Abstract
Caenorhabditis elegans is an ideal organism for the study of the molecular basis of fundamental biological processes such as germ-line development, especially because of availability of the whole genome sequence and applicability of the RNA interference (RNAi) technique. To identify genes involved in germ-line development, we produced subtracted cDNA pools either enriched for or deprived of the cDNAs from germ-line tissues. We then performed differential hybridization on the high-density cDNA grid, on which about 7,600 nonoverlapping expressed sequence tag (EST) clones were spotted, to identify a set of genes specifically expressed in the germ line. One hundred and sixty-eight clones were then tested with the RNAi technique. Of these, 15 clones showed sterility with a variety of defects in germ-line development. Seven of them led to the production of unfertilized eggs, because of defects in spermatogenesis (4 clones), or defects in the oocytes (3 clones). The other 8 clones led to failure of oogenesis. These failures were caused by germ-line proliferation defect (Glp phenotype), meiotic arrest, and defects in sperm–oocyte switch (Mog phenotype) among others. These results demonstrate the efficacy of the screening strategy using the EST library combined with the RNAi technique in C. elegans.
Germ cells play essential roles in transmission of parental genetic information to the next generation and have various characteristics that somatic cells do not. However, our knowledge of molecular mechanisms of germ-line development is still limited (1). Caenorhabditis elegans is an ideal organism for the study of such mechanisms, because of its powerful genetics and because it is hermaphroditic and transparent. The latter characteristics enable observation of spermatogenesis, oogenesis, fertilization, and early embryogenesis in a sequential manner in a single animal (2–4). However, the forward-genetic approach involving positional cloning, a common approach used so far in this organism, is laborious and time consuming. On the other hand, the whole genome sequence is now known and cDNA clones (“yk clones”) corresponding to about 9,500 genes have been cloned in the cDNA project. Furthermore, RNA interference (RNAi; ref. 5) is particularly effective in this organism, making a large-scale reverse-genetic screening possible (6–8). To identify genes involved in germ-line development efficiently, we took an approach to find genes expressed in a germ-line-specific manner by cDNA subtraction and differential hybridization. By performing RNAi screening through these genes, we could molecularly identify 15 genes essential for germ-line development, providing a platform for further understanding of the molecular mechanisms of germ-line development.
Materials and Methods
Strains and Culture.
C. elegans strains were maintained and handled basically as described (9). The glp-1(q224ts) strain and the glp-4(bn2ts) strain were provided by the Caenorhabditis Genetics Center (Univ. of Minnesota, St. Paul). For construction of the peIs1 strain that carries an integrated array of let-60∷gfp, the coding region of gfp was fused to the let-60 promoter and introduced into the wild-type N2. The extrachromosomal array was integrated into the chromosome by γ-ray irradiation. For synchronous cultures, worms were synchronized as embryos by treatment with alkaline bleach and grown in a liquid medium as described previously (10).
RNA Preparation and cDNA Subtraction by the Suppression Subtractive Hybridization Technique (SSH).
We prepared RNA samples from three sources: (A) To obtain worms with many meiotic germ nuclei, the glp-1(q224ts) mutant grown at the permissive temperature of 15°C was shifted at the L4 to young adult stage to the restrictive temperature of 25°C, then kept at this temperature for 6 h, and total RNA was extracted. At this stage, we could observe that most of the nuclei in the distal region of the gonad, where mitosis normally occurs, entered meiosis. (B) To obtain animals with few germ nuclei, the glp-4(bn2ts) mutant was grown at the permissive temperature, synchronized as embryos, and then grown at the restrictive temperature of 25°C to adulthood, where total RNA was isolated. (C) RNA was also isolated from N2 embryos. N2 adults grown at 15°C were shifted to and incubated at 25°C for 6 h and embryos were prepared by bleaching.
