Abstract
Background:
Emerging evidence has demonstrated that lysine-specific demethylase 1 (LSD1) has an important role in many pathological processes of cancer cells, such as carcinogenesis, proliferation and metastasis. In this study, we characterised the role and molecular mechanisms of LSD1 in proliferation and metastasis of colon cancer.
Methods:
We evaluated the correlation of LSD1, CDH-1 and CDH-2 with invasiveness of colon cancer cells, and investigated the roles of LSD1 in proliferation, invasion and apoptosis of colon cancer cells. We further investigated the mechanisms of LSD1-mediated metastasis of colon cancer.
Results:
Lysine-specific demethylase 1 was upregulated in colon cancer tissues, and the high LSD1 expression was significantly associated with tumour-node-metastasis (TNM) stages and distant metastasis. Functionally, inhibition of LSD1 impaired proliferation and invasiveness, and induced apoptosis of colon cancer cells in vitro. The LSD1 physically interacted with the promoter of CDH-1 and decreased dimethyl histone H3 lysine 4 (H3K4) at this region, downregulated CDH-1 expression, and consequently contributed to colon cancer metastasis.
Conclusion:
Lysine-specific demethylase 1 downregulates the expression of CDH-1 by epigenetic modification, and consequently promotes metastasis of colon cancer cells. The LSD1 antagonists might be a useful strategy to suppress metastasis of colon cancer.
Keywords: lysine-specific demethylase 1, CDH-1, CDH-2, colon cancer, metastasis, histone modifications
Colon cancer is the third most common malignancy worldwide (Center et al, 2009; Cui et al, 2011) and the second leading cause of cancer deaths in the United States (American Cancer Society, 2012), recurrence and metastasis are the leading cause of death among colon cancer patients. Epithelial-mesenchymal transition (EMT) is now considered to be the initial and necessary step in the metastatic cascade. During EMT, epithelial cells acquire fibroblast-like characteristics, including loss of cell polarity, reduced intercellular adhesion, increased motility and invasive capacity (Boyer et al, 2000). Transcription factors, such as snai1, slug, zeb1, zeb2 and twist, can promote EMT by suppressing CDH-1 (E-cadherin) expression (Kang and Massagué, 2004; Waldmann et al, 2008; Kato et al, 2010; Shah and Kakar, 2011) and consequently contribute to cancer metastasis.
Lysine-specific demethylase 1 (LSD1), the first discovered histone demethylase, is required in Snail/Slug-mediated transcriptional repression during EMT, in the absence of LSD1, Snail and Slug fail to repress CDH-1 transcription (Lin et al, 2010; Baron et al, 2011; Ferrari-Amorotti et al, 2013). The Snail/Gfi-1 (SNAG) domain of Snail/Slug assembles a histone H3-like structure and serves as a molecular ‘hook' to interact with the LSD1–CoREST complex, and brings this complex to its targeted gene promoters through the binding of the E-box through the zinc-finger motifs (Lin et al, 2010; Ferrari-Amorotti et al, 2013). The LSD1/CoREST complex functions as a reversible nanoscale binding clamp, recruits and anchors a variety of substrate peptides with high sequence similarity to the H3-histone tail (Hwang et al, 2011; Baron and Vellore, 2012a, 2012b).
As a lysine-specific demethylase belonging to the flavin-dependent amine oxidase family, LSD1 specifically catalysed the demethylation of mono- and di-methylated histone H3 lysine 4 (H3K4) and H3 lysine 9 (H3K9) through a redox process. Overexpression of LSD1 promotes proliferation, migration and invasion of various cancer cells (Cho et al, 2011; Lv et al, 2012). And knocking down of LSD1 with small-interfering RNAs (siRNAs) resulted in suppression of proliferation and metastasis of various cancer cells (Hayami et al, 2011; Lv et al, 2012; Pollock et al, 2012; Willmann et al, 2012; Jin et al, 2013; Zhao et al, 2013).
Our previous studies (Ding et al, 2013) have shown that LSD1 had significantly higher expression, in contrast to the significantly lower expression of CDH-1, in colon cancer at high tumour-node-metastasis (TNM) stages (Edge et al, 2010) and with distant metastasis. Positive expression of LSD1 and negative expression of CDH-1 may predict a worse prognosis of colon cancer. We speculated that LSD1 may promote metastasis of colon cancer by decreasing the level of dimethylated histone H3 lysine4 (H3K4m2) at the CDH-1 promoter and repressing CDH-1 transcription, which requires confirmation by further in vitro experiments. Therefore, we attempted to investigate the expression of LSD1, CDH-1 and CDH-2 (N-cadherin) in several colon cancer cell lines, and to analyse their relationship with proliferation and invasion abilities of colon cancer, we also aimed to determine the mechanism of colon cancer metastasis regulated by LSD1.
Material and methods
Immunohistochemical staining
The archival formalin-fixed and paraffin wax-embedded tissue blocks of 108 colon cancer and 30 normal colon mucosa removed by surgery from 2006 to 2008 were retrieved from the Department of Pathology, Xiangya Hospital, Central South University. Primary antibodies were directed towards LSD1 (rabbit monoclonal, 1 : 100; R&D Systems, Minneapolis, MN, USA). Serial sections of 5 μm were cut from the tissue blocks, deparaffinised in xylene, and hydrated in a graded series of alcohol. Staining was then performed using the EnVision+ anti-rabbit system (Dako Corporation, Carpinteria, CA, USA). Negative control staining was carried out by substituting non-immune rabbit and phosphate-buffered saline (PBS) for the primary antibodies. This study was conducted with the approval of the Ethics Committee of Xiangya Hospital, China.
Cell lines and cell culture
Colon cancer cell lines used in this study were purchased from American Type Culture Collection (Sigma-Aldrich Corp., St Louis, MO, USA). These cells include Lovo, SW620, HT-29, HCT-8 and HCT-116. HEK293 cell was obtained from Cancer Research Institute of Central South University, China. HT-29 cells and HCT-8 cells were cultured with RPMI-1640 medium (Sigma-Aldrich Corp.), Lovo cells were cultured with F-12K medium (Sigma-Aldrich Corp.), SW620 cells were cultured with L-15 medium (Sigma-Aldrich Corp.), HCT-116 cells were cultured with McCoy's5a medium (Sigma-Aldrich Corp.) and HEK293 cells were cultured with Modified Eagle's minimal essential medium (Sigma-Aldrich Corp.). All media were supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (Biowest, Nuaillé, France). All of the cell lines were grown in 5% CO2 at 37 °C in incubators with 100% humidity.
