −/−, no translocation was evident. In D4 +/+, more than half of the uteri showed complete nuclear localization of Stat3 in the entire LE but not in GE. In contrast, LIF −/− uteri showed little or no nuclear localization in the LE. In day-5 +/+ animals, the nuclear localization of Stat3 in LE is absent in the LE between implantation sites with some nuclear staining detected in GE. LIF −/− mice show a marked reduction and absence of Stat3 nuclear location in both LE and GE. WT, wild type.
LIF Receptor Expression in the LE Remains Constant Over the Implantation Period.
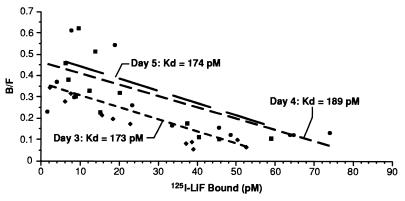
One possible route by which the responsiveness of the LE to LIF could be regulated temporally would be by changing the levels of the LIF receptor. Western analysis on LE extracts from days 3–5 revealed that LIFRα was detectable on all days, as were gp130 levels. To confirm these results, the binding capacity of the LE for iodinated LIF was determined for days 3–5. The conditions used were identical to those used to measure the binding of LIF to embryonic carcinoma cells (31). The equilibrium dissociation constants (Kd) for the LE for days 3, 4, and 5 were 63.06, 87.25, and 87.26 pM, respectively, which are equivalent to levels reported for LIF binding to other cell types (Fig. 5; ref. 32). Although there seemed to be slight differences in the mean binding capacity for LIF on different days, they were not statistically significant.
Figure 5.
LIF binding to the LE remains unchanged. Scatchard analysis of 125I-labeled LIF binding to LE cells on days 3 (⧫), 4 (■), and 5 (●). Specific binding was calculated by subtracting radioactivity bound in the presence of excess unlabeled LIF (nonspecific binding) from total binding. Points are the result of five separate experiments. Optimal (weighted least squares) estimates of affinity constants (K values), dissociation constants (Kd), and binding capacities (R) were analyzed by the LIGAND program (45). Alternative models were tested to determine the existence of multiple binding sites with the one-binding-site model providing the most parsimonious fit. Binding parameters for data corresponding to days 3, 4, and 5 were compared and found not to differ statistically. Apparent dissociation constants were as indicated, and binding capacities (Kd) for days 3, 4, and 5 were 63.06, 87.25, and 87.26 pM, respectively. F9 EC cells were used as a positive control as described (32). B/F, bound/free.
Discussion
In mice, blastocyst implantation depends on LIF expression in the uterus. In other mammalian species, including humans, primates, ungulates, rabbits, and mustelids, the onset of implantation also coincides with an increase in LIF in the uterus (8, 10, 12–14, 31–34). Therefore, LIF may be of general significance to all eutherian mammals at regulating implantation and possibly marsupials, where a LIF homologue recently has been identified (35). Previous results revealed LIF is required to induce a receptive state in the mouse uterus, thus initiating implantation and decidualization. A single injection of LIF into pregnant LIF-deficient females was sufficient to induce embryo implantation and normal development to term. Furthermore, injection of LIF could replace nidatory estrogen for inducing implantation (9).
Here we investigated the responsiveness of uterine tissues to LIF. By using a combination of procedures, we were able to detect transcripts for both the LIFRα and gp130 in the LE during the preimplantation period. In situ and immunohistochemical analysis on intact uteri confirmed these results, revealing the LIFRα was expressed most strongly in the LE, whereas gp130 was localized highly to the GE and to a lesser extent the myometrium. In other species, such as rabbits, primates, and humans, an identical distribution of the LIF receptor has also been reported, with LIFRα and gp130 being expressed predominantly in the uterine epithelia and being largely absent from the stromal tissues (10, 14, 33).
The majority of studies on signal transduction have been performed on established cell lines grown in vitro, which clearly simplifies such analyses. Here we have investigated LIF signaling pathways on tissue directly isolated from animals, potentially complicating such an analysis. In vivo, the tissues may well have been subjected to a multitude of exogenous factors activating or suppressing signaling pathways, in addition to the likelihood that “cross-talk” also may occur between the different pathways. Despite these caveats, it is clear that in purified LE, activation of the Jak–Stat pathway by LIF results in the phosphorylation and binding of p-Tyr-Stat3 to consensus recognition sequences. Of the different factors we tested, including Il-6 and ciliary neurotrophic factor, only LIF resulted in significant increases in p-Tyr-Stat3, although the soluble Il-6 receptor/Il-6 fusion protein was effective at inducing p-Tyr-Stat3, demonstrating the presence of sufficient gp130 in the LE to activate the Jak–Stat pathway. Other factors, such as EGF, previously shown to induce DNA synthesis in the LE (36), had minimal effects on Stat activation despite a 30–60% stimulation of MAPK.