For each sample, mRNA (2 μg each) was isolated by using Message Maker (GIBCO/BRL). cDNA synthesis and subtraction were performed with PCR-Select cDNA subtraction kit (CLONTECH) according to the manufacturer's instructions. Twenty-seven cycles of primary PCR and 12 cycles of secondary PCR were performed by using Advantage cDNA polymerase mix (CLONTECH). We generated forward-subtracted cDNA pool [“F cDNA pool,” cDNA A as tester and cDNA B plus cDNA C as driver: A − (B + C)] and reverse-subtracted cDNA pool [“R cDNA pool,” cDNA B plus cDNA C as tester and cDNA A as driver: (B + C) − A]. Subtraction of glp-4 cDNA from the glp-1 cDNA will enrich for germ-line-specific cDNAs. However, this procedure will include many cDNAs for maternal messages required for early embryogenesis rather than for meiosis and gametogenesis. In an effort to reduce these cDNAs (and also those for possible heat-shock-induced genes), embryonic cDNA (C) was added to the glp-4 cDNA (B) used for subtraction. To evaluate the efficiency of cDNA subtraction, we compared the content of the cDNA for ribosomal protein L10 by PCR in subtracted and unsubtracted cDNA pools. Detection of L10-encoding cDNA required 24 PCR cycles with either of the subtracted cDNAs as template, whereas only 18 cycles were sufficient to amplify it from unsubtracted cDNA pools, suggesting that housekeeping genes are efficiently eliminated. We also obtained the same result by Southern hybridization. On the other hand, abundance of the cDNA for the gld-1 gene, which is known to be gonad-specific, was tested by Southern analysis. As expected, the difference of signal intensity between glp-1 and glp-4 cDNA pools was magnified after subtraction.
Differential Hybridization.
Subtracted cDNAs were cut free of the primer sequences at both ends and labeled with [33P]dCTP by using the Megaprime DNA labeling system (Amersham Pharmacia). Probes were purified with the G-50 spin column (Amersham Pharmacia) and used for hybridization. High-density filters were preincubated in hybridization solution containing 5× SSC, 5× Denhardt's solution (0.1% polyvinylpyrrolidone/0.1% Ficoll/0.1% BSA), 0.5% SDS, and 100 μg/ml salmon sperm DNA at 65°C for 1 h, then incubated in the same solution containing a 33P-labeled probe and unlabeled genomic DNA from Escherichia coli OP50 (5 μg/ml) at 65°C for 20 h, the latter being added to avoid cross-hybridization with contaminating E. coli sequences in the probe. The hybridized filters were washed, and hybridization signals were detected and quantified with an imaging analyzer (Fuji BAS system) and expressed in photostimulated luminescence (PSL) units.
Evaluation of the Differential Hybridization.
To confirm the results of differential hybridization on high-density grids, the subtracted and unsubtracted cDNA pools were subjected to electrophoresis on agarose gels and Southern blots were probed with individual clones that showed various levels of signals in the differential hybridization (referred to as “virtual Northern blot” analysis). Twenty-two randomly chosen clones were used as probes. On the basis of this analysis, the cut-off standard for further analysis was set as follows:
(i) If the signal for R cDNA is detectable, the ratio of signal intensity values for F and R must be over 9-fold.
(ii) If a signal for R cDNA is not detected, the signal intensity value for F must be over 3 PSL.
Thirteen clones of 22 satisfied this standard. Of these, 11 (85%) were confirmed by virtual Northern analysis by giving the signals F > R. Of the remaining 9 clones that were below the standard, only 5 showed the signals F > R by virtual Northern.
In Situ Hybridization.
Whole-mount in situ hybridization was performed according to the protocol developed by Y.K. and described at the web site http://watson.genes.nig.ac.jp/db/method/insitu_embryo.html. Images were collected by Dual Mode Cooled CCD Camera (Hamamatsu Photonics, Hamamatsu City, Japan) from an Axioskop microscope (Zeiss).
RNAi Studies.