Real-time reverse transcription–PCR
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and cDNA synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania). Gene expression of LSD1 was monitored by real-time PCR using ‘Assays on Demand' (Applied Biosystems, Alameda, CA, USA). Expression values were normalised to the geometric mean of GAPDH. The primers that used to amplify the cDNA were as follows: F: 5′-CCTGAAGAACCATCGGGTGT-3′, R: 5′-CCTTCTGGGTCTGTTGTGGT-3′ for LSD1; F: 5′-TCATGAGTGTCCCCCGGTAT-3′, R: 5′-TCTTGAAGCGATTGCCCC AT-3′ for CDH-1; F: 5′-CCTTTCAAACACAGCCACGG-3′, R: 5′-TGTTTGGGT CGGTCTGGA TG-3′ for CDH-2; and F: 5′-CTCATGACCACAGTCCATGC-3′, R: 5′-TTCAGCTCTGGGATGACCTT-3′ for GAPDH.
Western blot analysis
Protein lysates were extracted from cells and blotted as described previously (Kahl et al, 2006). The membranes were incubated for 1–2 h using the following antibodies and dilutions: LSD1, 1 : 1000; CDH-1, 1 : 2000; CDH-2, 1 : 2000.
Small interference RNA and transfection
Cells were seeded with 5 × 104 cells in 24-well plates, then incubated for 2–4 days in standard medium in the presence of 10–20 nmol l−1 siRNA directed against LSD1. The siRNA LSD1 sequences used in this study were as follows: siLSD#1 (sense: 5′-GCCACCCAGAGAUAUUACUTT-3′, anti-sense: 5′-AGUAAUAUCUCUGGGUG GCTT-3′); siLSD#2 (sense: 5′-CCGGAUGACUUCUCAAGAATT-3′, anti-sense: 5′-UUCUUGAGAAGUCAUCCGGTT-3′); siLSD#3 (sense: 5′-CCACGAGUCAAAC CUUUAUTT-3′, anti-sense: 5′-AUAAAGGUUUGACUCGUGGTT-3′); or control siRNA (sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, anti-sense: 5′-ACGUGACACGUUCGGAGAATT-3′). Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Invasion assay
Invasion assays were performed as previously described (Wu et al, 2009; Lin et al, 2010). Briefly, invasion chambers were coated with BD Matrigel matrix according to the manufacturer's protocol (BD Biosciences, San Jose, CA, USA). Cancer cells were seeded on top of the Matrigel in the upper chamber, and the bottom chamber was filled with culture medium containing chemoattractant. Cells that invade through the Matrigel-coated membrane after 24 h were fixed with paraformaldehyde, followed by staining with crystal violet. All experiments were conducted at least three times in triplicate.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide growth inhibition assay
Cells were seeded at a density of 5 × 104 per well and cultured in standard medium, replaced daily. Treatment with siRNA directed against LSD1, and tranylcypromine (Sigma-Aldrich) was accomplished as indicated. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed according to the manufacturer's protocol (Bioassay Systems, Hayward, CA, USA).
Apoptosis assay
Apoptosis assay was performed as previously described (Gorczyca et al, 1998). According to the manufacturer's instructions, 2 × 105 cells were collected and washed twice in PBS. Cells were then resuspended in binding buffer and stained with AnnexinV-FITC and propidium iodide (Promab, Hunan, China). Stained cells were analysed with a flow cytometer (BD FACSAria; BD Biosciences). The measurements were performed independently for at least three times with similar results.
Chromatin immunoprecipitation and histone demethylation assay
Chromatin immunoprecipitation (ChIP) analysis was performed using Protein A and Protein G Dynabeads (Invitrogen) as previously reported (McGarvey et al, 2006). Cells were exposed to 1% formaldehyde to crosslink proteins, and 1.0 × 107 cells were used for each ChIP assay. The antibodies against H3 and LSD1 were from R&D Systems, and antibodies against H3K4m2 were from Invitrogen. Quantitative ChIP was performed using qPCR on the ABI PRISM 7900 real-time PCR detection system (Applied Biosystems). Primer sequences for qPCR of CDH-1 promoter for ChIP were as follows: F: 5′-AGTCCCACAACAGCATAGGG-3′, R: 5′-TTCTGAACTCAGGCGATCCT-3′. Sheared genomic DNA was used as a positive control (input) and for the normalisation of DNA immunoprecipitated by LSD1.
Statistical analysis
Statistical analysis was performed using SPSS software (version 17.0; SPSS Inc., Chicago, IL, USA). Statistical analysis was performed with Student's t-test and ANOVA. Values of P<0.05 were considered to be statistically significant.