LIF also induces the MAPK pathway. However, it was apparent that the MAPK was activated already in day-3 LE, probably by the action of another as yet unidentified growth factor or cytokine. By day 4, the overall levels of activated MAPK were reduced, and LIF treatment did not increase MAPK phosphorylation, whereas EGF was able to stimulate MAPK on all days. This result suggests that an increase in levels of MAPK by LIF are not essential in inducing a receptive state in the LE, consistent with the observation that mutated derivatives of gp130, which do not activate MAPK, have no effect on fertility (30). The reduced levels of MAPK at the time of uterine receptivity are particularly intriguing, as other studies have revealed cross-talk between the Jak–Stat and MAPK pathways. LIF induces both pathways in embryonic stem cells as well as in neuronal differentiation (28, 29). However, Stat activity is counteracted partially by MAPK in these cells, and if such antagonism occurs between the pathways in the LE, then down-regulation of the MAPK pathway on day 4 may be of significance at enhancing tyrosine phosphorylation or activity of Stat3.
The results on Stat3 phosphorylation from purified LE are supported by the immunohistochemical analysis of Stat3 localization in intact uteri over the periimplantation period. In day-3 uteri, Stat3 was localized to the cytoplasm of the LE. On day 4, localization was predominantly to the nuclei of the entire LE, and by day 5, Stat3 had relocalized to the cytoplasm in the LE between implantation sites. In addition, Stat3 nuclear localization also was detected in the GE on day 5. No significant levels of Stat3 nuclear localization were detected in either the stromal or myometrial tissues over this period except in cells starting to decidualize. Significantly, nuclear localization of Stat3 in the LE of LIF-deficient mice was not detectable in any sample. A few LE nuclei did exhibit localization, but these were isolated examples and did not show the uniform nuclear localization seen in all LE nuclei in the wild-type mice. These results support our in vitro analysis on isolated LE in that in the intact uterus LIF acts predominately on the LE and is primarily responsible for Stat activation in the LE. The stimulus for nuclear localization of Stat3 in the GE on day 5 is unclear, although it may be a result of activation by other factors that are up-regulated at the onset of implantation such as Il-11 (37) or a persistence of LIF expression.
A surprising result was the temporal regulation in responsiveness of the LE to LIF and that it was independent of LIF receptor expression levels. P-Tyr-Stat3 levels in the LE from day 4 of pregnancy uteri were induced maximally, whereas LE from day-5 and day-3 uteri were refractory to LIF stimulation. From these results, it is apparent that changes in uterine receptivity are tightly controlled. Not only does the induction of a state of receptivity in the uterus depend on the stimulus (LIF) being produced by the GE, the target tissue, namely the LE, must also change so that the receptors, although present and which bind LIF, respond by activating signaling pathways. The temporal regulation of both the ligand and the ability of the receptor to respond are key elements in establishing a receptive uterus.
The molecular basis for the change in LIFR activity remains to be established. The LE from mice on the different days of pregnancy binds LIF with the same high affinity, revealing that the temporal variation in responsiveness is not caused by changes in receptor binding. Rather, the mechanism regulating Stat activation must be downstream to the binding of LIF to its receptor. The possible molecular mechanisms regulating responsiveness of the LIFR are many, including cellular factors such as the suppressors of cytokine activity (SOCS) proteins inhibiting the activation of the Jak–Stat pathway (38), to interactions of P-Stat3 with other nuclear factors, such as the estrogen or progesterone receptors whose expression changes in the LE during implantation and which could modulate Stat3 activity (39). Our results indicate that the presence of unphosphorylated Stat3 at all times in the LE suggests that it is the activation/phosphorylation of Stat3 that is regulated. The basis for this is presently unclear, although preliminary results indicate that estrogen alone suppresses LIF responsiveness, but that P4 relieves E2-induced inhibition of Stat3 phosphorylation. However, we have been unable to correlate these changes with any known inhibitor of Stat phosphorylation, with the exception of SOCS3, which is elevated on days 4 and 5, suggesting it could be responsible for inhibiting Stat3 phosphorylation on day 5.