The inserts of expressed sequence tag (EST) clones were amplified by PCR using T3 and T7 primers, purified, and used as templates for RNA transcription with T3 RNA polymerase (Stratagene) and T7 RNA polymerase (Takara Shuzo, Kyoto). We injected double-stranded RNA (dsRNA) into the intestines by using the standard microinjection protocol. To assess the extent to which worms were affected by injected RNA, we used as a host the peIs1 strain, which carries an integrated array of let-60∷gfp and expresses green fluorescent protein (GFP) in a wide variety of tissues. A mixture of dsRNA derived from the EST clone and that from gfp was injected into this strain. Mixing two species of dsRNA did not reduce the effect of RNAi in the pilot experiments. We scored hatching rate, morphological abnormality, and sterility of F1 animals at 22°C. The hatching rate of the control worms that received only gfp dsRNA was 94.1% ± 9.6%. Clones that showed an average hatching rate of lower than 30% were regarded as causing embryonic lethality. Eight F1 progeny with the weakest GFP fluorescence were then picked at the L4 stage, and the number of embryos and unfertilized eggs these animals laid during a 10-h period on the next day was counted. Because 5.3% of the control animals showed complete sterility, probably caused by a weak toxicity of GFP, such clones that caused more than 2 of 8 F1 progeny to lay no fertilized embryos were regarded to cause sterility. For the clones that showed sterile phenotypes, dsRNA was also injected into N2 animals to confirm the phenotype. For examination of the somatic gonadal cells, we injected dsRNA into the strains carrying either an integrated array of lim-7∷gfp (11), which visualizes sheath cell pairs 1–5, or lag-2∷gfp (12), which visualizes the distal tip cells.
Results
Identification of Germ-Line-Specific Genes by cDNA Subtraction and Differential Hybridization.
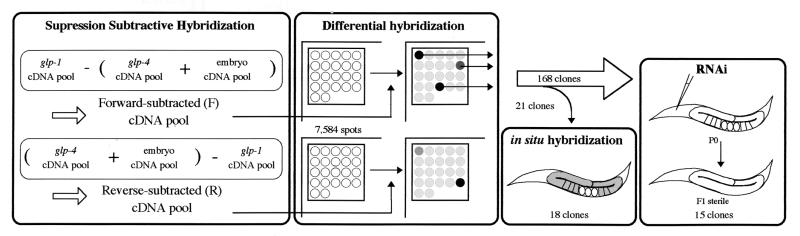
To identify genes that are expressed specifically in the gonad and function during meiosis and gametogenesis, we performed subtractive hybridization by SSH (13) between two temperature-sensitive mutants, glp-1(q224ts), which is enriched in meiotic nuclei (14), and glp-4(bn2ts), which essentially lacks a germ line (15) (Fig. 1). RNA from wild-type embryos was also included in an effort to remove cDNA of maternal messages that are expressed in the gonad and transmitted to the embryo. This subtraction, detailed in Materials and Methods, generated a forward-subtracted cDNA pool (called “F cDNA pool” hereafter) and a reverse-subtracted cDNA pool (called “R cDNA pool”). cDNA clones derived from germ-line-specific genes were expected to be enriched in the F cDNA pool. On the other hand, cDNAs for zygotic transcripts expressed only in the embryos are expected to be enriched in the R cDNA pool. The subtracted cDNA pools, along with unsubtracted cDNA pools for comparison, were used as probes for hybridization to high-density cDNA grids generated through the C. elegans cDNA project by Y.K. (16). Of the 7,584 spots on the membrane, which essentially correspond to separate genes, 510 spots were detected only with the F cDNA pool, 1,327 spots were detected only with R, and 2,937 spots were detected with both. When probed with the unsubtracted cDNA pools, many of the spots with strong intensity belonged to the class of spots that were commonly detected with the two probes. However, this class of spots disappeared when probed with the subtracted cDNA pools. By contrast, spots with signal intensities that differed between the two unsubtracted cDNA pools showed greater differences after subtraction, confirming the effectiveness of SSH. The radioactivity for each spot, quantified in photostimulated luminescence (PSL) units, ranged from 0 to >500 PSL. Reproducibility of hybridization was assessed by virtual Northern analysis using several selected clones, and on the basis of this analysis we determined the cut-off standard for choosing the clones to be analyzed further (see Materials and Methods).
Figure 1.
Summary of our screening scheme. cDNA subtraction was performed with cDNA from three mRNA sources as shown, to generate forward- (F) and reverse-subtracted (R) cDNA pools. These subtracted cDNA pools were used as probes for differential hybridization on high-density grid filters, on which 7,584 EST clones had been spotted. Expression pattern of several clones that gave differential signals was determined by in situ hybridization. One hundred and sixty-eight clones were subjected to RNAi, of which 15 gave a sterile phenotype.
Validation of the Differential Hybridization Results.