Results
LSD1 expression is upregulated in colon cancer tissues
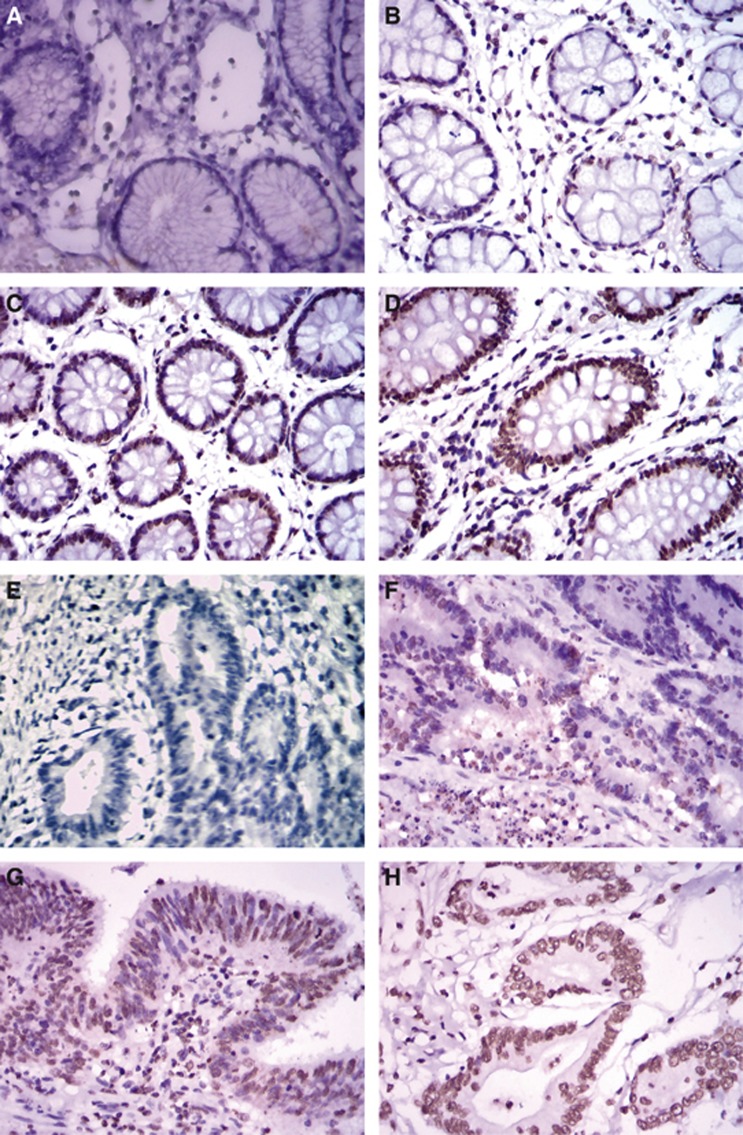
We first examined the expression levels of LSD1 in 108 colon cancer tissues and 30 normal colon tissues by immunohistochemistry (IHC) obtained in Xiangya Hospital, Central South University. High LSD1 expression was detected in the nuclei of malignant cells, while low staining was observed in any of non-neoplastic tissues (Figure 1). Specifically, the expression of LSD1 was observed in 67% (72 out of 108) of colon cancer tissues and 37% (11 out of 30) of normal colon mucosa, indicating a significant elevation of LSD1 expression in tumours compared with normal control tissues (P=0.005, Table 1). High levels of LSD1 expression (score: ‘++' to ‘+++') were detected in 52 (48%) tumour tissues and 5 (17%) benign colon tissues, respectively. As shown in Table 1, there was no significant correlation of LSD1 expression with gender, age and Broders' classification (P>0.05). However, the expression of LSD1 was significantly more in colon cancer with high TNM stages and distant metastasis (P<0.05). Therefore, we hypothesised that LSD1 may have an important role in metastasis of colon cancer.
Figure 1.
(A–D) Representative LSD1 staining in normal control specimens. (A) Negative, (B) weak, (C) moderate and (D) strong. (E–H) Representative LSD1 staining in colon cancer specimens. (E) Negative, (F) weak and (G) moderate and (H) strong. LSD1 was positively expressed in 37% (11 out of 30) of control specimens and 67% (72 out of 108) of colon cancer specimens, respectively.
Table 1. Statistical analysis of LSD1 expression levels in colon cancer and control specimens.
| |
|
LSD1 (+) |
|
||
|---|---|---|---|---|---|
| Item | Case (n) | n | % | χ2 | P-value |
|
Origin of specimens | |||||
| Colon cancer | 108 | 72 | 67 | ||
| Normal control tissues |
30 |
11 |
37 |
7.61 |
0.005 |
|
Gender | |||||
| Male | 65 | 42 | 72 | ||
| Female |
43 |
30 |
81 |
0.31 |
0.578 |
|
Age | |||||
| <50 years | 37 | 26 | 84 | ||
| ⩾50 years |
71 |
46 |
72 |
0.33 |
0.566 |
|
TNM stage | |||||
| Stage I | 19 | 7 | 37 | ||
| Stage II | 34 | 23 | 68 | ||
| Stage III | 47 | 34 | 72 | ||
| Stage IV |
8 |
8 |
100 |
12.3 |
0.006 |
|
Broders' classification | |||||
| Grade I | 18 | 8 | 44 | ||
| Grade II | 58 | 41 | 71 | ||
| Grade III | 26 | 18 | 69 | ||
| Grade IV |
6 |
5 |
83 |
5.25 |
0.154 |
|
Distant metastasis | |||||
| Yes | 8 | 8 | 100 | ||
| No | 100 | 64 | 64 | 4.32 | 0.038 |
Abbreviations: LSD1=lysine-specific demethylase 1; TNM=tumour-node-metastasis.
Correlation of LSD1, CDH-1 and CDH-2 with invasiveness of colon cancer cell lines
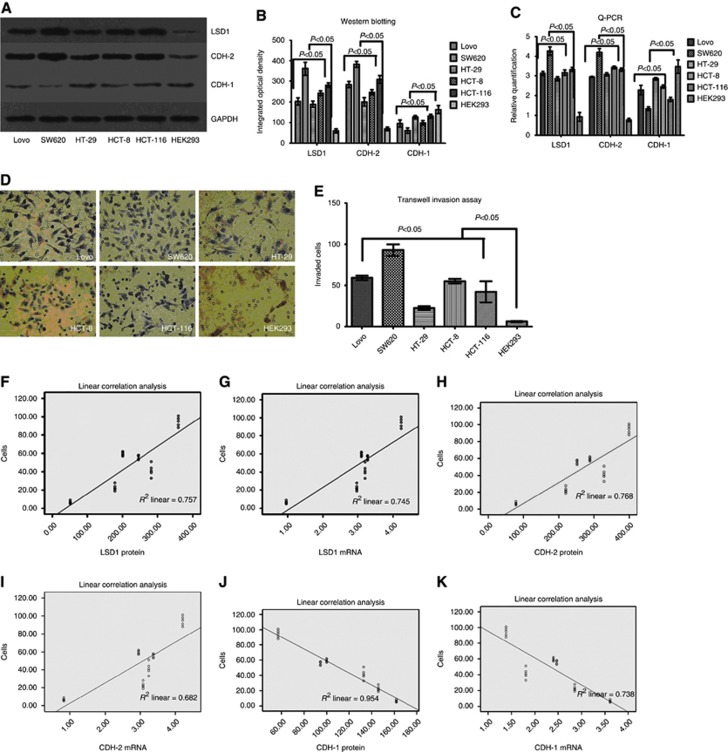
To evaluate the correlation of LSD1, CDH-1 and CDH-2 with invasiveness of colon cancer, we detected their expression in colon cancer cells by RT–PCR and western blot. The results showed that the mRNA and protein levels of LSD1 and CDH-2 in SW620 cell line were significantly higher than those in the Lovo, HT-29, HCT-8, HCT-116 and HEK293 (control) cell lines. However, the levels of CDH-1 in SW620 cell line were evidently lower than the other five cell lines (P<0.05, Figure 2A–C).