Determining the role of different factors in the many signaling pathways involved in regulating such a process as implantation is complicated by our current inability to culture such tissues as the LE without losing the ability of the tissue to respond in a biologically meaningful manner (40). An alternative approach is to use genetics, and in mice this has been productive in identifying key factors, particularly in the events after the onset of embryo implantation (37, 41–43). Understanding the requirement for such factors as LIFRα, gp130, and Stat3 has been frustrating in that their constitutive ablation results in embryonic lethality. Nevertheless, alternative strategies are available. Mice carrying a mutation of gp130, in which the receptor is truncated and does not activate Stat3, are viable with females being infertile because of implantation failure (44). The uterine levels of P-Stat3 were markedly reduced, resulting in a phenotype indistinguishable from that of LIF deficiency, thus supporting our observations that up-regulation of P-Stat3 is essential to the LE in regulating implantation. An alternative approach to addressing these questions is tissue-specific gene ablation. Application of such procedures and the current techniques for analyzing changes in the complex patterns of gene expression in tissues should provide further insights into how such a critical event as embryo implantation is regulated during the mammalian life cycle.
Acknowledgments
We thank Stefan Rose-John for a generous gift HIl-6, and Lori Sewell and Teresa Shatzer for excellent maintenance of our mouse colony. This research was supported by the National Cancer Institute and the Department of Health and Human Services (C.L.S.).
Abbreviations
- Stat
signal transducer and activator of transcription
- MAPK
mitogen-activated protein kinase
- LIF
leukemia inhibitory factor
- LIFRα
LIF receptor α
- EGF
epidermal growth factor
- LE
luminal epithelium
- GE
glandular epithelium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Finn C A, Martin L. J Reprod Fertil. 1974;39:195–206. doi: 10.1530/jrf.0.0390195. [DOI] [PubMed] [Google Scholar]
- 2.Abrahamsohn P A, Zorn T M. J Exp Zool. 1993;266:603–628. doi: 10.1002/jez.1402660610. [DOI] [PubMed] [Google Scholar]
- 3.Psychoyos A. In: Handbook of Physiology. Greep R O, Astwood E B, editors. Vol. 2. Baltimore: Williams & Wilkins; 1973. pp. 187–215. [Google Scholar]
- 4.Renfree M B, Shaw G. Annu Rev Physiol. 2000;62:353–375. doi: 10.1146/annurev.physiol.62.1.353. [DOI] [PubMed] [Google Scholar]
- 5.Cross J C, Werb Z, Fisher S J. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 6.Stewart C L, Cullinan E B. Dev Genet. 1997;21:91–101. doi: 10.1002/(SICI)1520-6408(1997)21:1<91::AID-DVG11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 7.Stewart C L, Kaspar P, Brunet L J, Bhatt H, Gadi I, Kontgen F, Abbondanzo S J. Nature (London) 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 8.Bhatt H, Brunet L J, Stewart C L. Proc Natl Acad Sci USA. 1991;88:11408–11412. doi: 10.1073/pnas.88.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J R, Cheng J G, Shatzer T, Sewell L, Hernandez L, Stewart C L. Endocrinology. 2000;141:4365–4372. doi: 10.1210/endo.141.12.7855. [DOI] [PubMed] [Google Scholar]
- 10.Cullinan E B, Abbondanzo S J, Anderson P S, Pollard J W, Lessey B A, Stewart C L. Proc Natl Acad Sci USA. 1996;93:3115–3120. doi: 10.1073/pnas.93.7.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirzel D J, Wang J, Das S K, Dey S K, Mead R A. Biol Reprod. 1999;60:484–492. doi: 10.1095/biolreprod60.2.484. [DOI] [PubMed] [Google Scholar]
- 12.Kholkute S D, Katkam R R, Nandedkar T D, Puri C P. Mol Hum Reprod. 2000;6:337–343. doi: 10.1093/molehr/6.4.337. [DOI] [PubMed] [Google Scholar]
- 13.Vogiagis D, Fry R C, Sandeman R M, Salamonsen L A. J Reprod Fertil. 1997;109:279–288. doi: 10.1530/jrf.0.1090279. [DOI] [PubMed] [Google Scholar]
- 14.Yue Z P, Yang Z M, Wei P, Li S J, Wang H B, Tan J H, Harper M J. Biol Reprod. 2000;63:508–512. doi: 10.1095/biolreprod63.2.508. [DOI] [PubMed] [Google Scholar]
- 15.Auernhammer C J, Melmed S. Endocr Rev. 2000;21:313–345. doi: 10.1210/edrv.21.3.0400. [DOI] [PubMed] [Google Scholar]