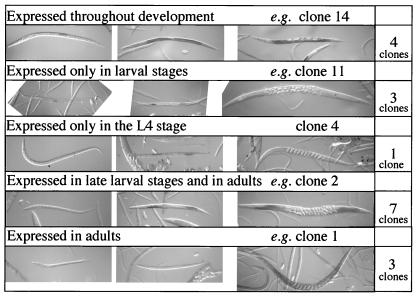
We regarded 199 clones meeting this standard as candidate germ-line-specific clones. Their sequences, already obtained through the cDNA project, revealed that genes previously identified to be expressed and/or function in the germ line were included among them: 22 major sperm protein genes (17), tra-2 (18), glh-1 (19), mex-1 (20), and pos-1 (21). To see whether our candidate genes are actually expressed in the gonad, 21 clones were arbitrarily selected and subjected to in situ hybridization. Eighteen were expressed specifically in the gonad as expected. Other clones were either expressed in other tissues (1 clone) or showed weak signals in the whole body (2 clones). The expression of each gonad-specific clone was usually stage-specific (Fig. 2).
Figure 2.
Whole-mount in situ hybridization showing typical expression patterns of the genes identified by cDNA subtraction/differential hybridization. Of the 21 genes analyzed, the expression of 18 genes was restricted to the germ line with various stage specificity. (Left) L1 or L2 larvae. (Center) L3 or L4 larvae. (Right) Adults observed under Nomarski optics. The ID numbers for the cDNA clones were arbitrarily given in this study and their correspondence to the original yk numbers is given on our web site (http://park.ecc.u-tokyo.ac.jp/mgrl/germline/). The numbers of clones, among the 18, showing expression patterns similar to the examples shown are indicated on the right.
Functional Screening by RNAi.
Because the in situ hybridization experiment suggested that most, although not all, of the candidate genes are expressed specifically in the germ line, we subsequently assessed the functions of these candidates by using the RNAi technique (see Materials and Methods for details). RNAi was performed with 168 clones, omitting those for already characterized genes. We hereafter refer to these yk clones by serial numbers for convenience. We observed embryonic lethality (13 clones), larval lethality (8 clones), morphological abnormality (Dumpy phenotype: 1 clone), and sterile phenotypes (15 clones). These phenotypes are summarized in Tables 1 and 2. Details of the RNAi results can be viewed at our web site, http://park.ecc.u-tokyo.ac.jp/mgrl/germline/.
Table 1.
Nongonadal RNAi phenotype
| Clone no. | Clone name | CDS ID | Phenotype |
|---|---|---|---|
| Embryonic lethal | |||
| 1 | yk37e9 | C26E6.4 | |
| 15 | yk499g10 | W06F12.1 | |
| 16 | yk538a12 | F26B1.3 | |
| 20 | yk602f11 | F52E1.1 | |
| 23 | yk41h6 | T22F3.3 | |
| 26 | yk118h8 | M03F8.3 | |
| 112 | yk507f10 | T05G5.7 | |
| 123 | yk391b7 | F20D12.4 | |
| 126 | yk486e10 | T20G5.1 | |
| 134 | yk540f8 | — | |
| 157 | yk585e12 | W02A2.h | |
| 161 | yk595h12 | C28C12.2 | |
| 164 | yk605b2 | M28.5 | |
| Larval lethal | |||
| 14 | yk490e12 | K02F2.2 | |
| 17 | yk556g5 | F09F7.3 | |
| 51 | yk293a7 | C47D12.6 | |
| 123 | yk391b7 | F20D12.4 | |
| 153 | yk578a1 | C56A3.8 | |
| 156 | yk585c12 | T01C3.7 | |
| 164 | yk605b2 | M28.5 | |
| 168 | yk670b1 | K07C5.4 | |
| Others | |||
| 32 | yk199f3 | C31C9.2 | Slow growing |
| 50 | yk286h12 | — | Bursting |
| 80 | yk383f7 | F11G11.8 | Dpy |
| 111 | yk506a7 | — | Pvu |
| 128 | yk529f3 | ZK1151.2 | Bursting |
Major phenotypes are listed for each clone. Clone numbers were arbitrarily given in this study. CDS IDs are as defined in the Wormpep database (http://www.sanger.ac.uk/Projects/C_elegans/wormpep/). Dpy, Dumpy; Pvu, Protruding vulva.