Figure 2.
LSD1 and CDH-2 expression correlated positively with the invasiveness of colon cancer cells, whereas the CDH-1 exhibited a negative correlation with the invasiveness of colon cancer cells. (A–C) LSD1 and CDH-2 had significantly higher expression, in contrast to the significantly lower expression of CDH-1 in SW620 cells than the other cells. (D and E) SW620 cells exhibited significantly higher invasive ability. Bars represent mean±standard deviation of three independent experiments. (F–K) The protein and mRNA levels of LSD1 and CDH-2 correlated positively with the invasiveness of colon cancer cells, whereas that of CDH-1 exhibited a negative correlation with the invasiveness of colon cancer cells.
Using Transwell Invasion Assay, we detected the invasiveness of six cell lines. The ability to cross matrigel indicated the invasiveness of cancer cells. Interestingly, the amount of invading cells crossing the matrigel basement membrane was evidently higher in SW620 cell line than that in the other five cell lines (P<0.05, Figure 2D and E). Linear correlation was used to analyse the correlation between expression of LSD1, CDH-1, CDH-2 in six cell lines and invasiveness of the six cell lines due to normal distribution data (Z=0.727, P=0.665), and the results showed that the protein and mRNA levels of LSD1 and CDH-2 correlated positively with the invasiveness of colon cancer cells, whereas that of CDH-1 exhibited a negative correlation with the invasiveness of colon cancer cells (P<0.05, Tables 2 and 3; Figure 2F–K).
Table 2. Correlation analysis (Linear correlation) between the protein levels of LSD1, CDH-1 and CDH-2 and invasiveness of six cell lines.
| Item | β | F | P |
|---|---|---|---|
| LSD1 |
0.87 |
87.28 |
<0.05 |
| CDH-2 |
0.88 |
92.48 |
<0.05 |
| CDH-1 | −0.98 | 87.28 | <0.05 |
Abbreviation: LSD1=lysine-specific demethylase 1.
Table 3. Correlation analysis (Linear correlation) between the mRNA levels of LSD1, CDH-1 and CDH-2 and invasiveness of six cell lines.
| Item | β | F | P |
|---|---|---|---|
| LSD1 |
0.86 |
81.74 |
<0.05 |
| CDH-2 |
0.83 |
60.06 |
<0.05 |
| CDH-1 | −0.86 | 79.05 | <0.05 |
Abbreviation: LSD1=lysine-specific demethylase 1.
Inhibition of LSD1 impairs proliferation and invasiveness, and induces apoptosis of colon cancer cells in vitro
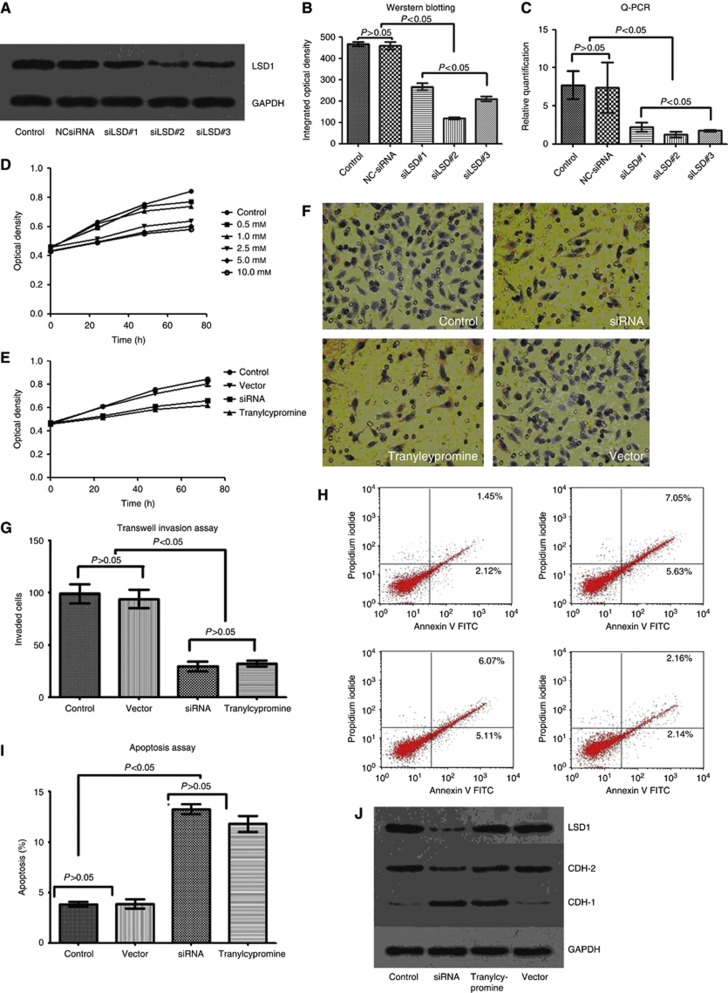
We performed a knockdown experiment using siRNAs targeting LSD1, and tranylcypromine, a chemical inhibitor for LSD1 (Karytinos et al, 2009; Culhane et al, 2010; Schenk et al, 2012), to further investigate the roles of LSD1 in proliferation and invasion of colon cancer cells. Considering SW620 cell line was more aggressive than the other cell lines, it was used to perform the following experiment. First, three independent siRNAs targeting LSD1 (siLSD#1, #2 and #3) were transfected to SW620 cells to detect gene-silencing efficiency. As shown in Figure 3, the knockdown effect of siLSD#2 were better than that of the other two siRNAs at both mRNA and protein levels. Then, the MTT Cell Proliferation Assay was used for evaluation of optimal drug concentration in response to tranylcypromine. The cellular proliferation declined along with tranylcypromine treatment in a concentration-dependent manner. And it was notably lower in the 2.5 mM-, 5 mM- and 10 mM-treated groups than in the 0.5 mM- or 1 mM-treated groups (P<0.05). However, there was no significant difference between the 0.5 mM- and 1 mM-treated groups, or the 2.5 mM-, 5 mM- and 10 mM-treated groups (P>0.05, Figure 3D).