- 16.Bigsby R M, Cooke P S, Cunha G R. Am J Physiol. 1986;251:E630–E636. doi: 10.1152/ajpendo.1986.251.5.E630. [DOI] [PubMed] [Google Scholar]
- 17.Duncan S A, Zhong Z, Wen Z, Darnell J E., Jr Dev Dyn. 1997;208:190–198. doi: 10.1002/(SICI)1097-0177(199702)208:2<190::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Nicola N A, Metcalf D. J Cell Physiol. 1985;124:313–321. doi: 10.1002/jcp.1041240222. [DOI] [PubMed] [Google Scholar]
- 19.Lim H, Dey S K. Endocrinology. 1997;138:4599–4606. doi: 10.1210/endo.138.11.5528. [DOI] [PubMed] [Google Scholar]
- 20.Saito M, Yoshida K, Hibi M, Taga T, Kishimoto T. J Immunol. 1992;148:4066–4071. [PubMed] [Google Scholar]
- 21.Tomida M, Yamamoto-Yamaguchi Y, Hozumi M. J Biochem (Tokyo) 1994;115:557–562. doi: 10.1093/oxfordjournals.jbchem.a124375. [DOI] [PubMed] [Google Scholar]
- 22.Lai C F, Ripperger J, Morella K K, Wang Y, Gearing D P, Horseman N D, Campos S P, Fey G H, Baumann H. J Biol Chem. 1995;270:23254–23257. doi: 10.1074/jbc.270.40.23254. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer M, Goldschmitt J, Peschel C, Brakenhoff J P, Kallen K J, Wollmer A, Grotzinger J, Rose-John S. Nat Biotechnol. 1997;15:142–145. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- 25.Leaman D W, Pisharody S, Flickinger T W, Commane M A, Schlessinger J, Kerr I M, Levy D E, Stark G R. Mol Cell Biol. 1996;16:369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong Z, Wen Z, Darnell J E., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 27.Paria B C, Lim H, Das S K, Reese J, Dey S K. Semin Cell Dev Biol. 2000;11:67–76. doi: 10.1006/scdb.2000.0153. [DOI] [PubMed] [Google Scholar]
- 28.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 29.Ihara S, Nakajima K, Fukada T, Hibi M, Nagata S, Hirano T, Fukui Y. EMBO J. 1997;16:5345–5352. doi: 10.1093/emboj/16.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtani T, Ishihara K, Atsumi T, Nishida K, Kaneko Y, Miyata T, Itoh S, Narimatsu M, Maeda H, Fukada T, et al. Immunity. 2000;12:95–105. doi: 10.1016/s1074-7613(00)80162-4. [DOI] [PubMed] [Google Scholar]
- 31.Anegon I, Cuturi M C, Godard A, Moreau M, Terqui M, Martinat-Botte F, Soulillou J P. Cytokine. 1994;6:493–499. doi: 10.1016/1043-4666(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 32.Williams R L, Hilton D J, Pease S, Willson T A, Stewart C L, Gearing D P, Wagner E F, Metcalf D, Nicola N A, Gough N M. Nature (London) 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z M, Le S P, Chen D B, Yasukawa K, Harper M J. J Reprod Fertil. 1995;103:249–255. doi: 10.1530/jrf.0.1030249. [DOI] [PubMed] [Google Scholar]
- 34.Song J H, Houde A, Murphy B D. Mol Reprod Dev. 1998;51:13–21. doi: 10.1002/(SICI)1098-2795(199809)51:1<13::AID-MRD2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 35.Cui S, Selwood L. Gene. 2000;243:167–178. doi: 10.1016/s0378-1119(99)00513-2. [DOI] [PubMed] [Google Scholar]
- 36.Nelson K G, Takahashi T, Bossert N L, Walmer D K, McLachlan J A. Proc Natl Acad Sci USA. 1991;88:21–25. doi: 10.1073/pnas.88.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robb L, Li R, Hartley L, Nandurkar H H, Koentgen F, Begley C G. Nat Med. 1998;4:303–308. doi: 10.1038/nm0398-303. [DOI] [PubMed] [Google Scholar]
- 38.Starr R, Hilton D J. BioEssays. 1999;21:47–52. doi: 10.1002/(SICI)1521-1878(199901)21:1<47::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Tan J, Paria B C, Dey S K, Das S K. Endocrinology. 1999;140:5310–5321. doi: 10.1210/endo.140.11.7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glasser S R, Mulholland J. Microsc Res Tech. 1993;25:106–120. doi: 10.1002/jemt.1070250204. [DOI] [PubMed] [Google Scholar]
- 41.Lim H, Paria B C, Das S K, Dinchuk J E, Langenbach R, Trzaskos J M, Dey S K. Cell. 1997;91:197–208. doi: 10.1016/s0092-8674(00)80402-x. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Van De Water T, Lufkin T. Development (Cambridge, UK) 1998;125:621–634. doi: 10.1242/dev.125.4.621. [DOI] [PubMed] [Google Scholar]
- 43.Benson G V, Lim H, Paria B C, Satokata I, Dey S K, Maas R L. Development (Cambridge, UK) 1996;122:2687–2696. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- 44.Ernst, M. (2001) J. Exp. Med., in press.
- 45.Munson P J. Methods Enzymol. 1983;92:543–576. doi: 10.1016/0076-6879(83)92044-x. [DOI] [PubMed] [Google Scholar]