Table 2.
Gonadal RNAi phenotype
| Clone no. | Clone name | CDS ID | Homologue | Phenotype | Penetrance, % |
|---|---|---|---|---|---|
| Sterile (affected worms laid unfertilized eggs) | |||||
| 9 | yk385e11 | W09C3.6 | Serine/threonine protein phosphatase PP1 | Type I | 100 |
| 10 | yk437b3 | C25G4.6 | PDZ domains | Type I | 100 |
| 42 | yk233g4 | F32A11.3 | — | Type I | 58 |
| 47 | yk270c1 | T25G3.2 | Chitin synthase | Type II | 100 |
| 121 | yk292d2 | T21E3.1 | Protein tyrosine phosphatase | Type II | 100 |
| 131 | yk534e2 | F07A11.2 | Glutamine:fructose-6-phosphate aminotransferase | Type II | 67 |
| 148 | yk574e1 | H02I12.1 | — | Type III | 71 |
| Sterile (affected worms did not lay eggs) | |||||
| 12 | yk445a8 | T05G5.10 | Eukaryotic initiation factor 5A | Glp | 100 |
| 50 | yk286h12 | — | — | Glp | 54 |
| 111 | yk506a7 | — | — | Degenerative nuclei | 100 |
| 116 | yk519f1 | F35G12.10 | ATP synthase b chain | Pachytene arrest | 100 |
| 127 | yk520f4 | C05D2.5 | — | Degenerative nuclei | 75 |
| 134 | yk540f8 | — | Pumilio repeat family | Mog, abnormal diakinesis | 43 |
| 153 | yk578a1 | C56A3.8 | — | Glp | 57 |
| 160 | yk590b8 | C07H6.5 | ATP-dependent RNA helicase | Diakinesis defects | 100 |
Major phenotypes are listed for each clone. Glp, germ-line proliferation defect; Mog, masculinized germ line. Penetrance indicates the percentage of the F1 animals that laid fewer than 10 fertilized eggs. For the sterility class with unfertilized eggs, it was described whether the defect was seen in the injected animals (P0) or their progeny (F1), and whether their fertility recovered after cross with wild-type males. Type I, only F1 defect, recovers after cross; type II, P0 and F1 defects, does not recover after cross; type III, P0 and F1 defects, recovers after cross.
Analysis of the Sterile Phenotypes: Clones That Cause the Affected Animals to Lay Unfertilized Eggs.
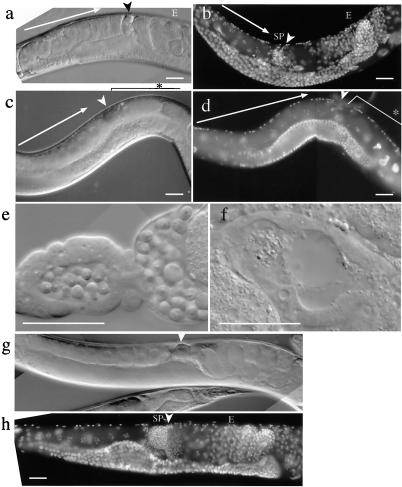
The sterile phenotypes, on which we focused, were classified broadly into two classes. In the first class, affected worms laid eggs with an appearance typical of unfertilized eggs under the dissecting microscope. Seven clones caused this phenotype. Inspection under the Nomarski microscope showed essentially the same phenotype for all of the members in this class: oocytes appeared to develop normally, but after ovulation into the uterus through the spermatheca, where sperm are usually stored, essentially no embryonic cleavages were observed (Fig. 3 c and d). This class was further classified into two subclasses by the criterion of male rescue. In one subclass, namely clones 9, 10, 42, and 148, the RNAi hermaphrodites started to lay normal embryos after the cross with wild-type (N2) males (e.g., Fig. 3 g and h), whereas those in the other subclass, namely clones 47, 121, and 131, were not rescued by the wild-type male sperm. Sterility in the former subclass is likely to be caused by the defect or absence of sperm because it is rescued by introduction of male sperm, whereas sterility in the latter subclass is more likely to be caused by the defects in oocytes. Direct observation of spermatheca of RNAi-affected hermaphrodites in the first subclass revealed that sperm were in fact absent or very scarce for clones 10, 42, and 148 (e.g., Fig. 3f), indicating that these genes are required for spermatogenesis. Roughly normal numbers of sperm were observed for clone 9. These sperm may have defects in motility or in the ability to fertilize oocytes, like some of the fer and spe mutants (22, 23). The latter subclass included genes encoding enzymes probably involved in eggshell synthesis: clones 47 and 131 code for chitin synthase and glutamine:fructose-6-phosphate aminotransferase, respectively. The affected worms are likely to be defective in synthesis of eggshell and lay eggs morphologically resembling unfertilized eggs. Clone 121, which also belongs to this phenotypic class, encodes a protein tyrosine phosphatase. It may be also a component of the pathway for eggshell synthesis.