Figure 3.
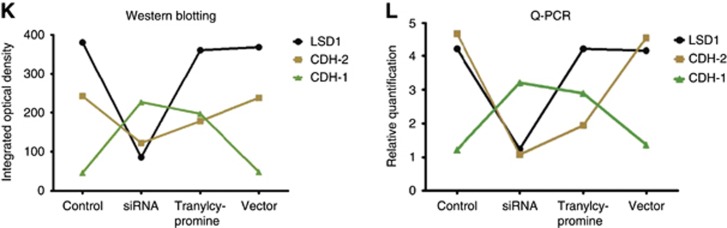
Inhibition of LSD1 impairs proliferation and invasiveness of colon cancer cells. (A–C) The knockdown effect of siLSD#2 was more efficient than the other two siRNAs at protein and mRNA levels. (D) Cellular proliferation declined along with tranylcypromine treatment in a concentration-dependent manner, and it was notably lower in the 2.5 mM-, 5 mM- and 10 mM-treated groups than in the 0.5 mM- or 1 mM-treated groups. (E) Cell Proliferation Assay showed that siLSD#2 and tranylcypromine effectively suppressed proliferation of colon cancer cells. (F and G) Transwell Invasion Assay revealed that siLSD#2 and tranylcypromine effectively inhibited invasiveness of colon cancer cells. (H and I) Apoptosis Assay showed that suppression of LSD1 with siRNA and tranylcypromine induced apoptosis of colon cancer cell. (J–L) Downregulation of LSD1 and CDH-2 and upregulation of CDH-1 were observed after treated with siLSD#2. Bars represent mean±standard deviation of three independent experiments.
Using the siLSD#2 and tranylcypromine (2.5 mM), we performed Transwell Invasion Assay, Cell Proliferation Assay and Apoptosis Assay in SW620 cell lines, and found a significant suppression of invasion and growth, and induced cell apoptosis by siRNA and tranylcypromine (P<0.05, Figure 3E–I). No suppressive effect was found when we used control siRNAs (Figure 3J–L). Interestingly, upregulation of CDH-1 and downregulation of CDH-2 were observed after treated with siLSD#2 and tranylcypromine for 72 h (P<0.05, Figure 3J–L). However, the LSD1 level of SW620 cells remained almost the same after treated with tranylcypromine, which might demonstrate that tranylcypromine cannot downregulate the expression level, but inhibit the enzymatic activity of LSD1.
LSD1 regulates EMT via demethylation of CDH-1 gene
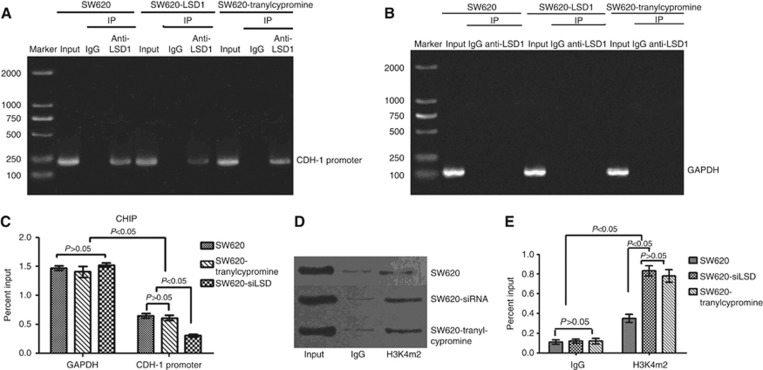
Given that inhibition of LSD1 accompanied by upregulation of CDH-1 and downregulation of invasiveness of colon cancer, we speculated that LSD1 can promote metastasis of colon cancer by downregulating CDH-1 expression. To assess whether the promoter of CDH-1 is directly regulated by LSD1, which consequently would lead to an enrichment of activating histone marks, ChIP analysis was performed using anti-LSD1 and anti-H3K4m2 antibodies in SW620 cells, LSD1-silenced SW620 cells and tranylcypromine-treated SW620 cells (transfected with siLSD#2 or treated with tranylcypromine for 48 h). The results confirmed that LSD1 is present at the proximal promoter of CDH-1 in cells of all the three groups, and quantitative analysis revealed that the enrichment of LSD1 at the proximal promoter of CDH-1 was significantly higher in SW620 and tranylcypromine-treated SW620 cells than in LSD1-silenced SW620 cells (Figure 4A–C), which was in accordance with invasiveness of SW620 cells and LSD1-silenced SW620 cells, but was inconsistent with invasiveness of tranylcypromine-treated SW620 cells. We further investigate whether tranylcypromine influenced the enzymatic activity of LSD1.
Figure 4.
LSD1 reduces H3K4 dimethylation at the promoter of CDH-1. (A–C) The enrichment of LSD1 at the proximal promoter of CDH-1 (immunoprecipitated with anti-LSD1) was significantly lower in LSD1-silenced SW620 cells than in SW620 cells and tranylcypromine-treated SW620 cells. (D and E) The level of H3K4m2 at the promoter of the CDH-1 gene in LSD1-silenced SW620 cells and tranylcypromine- treated SW620 cells was significantly higher than that in SW620 cells. Values represent mean±standard deviation of three independent experiments.
Both di- and tri-methylated H3K4 are associated with active transcription (Kouzarides, 2007; Li et al, 2007). Using an antibody specific for dimethyl H3K4 (H3K4m2), we detected relatively high levels of H3K4m2 at the promoter of the CDH-1 gene in LSD1-silenced SW620 cells and tranylcypromine-treated SW620 cells, and a significant decrease in this active mark specifically at the promoter region in SW620 cells (Figure 4D and E). Therefore, we conclude that the expression of LSD1 leads to a specific decrease in H3K4m2 at CDH-1 promoters, downregulates the CDH-1 expression, and consequently contributes to colon cancer metastasis; Tranylcypromine cannot downregulate the protein level, but suppress the enzymatic activity of LSD1 at the proximal promoter of CDH-1. Downregulation of protein level or inhibition of enzymatic activity of LSD1 can prevent metastasis of colon cancer effectively. Since LSD1 cannot remove trimethylation of H3K4 (H3K4m3) (Shi et al, 2004), we did not perform ChIP assay for the abundance of H3K4m3 at the CDH-1 promoter.
Discussion
Metastasis is the major cause of mortality among colon cancer patients. Therefore, it is important for us to understand mechanisms that help colon cancer cells to acquire invasive/metastatic potential (Singh et al, 2011). This study has demonstrated that LSD1 was significantly upregulated in colon cancer, by IHC, western blot and real-time PCR. With high levels of LSD1 and CDH-2, and low levels of CDH-1, SW620 cells showed significantly higher invasive potential than the other colon cancer cells, indicating that LSD1 and CDH-2 correlated positively with the invasiveness of colon cancer cell lines, whereas the CDH-1 exhibited a negative correlation with the invasiveness of colon cancer cell lines.
Lysine-specific demethylase 1 is composed of three domains, including an N-terminal SWIRM domain, a conserved motif shared by many chromatin regulatory complexes, an amine oxidase domain (AO domain) and a C-terminal Tower domain (Forneris et al, 2005; Shi and Whetstine, 2007; Yang et al, 2007). The LSD1 cooperates with CoREST, CtBP24 corepressor complex, demethylates histone H3-K4 and H3-K9 through this interaction (Lee et al, 2005; Gatta and Mantovani, 2008), and regulates the expression of its target gene by this epigenetic modification. Lysine-specific demethylase 1 is found to participate in development and differentiation (Lan et al, 2008; Adamo et al, 2011; Whyte et al, 2012), regulation of chromatin remodelling, cell apoptosis and methylation of DNA and histone (Ouyang and Gill, 2009; Wang et al, 2009; Amente et al, 2010; Deleris et al, 2010; Hayami et al, 2011; He et al, 2011; Yeh et al, 2011). More importantly, LSD1 is involved in many pathological processes of cancer, such as carcinogenesis, proliferation, metastasis and apoptosis (Huang et al, 2007; Scoumanne and Chen, 2007; Schulte et al, 2009; Hayami et al, 2011; Bennani-Baiti et al, 2012).
The LSD1/CoREST complex dynamics works as a nanoscale clamp opening/closing on several hundred nanosecond time scales reversibly (Baron and Vellore, 2012a, 2012b), making LSD1/CoREST a potential docking site for multiple protein partners sharing high N-terminus sequence similarity with H3 histone tail (Baron et al, 2011; Hwang et al, 2011; Baron and Vellore, 2012a, 2012b; Laurent et al, 2012). The H3 histone tail binding pocket is considered to be a potential allosteric site regulating the opening/closing motion of the clamp (Hwang et al, 2011; Baron and Vellore, 2012a, 2012b). Binding of CoREST can not only protect LSD1 from proteasomal degradation, but also regulate the structure of the AO domain of LSD1 to control the interaction of LSD1 with its substrate (Lin et al, 2010). In fact, LSD1/CoREST complex does not have the function of cadherin recognition, which is mainly based on the binding of the E-box through the zinc-finger motifs of transcription factor (e.g., Snail and slug) (Lin et al, 2010; Ferrari-Amorotti et al, 2013).
Our experiment revealed that inhibition of LSD1 impaired proliferation and invasiveness, and induced apoptosis of colon cancer cells in vitro. The LSD1 is required for cell proliferation in both p53-dependent and -independent manners, deficiency in LSD1 can lead to a partial cell-cycle arrest in G(2)/M and sensitises cells to growth suppression induced by DNA damage or murine double minute 2 (MDM2) inhibition (Scoumanne and Chen, 2007). Through enhancement of cell-cycle progression, LSD1 could promote growth of cancer cells, whereas inhibition of LSD1 could suppress the G1-to-S progression (Hayami et al, 2011), and even induced cells apoptosis (Wang et al, 2009; Wen et al, 2012).
Cancer invasion and metastasis are landmark events that transform a locally growing tumour into a systemic, metastatic and live-threatening disease (Christofori, 2003). As an important regulator of cell shape, growth and polarity, CDH-1 has a crucial role in epithelial cell–cell adhesion and in the maintenance of tissue architecture (Angst et al, 2001; Greenspon et al, 2011). Indeed, CDH-1 serves as a widely acting suppressor of invasion and growth of epithelial cancers (Hazan et al, 2004). The loss of CDH-1-mediated cell–cell adhesion is a prerequisite for tumour cell invasion and metastasis formation (Friedl and Alexander, 2011; Yip and Seow, 2012). Transcriptional factor Snail can repress the expression of CDH-1 by epigenetic mechanisms dependent on the interaction of its N-terminal SNAG domain with LSD1 (Ferrari-Amorotti et al, 2013). Di- and tri-methylation of H3K4 (H3K4m2/m3) is associated with actively transcribed genes (Santos-Rosa et al, 2002; Miao and Natarajan, 2005; Morillon et al, 2005). In the process of EMT, LSD1 removes dimethylation of lysine 4 on histone H3 (H3K4m2) at the CDH-1 promoter, and downregulates the CDH-1 expression (Lin et al, 2010; Huang et al, 2011), which is also proved by our experiment.