Figure 3.
Comparison of germ-line structures between control hermaphrodites and hermaphrodites caused to lay unfertilized eggs by RNAi with clone 42. (a and b) F1 progeny of the N2 (wild-type) animals injected with gfp dsRNA alone. (c and d) F1 progeny of the N2 animals injected with gfp dsRNA and clone 42 dsRNA. (e) Dissected spermatheca from an N2 adult hermaphrodite, which has many sperm. (f) Dissected spermatheca from an RNAi-affected adult hermaphrodite, which has no identifiable sperm. (g and h) RNAi-affected hermaphrodites after cross with wild-type males. Although the uterus was filled with unfertilized eggs before the cross, normal embryos were observed after the cross with wild-type males. a, c, e, f, and g were observed under Nomarski optics; b, d, and h were fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI). Arrows, developing oocytes; arrowheads, spermatheca; SP, mature sperm; E, embryos; ∗, unfertilized eggs in the uterus. Ventral is up in all panels. (Scale bars: 0.05 mm).
Analysis of the Sterile Phenotypes: Clones That Cause Failure of Egg Production.
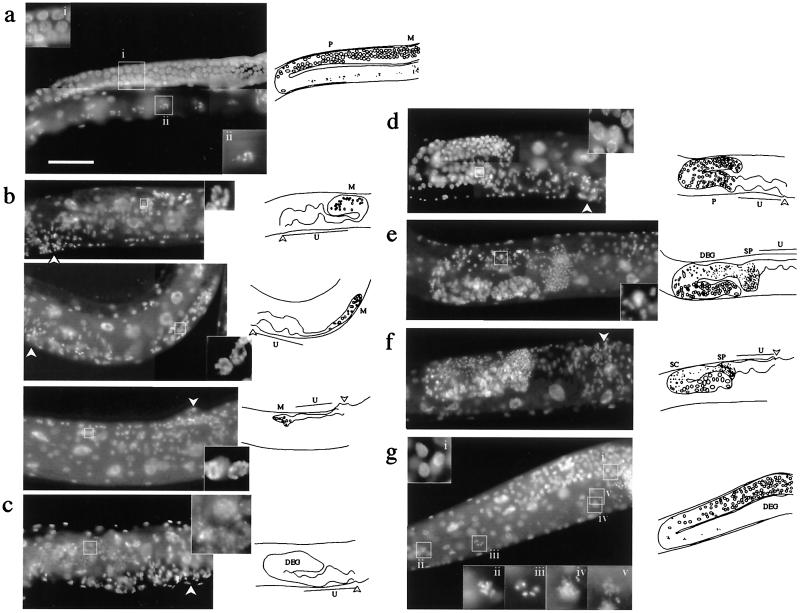
The second class was composed of 8 clones. Worms injected with dsRNA derived from these clones failed to lay eggs. Upon examination under Nomarski optics and with 4′,6-diamidino-2-phenylindole (DAPI) staining, the members of this class showed a variety of phenotypes, which are summarized in Table 2 and examples are shown in Fig. 4. Germ nuclei in C. elegans usually proliferate in the distal region of the gonad and enter meiosis as they move toward the proximal end and gain a certain distance from the distal tip. Several clones (clones 12, 50, and 153) in this latter class caused the Glp phenotype with underproliferative germ nuclei. Intercellular communication involving the GLP-1 receptor is known to control mitosis-to-meiosis switch of germ nuclei (24, 25), and its failure leads to underproliferation and premature entry into meiosis and gametogenesis. Other genes such as glp-4 are known to be required for mitotic proliferation per se (15). No mature sperm were observed in affected hermaphrodites for clones 12, 50, and 153, suggesting that they all belong to the glp-4 class. Excess cell death could cause a similar reduction in germ nuclei, but no extra cell death was detected by using SYTO 12 (Molecular Probes), a vital dye that preferentially stains apototic germ cells (data not shown). Meiotic arrest was observed in clone 116, where meiotic prophase was arrested at the pachytene stage. The germ line of the mutants affected in the Ras–mitogen-activated protein kinase (MAPK) pathway are also known to arrest at the pachytene stage (26). The functional relationship of this gene with the Ras–MAPK pathway is of interest. Clone 134 showed a Mog phenotype in which the sperm-to-oocyte switch in the germ line, which usually occurs at the late L4 stage, fails to occur and the affected adult hermaphrodites continued to produce sperm. Because abnormality in somatic gonadal tissues such as absence of gonadal sheath cells or the distal tip cells can cause germ-line phenotypes (27), we examined the presence of the sheath cells and the distal tip cells in the dsRNA-treated animals (see Materials and Methods). No large decrease in the number of either type of cells was detected for the eight genes of this subclass (data not shown). The predicted products of some of these sterility genes showed similarity to proteins known to be involved in translational control or mRNA metabolism, such as eukaryotic translation initiation factor 5A, RNA helicase family, and pumilio family (19, 28, 29). These genes may therefore regulate the translation or metabolism of germ-line-specific transcripts, possibly in the P granules, which is the C. elegans germ granule and is already known to include RNA-binding proteins and RNA (30, 31).
Figure 4.
Sterile phenotypes caused by RNAi. All of the RNAi adults in this figure lacked embryos in the uterus. (Left) Micrographs. (Right) Diagrams of the micrographs. (a) An uninjected N2 hermaphrodite shown for comparison. (Insets) Enlargement of the boxed regions: pachytene nuclei (i), diakinesis nucleus (ii). (b) Glp phenotypes caused by dsRNA injection. (Top) Clone 12. (Middle) Clone 50. (Bottom) Clone 153. Only a few germ nuclei with the appearance of mitotic interphase (Insets show enlargements) were found in the gonad. No sperm were found. (c) Degenerative nuclei (Inset) in the distal region of the gonad caused by clone 111 dsRNA. (d) Pachytene arrest phenotype caused by clone 116. Germ nuclei arrested at the pachytene stage of meiotic prophase (Inset) fill the proximal part of the gonad. (e) Degenerative nuclei (Inset) caused by clone 127. (f) Mog (masculinization of germline) phenotype caused by clone 134. The proximal half of the gonad was filled with excess sperm and presumptive spermatocytes, the latter of which are usually seen only in L3–L4. (g) Diakinesis defect caused by clone 160. Meiotic prophase appears to progress normally to diakinesis (Insets i, ii, and iii), but the oocyte nuclei degenerate thereafter (Insets iv and v). The photographs show 4′,6-diamidino-2-phenylindole (DAPI)-stained adult hermaphrodites. SP, sperm; U, uterus; M, mitotic germ nuclei; DEG, degenerative germ nuclei; SC, spermatocytes; P, germ nuclei at pachytene stage; arrowhead, vulva. (Scale bar: 0.1 mm.)
Discussion
We performed cDNA subtraction to generate probes for differential hybridization against high-density grid with 7,584 arrayed EST clones and selected 168 germ-line-specific candidate clones. As a result of reverse-genetic approach for these clones, we identified 15 genes essential for fertility, which will each contribute to expand our knowledge on germ-line development in this organism. In this study, we adopted a relatively conservative cut-off standard in evaluating the differential hybridization results. Nevertheless, the set of genes identified in this study expands greatly the current list of molecularly identified sterility genes, which have been obtained mostly through efforts involving forward-genetic approaches. Recently, large-scale RNAi analyses of nonselected C. elegans genes were reported (6–8). Small overlaps were found between the gene sets included in our analysis and those in such studies. In these cases of overlap, the two results were mostly consistent, but differences were occasionally found, probably because of the difference in test conditions. The fraction of genes causing F1 sterility was 9% in our analysis, whereas it ranged from 0.6% (6) to 1% (8) in the RNAi screen with nonselected clones, indicating an enrichment of functional genes by our subtraction-differential hybridization as one would expect (P0 sterility was observed by others at 0.4% (7), 3.4% (6), and 6.7% (8), but many genes with non-germ-line-specific functions are probably included in these). A detailed analysis of germ-line gene expression by using DNA microarray has recently been reported for C. elegans (32). Our results demonstrate that a combination of such expression analyses and reverse genetics employing RNAi will be a powerful approach of functional genomics in the postgenomic era.