Since LSD1 has been demonstrated to be overexpressed in colon cancer with higher TNM stages and distant metastasis, inhibition of LSD1 impaired proliferation and invasiveness, and induced apoptosis of colon cancer cells in vitro, the use of LSD1 inhibitors may provide an important potential therapy of cancer. Due to the high structural and mechanistic similarities between LSD1 and amine oxidases, monoamine oxidase (MAO) covalent inhibitors such as pargyline, tranylcypromine and polyamine analogues have been shown to inhibit LSD1 enzymatic activity (Huang et al, 2007, 2009). In our experiment, tranylcypromine inhibited LSD1 enzymatic activity and suppressed the growth and invasiveness of colon cancer cells effectively. Inhibitors of LSD1 could also induce the reexpression of the aberrantly silenced gene and result in significant inhibition of the growth of colon cancer xenograft model in vivo (Santos-Rosa et al, 2002).
However, most of the MAO inhibitors do not selectively target LSD1 and therefore, limits their use as therapeutics owing to potential side effects (Willmann et al, 2012). Unlike the non-selective MAO inhibitors that form a covalent bond between FAD and the compounds, the selective and reversible inhibitors such as Namoline (Willmann et al, 2012) and CBB compounds (Wang et al, 2011) can specifically interact with LSD1 and inhibit its activity without forming a covalent bond, and impair proliferation of cancer cells in vitro and in vivo (Wang et al, 2011; Willmann et al, 2012). Unfortunately, these reversible LSD1 inhibitors are either modest in activity or polycationic in nature. Therefore, more effective, high-affinity, non-covalent and fully reversible LSD1 inhibitors still have yet to be found.
In conclusion, LSD1 was expressed significantly higher in colon cancer with higher TNM stages and distant metastasis. Inhibition of LSD1 impairs proliferation and invasiveness, and induces apoptosis of colon cancer cells in vitro. By removing dimethylation of lysine 4 on histone H3 (H3K4m2) at the CDH-1 promoter, LSD1 downregulates the CDH-1 expression, and contributes to metastasis of colon cancer.
Acknowledgments
We authors are grateful to Dr Hecheng Zhu (Cancer Research Institute, Central South University, Changsha, China) for providing technical guidance. This study was supported by Science and Technology Fund of Guizhou Province. Grant Number: [2013]2178.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Adamo A, Sesé B, Boue S, Castaño J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13 (6:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29 (25:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- American Cancer Society . Cancer Facts and Figures 2012. American Cancer Society: Atlanta, GA; 2012. [Google Scholar]
- Angst BD, Marcozzi C, Magee AI. The cadherin superfamily. J Cell Sci. 2001;114 (Pt 4:625–626. doi: 10.1242/jcs.114.4.625. [DOI] [PubMed] [Google Scholar]
- Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure. 2011;19 (2:212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R, Vellore NA. LSD1/CoREST is an allosteric nanoscale clamp regulated by H3-histone-tail molecular recognition. Proc Natl Acad Sci USA. 2012a;109 (31:12509–12514. doi: 10.1073/pnas.1207892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R, Vellore NA. LSD1/CoREST reversible opening-closing dynamics: Discovery of a nanoscale clamp for chromatin and protein binding. Biochemistry. 2012b;51 (5:3151–3153. doi: 10.1021/bi300068r. [DOI] [PubMed] [Google Scholar]
- Bennani-Baiti IM, Machado I, Llombart-Bosch A, Kovar H. Lysine- specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing's sarcoma, osteosarcoma, and rhabdomyosarcoma. Hum Pathol. 2012;43 (8:1300–1307. doi: 10.1016/j.humpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Boyer B, Valles AM, Edme N. Induction and regulation of epithelial-mesenchymal transitions. Biochem Pharmacol. 2000;60 (8:1091–1099. doi: 10.1016/s0006-2952(00)00427-5. [DOI] [PubMed] [Google Scholar]
- Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59 (1:366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- Cho HS, Suzuki T, Dohmae N, Hayami S, Unoki M, Yoshimatsu M, Toyokawa G, Takawa M, Chen T, Kurash JK, Field HI, Ponder BA, Nakamura Y, Hamamoto R. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71 (3:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- Christofori G. Changing neighbours, changing behaviour: cell adhesion molecule-mediated signalling during tumour progression. EMBO J. 2003;22 (10:2318–2323. doi: 10.1093/emboj/cdg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R, Okada Y, Jang SG, Ku JL, Park JG, Kamatani Y, Hosono N, Tsunoda T, Kumar V, Tanikawa C, Kamatani N, Yamada R, Kubo M, Nakamura Y, Matsuda K. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60 (6:799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J Am Chem Soc. 2010;132 (9:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Greenberg MV, Ausin I, Law RW, Moissiard G, Schubert D, Jacobsen SE. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2- mediated maintenance of DNA methylation. EMBO Rep. 2010;11 (12:950–955. doi: 10.1038/embor.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Liao G, Zhang Z, Wen J, Zeng L, Liu S, Zhang Y. Positive expression of LSD1 and negative expression of E-cadherin correlates with metastasis and poor prognosis of colon cancer. Dig Dis Sci. 2013;58 (6:1581–1589. doi: 10.1007/s10620-012-2552-2. [DOI] [PubMed] [Google Scholar]
- Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A.2010AJCC Cancer Staging Manual7th ednSpringer: New York, NY [Google Scholar]
- Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S, Manzotti G, Grisendi G, Dominici M, Pieraccioli M, Raschellà G, Claudia C, Colombo MP, Calabretta B. Inhibiting interactions of lysine demethylase LSD1 with Snail/Slug blocks cancer cell invasion. Cancer Res. 2013;73 (1:235–245. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579 (10:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147 (5:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Gatta R, Mantovani R. NF-Y substitutes H2A-H2B on active cell-cycle promoters: recruitment of CoREST-KDM1 and fine-tuning of H3 methylations. Nucleic Acids Res. 2008;36 (20:6592–6607. doi: 10.1093/nar/gkn699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gorczyca W, Melamed MR, Darzynkiewicz Z. Analysis of apoptosis by flow cytometry. Methods Mol Biol. 1998;91:217–238. doi: 10.1385/0-89603-354-6:217. [DOI] [PubMed] [Google Scholar]
- Greenspon J, Li R, Xiao L, Rao JN, Sun R, Strauch ED, Shea-Donohue T, Wang JY, Turner DJ. Sphingosine-1-phosphate regulates the expression of adherens junction protein E-cadherin and enhances intestinal epithelial cell barrier function. Dig Dis Sci. 2011;56 (5:1342–1353. doi: 10.1007/s10620-010-1421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128 (3:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann NY Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- He S, Huang K, Zhang X, Yu X, Huang P, An C. The LSD1-type zinc finger motifs of Pisum sativa LSD1 are a novel nuclear localization signal and interact with importin alpha. PLoS One. 2011;6 (7:e22131. doi: 10.1371/journal.pone.0022131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449 (7158:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Huang PH, Chen CH, Chou CC, Sargeant AM, Kulp SK, Teng CM, Byrd JC, Chen CS. Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol Pharmacol. 2011;79 (1:197–206. doi: 10.1124/mol.110.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA. 2007;104 (19:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15 (23:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Schmitt AA, Luteran AE, Toone EJ, McCafferty DG. Thermodynamic characterization of the binding interaction between the histone demethylase LSD1/KDM1 and CoREST. Biochemistry. 2011;50 (4:546–557. doi: 10.1021/bi101776t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hanigan CL, Wu Y, Wang W, Park BH, Woster PM, Casero RA., Jr Loss of Lysine-Specific Demethylase 1 (LSD1) suppresses growth and alters gene expression of human colon cancer cells in a p53 and DNA methyltransferase 1 (DNMT1) independent manner. Biochem J. 2013;449 (2:459–468. doi: 10.1042/BJ20121360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schüle R, Buettner R. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118 (3:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284 (26:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Yashiro M, Noda S, Tendo M, Kashiwagi S, Doi Y, Nishii T, Matsuoka J, Fuyuhiro Y, Shinto O, Sawada T, Ohira M, Hirakawa K. Establishment and characterization of a new hypoxia-resistant cancer cell line, OCUM-12/Hypo, derived from a scirrhous gastric carcinoma. Br J Cancer. 2010;102 (5:898–907. doi: 10.1038/sj.bjc.6605543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128 (4:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20 (3:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B, Randrianarison-Huetz V, Frisan E, Andrieu-Soler C, Soler E, Fontenay M, Dusanter-Fourt I, Duménil D. A short Gfi-1B isoform controls erythroid differentiation by recruiting the LSD1-CoREST complex through the dimethylation of its SNAG domain. J Cell Sci. 2012;125 (Pt 4:993–1002. doi: 10.1242/jcs.095877. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437 (7057:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128 (4:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29 (35:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29 (11:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS One. 2012;7 (4:e35065. doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66 (7:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Miao F, Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Mol Cell Biol. 2005;25 (11:4650–4661. doi: 10.1128/MCB.25.11.4650-4661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell. 2005;18 (6:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription. Epigenetics. 2009;4 (7:440–444. doi: 10.4161/epi.4.7.9807. [DOI] [PubMed] [Google Scholar]
- Pollock JA, Larrea MD, Jasper JS, McDonnell DP, McCafferty DG. Lysine- specific histone demethylase 1 inhibitors control breast cancer proliferation in ERα-dependent and -independent manners. ACS Chem Biol. 2012;7 (7:1221–1231. doi: 10.1021/cb300108c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419 (6905:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, Casero RA, Jr, Marton L, Woster P, Minden MD, Dugas M, Wang JC, Dick JE, Müller-Tidow C, Petrie K, Zelent A. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18 (4:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, Metzger E, Schüle R, Eggert A, Buettner R, Kirfel J. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69 (5:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and -independent manners. J Biol Chem. 2007;282 (21:15471–15475. doi: 10.1074/jbc.M701023200. [DOI] [PubMed] [Google Scholar]
- Shah PP, Kakar SS. Pituitary tumor transforming gene induces epithelial to mesenchymal transition by regulation of Twist, Snail, Slug, and E-cadherin. Cancer Lett. 2011;311 (1:66–76. doi: 10.1016/j.canlet.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119 (7:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25 (1:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S, Washington MK, Yeatman TJ, Beauchamp RD, Dhawan P. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. 2011;141 (6:2140–2153. doi: 10.1053/j.gastro.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann J, Feldmann G, Slater EP, Langer P, Buchholz M, Ramaswamy A, Saeger W, Rothmund M, Fendrich V. Expression of the zinc-finger transcription factor Snail in adrenocortical carcinoma is associated with decreased survival. Br J Cancer. 2008;99 (11:1900–1907. doi: 10.1038/sj.bjc.6604755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41 (1:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, Zhang H. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71 (23:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138 (4:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wen L, Chen Y, Zeng LL, Zhao F, Li R, Liu Y, Zhang C. Triptolide induces cell-cycle arrest and apoptosis of human multiple myeloma cells in vitro via altering expression of histone demethylase LSD1 and JMJD2B. Acta Pharmacol Sin. 2012;33 (1:109–119. doi: 10.1038/aps.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482 (7384:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, Kirfel J, Waldmann H, Schüle R, Buettner R. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. Int J Cancer. 2012;131 (11:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15 (5:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14 (6:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- Yeh SH, Lin CS, Wu FH, Wang AY. Analysis of the expression of BohLOL1, which encodes an LSD1-like zinc finger protein in Bambusa oldhamii. Planta. 2011;234 (6:1179–1189. doi: 10.1007/s00425-011-1467-z. [DOI] [PubMed] [Google Scholar]
- Yip WK, Seow HF. Activation of phosphatidylinositol 3-kinase/Akt signaling by EGF downregulates membranous E-cadherin and β-catenin and enhances invasion in nasopharyngeal carcinoma cells. Cancer Lett. 2012;318 (2:162–172. doi: 10.1016/j.canlet.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Zhao ZK, Dong P, Gu J, Chen L, Zhuang M, Lu WJ, Wang DR, Liu YB. Overexpression of LSD1 in hepatocellular carcinoma: a latent target for the diagnosis and therapy of hepatoma. Tumour Biol. 2013;34 (1:173–180. doi: 10.1007/s13277-012-0525-x. [DOI] [PubMed] [Google Scholar]