Acknowledgments
We thank David Greenstein for the lim-7∷gfp strain, Judith Kimble for the lag-2∷gfp strain, and Takaaki Hirotsu for constructing the let-60∷gfp strain. We also thank Hirofumi Kunitomo for technical advice and discussions, Ikuma Maeda and Asako Sugimoto for their comments and discussions, Tim Doyle for critical reading of the manuscript, and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources, for other C. elegans strains. We express special thanks to Masayuki Yamamoto for his courteous support throughout the course of the study.
Abbreviations
- RNAi
RNA interference
- SSH
suppression subtractive hybridization technique
- EST
expressed sequence tag
- dsRNA
double-stranded RNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Saffman E E, Lasko P. Cell Mol Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.L'Hernault S W. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 271–294. [Google Scholar]
- 3.Shedl T. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 241–269. [Google Scholar]
- 4.Albertson D G, Rose A M, Villeneuve A M. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 47–78. [Google Scholar]
- 5.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Fraser A G, Kamath R S, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature (London) 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 7.Gonczy P, Echeverri G, Oegema K, Coulson A, Jones S J, Copley R R, Duperon J, Oegema J, Brehm M, Cassin E, et al. Nature (London) 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 8.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 9.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood W B. The nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 11.Hall D H, Winfrey V P, Blaeuer G, Hoffman L H, Furuta T, Rose K L, Hobert O, Greenstein D. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- 12.Blelloch R, Anna-Arriola S S, Gao D, Li Y, Hodgkin J, Kimble J. Dev Biol. 1999;216:382–393. doi: 10.1006/dbio.1999.9491. [DOI] [PubMed] [Google Scholar]
- 13.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austin J, Kimble J. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 15.Beanan M J, Strome S. Development (Cambridge, UK) 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 16.Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. Proc Natl Acad Sci USA. 1999;96:15020–15025. doi: 10.1073/pnas.96.26.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith H E, Ward S. J Mol Biol. 1998;279:605–619. doi: 10.1006/jmbi.1998.1793. [DOI] [PubMed] [Google Scholar]
- 18.Kuwabara P E, Kimble J. Development (Cambridge, UK) 1995;121:2995–3004. doi: 10.1242/dev.121.9.2995. [DOI] [PubMed] [Google Scholar]
- 19.Gruidl M E, Smith P A, Kuznicki K A, McCrone J S, Kirchner J, Roussell D L, Strome S, Bennett K L. Proc Natl Acad Sci USA. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guedes S, Priess J R. Development (Cambridge, UK) 1997;124:731–739. doi: 10.1242/dev.124.3.731. [DOI] [PubMed] [Google Scholar]
- 21.Tabara H, Hill R J, Mello C C, Priess J R, Kohara Y. Development (Cambridge, UK) 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- 22.L'Hernault S W, Shakes D C, Ward S. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singson A, Hill K L, L'Hernault S W. Genetics. 1999;152:201–208. doi: 10.1093/genetics/152.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Austin J, Kimble J. Cell. 1989;58:565–571. doi: 10.1016/0092-8674(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 25.Yochem J, Greenwald I. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- 26.Church D L, Guan K L, Lambie E J. Development (Cambridge, UK) 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- 27.McCarter J, Bartlett B, Dang T, Schedl T. Dev Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- 28.Zuk D, Jacobson A. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wharton R P, Sonoda J, Lee T, Patterson M, Murata Y. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- 30.Pitt J N, Schisa J A, Priess J R. Dev Biol. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- 31.Jan E, Motzny C K, Graves L E, Goodwin E B. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinke V, Smith H E, Nance J, Wang J, Van Doren C, Begley R, Jones S J M, Davis E B, Scherer S, Ward S, Kim S K. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]